Abstract
Background
Previous linkage studies, including a study of the Native American population described in the present report, have provided evidence for linkage of alcohol dependence and related traits to chromosome 4q near a cluster of alcohol dehydrogenase (ADH) genes, which encode enzymes of alcohol metabolism.
Methods
The present study tested for associations between alcohol dependence and related traits and 22 single nucleotide polymorphisms (SNPs) spanning the seven ADH genes. Participants included 586 adult men and women recruited from eight contiguous Native American reservations. A structured interview was used to assess DSM-III-R alcohol dependence criteria as well as a set of severe alcohol misuse symptoms and alcohol withdrawal symptoms.
Results
No evidence for association with the alcohol dependence diagnosis was observed, but a SNP in exon 9 of ADH1B (rs2066702; ADH1B*3) and a SNP at the 5' end of ADH4 (rs3762894) showed significant evidence of association with the presence of withdrawal symptoms (p=0.0018 and 0.0012, respectively). Further, a haplotype analysis of these two SNPs suggested haplotypes containing either of the minor alleles were protective against alcohol withdrawal relative to the ancestral haplotype (p=0.000006).
Conclusions
These results suggest that variants in the ADH1B and ADH4 genes may be protective against the development of some symptoms associated with alcohol dependence.
Keywords: alcohol dehydrogenase, alcohol dependence, alcohol withdrawal, genetic association, candidate gene analysis
Introduction
Numerous family, twin, and adoption studies have suggested that alcohol dependence represents a heritable condition, with approximately 50–60% of the variance in the development of alcohol dependence explained by genetic influences (e.g., Allgulander et al., 1991; Goodwin et al., 1973; Heath et al., 1997; Kendler et al., 1997; Reich et al., 1988). Most of these studies, as well as molecular genetic studies of alcohol dependence, have been conducted using Caucasian samples, despite similar or higher prevalence rates in some minority groups, including some Native American communities. Notably, there may be specific advantages in conducting genetic studies of complex diseases, such as alcoholism, in well-defined ethnic populations such as Native American tribes (Lander and Schork, 1994). Often such populations are more environmentally and genetically homogeneous, more geographically restricted, and frequently have large extended pedigrees, all of which can allow for greater power to detect genetic linkage and/or association. Further, specific facets of the studied trait may be more prevalent within the ethnic population under study than the general population thus aiding gene identification (Burchard et al., 2003). In an attempt to capitalize on these advantages, the present study sought to investigate the relations between alcohol dependence and genes involved in alcohol metabolism within a Native American sample.
Genetically influenced metabolic factors have been implicated in the etiology of alcoholism in a number of ethnic groups. The major enzyme families involved in alcohol metabolism, alcohol dehydrogenase (ADH), which is responsible for the oxidation of alcohol to acetaldehyde, and aldehyde dehydrogenase (ALDH), which is responsible for the oxidation of acetaldehyde to acetate, exist as multiple isozymes that differ in their kinetic properties. The genes that encode them have been considered candidate genes that are likely to contribute to variation in alcohol metabolism, variability in response to alcohol, and differences in individual vulnerability for developing alcohol dependence and alcohol-related disability (see Bosron et al., 1993; Chen et al., 2009; Crabb, 1995; Edenberg, 2007; Li, 2000). A large proportion of individuals of Far East Asian descent (often in the range of 30%), possess a mutation in the ALDH2 gene, which produces a largely inactive form of the enzyme resulting in elevated acetaldehyde levels, an alcohol induced flushing reaction, an increased level of response to alcohol, and lower rates of alcohol use and alcoholism (Higuchi et al., 1995; Luczak et al., 2002; Takeshita et al., 1994; Thomasson et al., 1991; Shen et al., 1997; Wall et al., 1992, 1993, 1999).
In addition to ALDH2, one of the more replicable findings described in the genetics of alcohol dependence literature has been evidence of an association for alcohol dependence and related behaviors to chromosome 4 near the ADH gene cluster. This gene cluster is approximately 364 kilobases (kb) in length, and the genes are all transcribed from the same DNA strand (4qter to 4pter). The order of genes from qter to pter is ADH7, ADH1C, ADH1B, ADH1A, ADH6, ADH4, and ADH5, with each ADH gene coding for a unique isozyme. The relation between this chromosomal region and alcohol dependence has been reported in a number of linkage studies of diverse ethnic groups (e.g., Corbett et al., 2005; Long et al., 1998; Prescott et al., 2006; Williams et al., 1999) including the Native American sample presented in this report (Ehlers et al., 2004b). Additionally, genome screens for both the “unaffected by alcoholism” (Reich et al., 1998) and “maximum drinks ever consumed in a 24 hour period,” (Saccone et al., 2000) phenotypes were found to yield evidence of linkage to chromosome 4 in the region of the ADH gene cluster in the Collaborative Study of the Genetics of Alcoholism (COGA).
Given that the ADH cluster consists of seven genes, researchers have sought to identify which of the ADH genes might be involved in the etiology of alcohol dependence. Because the class 1 ADH isozymes account for the majority of alcohol metabolism in the liver and have been shown to contain nonsynonymous coding SNPs that alter the kinetic properties of ADH, the genes encoding these isozymes, ADH1A, ADH1B, and ADH1C, have received the most initial attention with each gene showing evidence of association with alcohol dependence and related phenotypes (Borras et al., 2000; Chen et al., 1996; Chen et al., 1999; Edenberg et al., 2006; Hasin et al. 2002; Ma et al., 2005; Mulligan et al., 2003; Neumark et al., 2004; Nishimura et al., 2009; Spivak et al., 2007; Thomasson et al., 1991; Wall et al., 2005). For example, the ADH1B*2 allele (rs1229984, A allele) located in exon 3 of ADH1B results in an arginine to histidine amino acid change that alters the kinetics of the enzyme (Hurley et al., 1990) and has demonstrated a protective relation with alcohol dependence and related phenotypes (e.g., MacGregor et al., 2009; Shen et al., 1997; Thomasson et al., 1991, 1994; Whitfield, 1997). Similarly, the ADH1B*3 allele (rs2066702, located in exon 9 of ADH1B) results in an arginine to cysteine change (Carr et al., 1989) that has shown a protective association in the development of alcohol dependence in samples of African descent (Edenberg et al., 2006; Ehlers et al., 2001a, 2007; Luo et al., 2006; McCarthy et al., 2010) and a subset of the Native American population described in the present study (Wall et al., 2003). More recently, researchers have begun to study the remaining ADH genes to evaluate whether variants in these genes might also be related to alcohol dependence and related phenotypes (e.g., Birley et al., 2009; Hall et al., 2007; Han et al., 2007; Kuo et al., 2008; Luo et al., 2007; Sherva et al., 2009; van Beek et al., 2010), with several of these studies reporting evidence of association with ADH4 (Edenberg et al., 2006; Guindalini et al., 2005; Kimura et al., 2009; Luo et al., 2005; MacGregor et al., 2009; Preuss et al., 2011) as well as with ADH1A and ADH1B (Edenberg et al., 2006).
The present report is part of a larger study exploring risk factors for alcoholism in a Native American community (Ehlers and Wilhelmsen, 2005; Ehlers et al., 1998, 1999, 2001a,2001b, 2004a, 2004b; Garcia-Andrade et al., 1996, 1997; Gilder et al., 2002; 2004; Wall et al., 1996, 2000, 2003). In previous studies, we have demonstrated the utility of examining evidence of linkage and association using the alcohol dependence diagnosis as well as a severe use and a withdrawal phenotype, with the latter phenotypes selected to identify a more severe form of alcohol dependence given the high prevalence rate of alcohol dependence in this sample. Specifically, the severe use phenotype consists of four alcohol use items that indicate an advanced clinical course in this population (Ehlers et al., 2004a) and has previously shown evidence of linkage to the chromosome 4q ADH gene cluster. The withdrawal phenotype was selected given studies suggesting that withdrawal symptoms are late developing and indicate a particularly severe form of the disorder (Bucholz et al., 1996; Gilder et al., 2011; Martin et al., 2006; Nelson et al., 1996; Saha et al., 2006). A previous study conducted in a subset of the present Native American sample yielded evidence of association between ADH1B and DSM-III-R defined alcohol dependence (Wall et al., 2003). Thus, the present study sought to extend these findings by testing for associations between alcohol dependence as well as severe use and withdrawal phenotypes and SNPs in ADH genes in an expanded Native American sample.
Methods and Materials
The protocol for the study was approved by the Scripps Institutional Internal Review board and Indian Health Council, a tribal review group overseeing health issues for the reservations where recruitments took place. Written informed consent was obtained from each participant after study procedures had been fully explained. Participants were compensated for their time spent in the study.
Participants
Participants who were of at least one-sixteenth Native American Heritage were targeted for study and recruited from eight geographically contiguous reservations with a total population of about 3,000 individuals. Participants who were mobile and between the ages of 18–82 years were recruited using a combination of a venue-based method for sampling hard-to-reach populations (Kalton and Anderson, 1986; Muhib et al., 2001) and a respondent-driven procedure (Heckathorn, 1997), as reported previously (Ehlers et al., 2004a). Women were intentionally over-sampled to provide adequate numbers for meaningful analyses.
Demographic characteristics of this Native American population have been reported previously (Ehlers et al. 2004a). Individuals with blood samples genotyped for the present study (N=586) had a mean age of 31 (range 18–82) years (SD= 12 years), with 42% of the sample being male. Participants had a mean of 11.6 years of education (SD=1.6), 47% of the sample by self report had at least 50% Native American heritage as indicated from their federal Indian blood quantum. Among participants, 59% (n = 346) had a lifetime diagnosis of alcohol dependence, 70% (n = 410) reported a first degree family history of alcoholism, 42% (n = 246) reported experiencing one or more of the severe use symptoms, and 32% (n = 185) reported experiencing one or more withdrawal symptoms.
Participants completed an interview with the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) which was used to gather demographic information and make a lifetime diagnosis of alcohol dependence according to DSM-III-R criteria (American Psychiatric Association, 1980). The SSAGA is a polydiagnostic psychiatric interview that has undergone both reliability and validity testing (Bucholz et al., 1994; Hesselbrock et al., 1999). It has been successfully used in Native American populations previously (Hesselbrock et al., 2000; Wall et al., 2003). Interviewers were all trained by Collaborative Study of the Genetics Alcoholism (COGA) personnel. All best final diagnoses were made by one research psychiatrist/addiction specialist (DAG) using a best estimate procedure.
The SSAGA groups individual interview response items into nine categories that correspond to the nine DSM-III-R criteria used for making a dependence diagnosis. These items include: alcohol use severity items, legal, family, work and medical problems, tolerance, wanting/unable to quit and withdrawal. Two phenotypes hypothesized to most likely have a heritable basis were defined using these categories. The first phenotype grouped responses only on four alcohol use severity items: (1) drank more than intended/more days in a row or when promised self wouldn’t for three or more times, (2) drank when didn’t want to three or more times, (3) during drinking or recovering from the effects of drinking had little time for anything else, and (4) given up or greatly reduced important activities to drink. Each subject was scored as having 1 or more alcohol severity symptoms. The second phenotype was DSM-III-R physiological alcohol withdrawal. The diagnosis of withdrawal was made if the participant had a history of any of the following occurring when he or she stopped, cut down or went without drinking after a prolonged period of steady drinking: (1) the shakes (trembling of the hands), unable to sleep, anxiety or depression, sweating, rapid heart rate, nausea or vomiting, feeling physically weak, headache, auditory or visual hallucinations; (2) seizures; and (3) delirium tremens (DT’s).
Genotyping
A blood sample was obtained by venipuncture from each participant and DNA was isolated from leukocytes. Samples were sent to Indiana University for genotyping (see Edenberg et al., 2006). The genotyped sample of 586 participants was collected from 174 families. Sixty-eight of these families consisted of a single participant, and thus were only included in the linkage disequilibrium block analyses described below. The remaining 518 participants came from 104 families that contained between 2 and 16 genotyped individuals (average family size = 5.2, standard deviation = 3.6).
A set of 22 SNPs that have either shown prior evidence of a functional effect on gene expression or evidence of association with alcohol dependence in a previous study (Edenberg et al., 2006) were genotyped (see Table 1 for details regarding each SNP). An additional 47 SNPs were genotyped in a subset of 184 participants thus providing a more complete assessment of the haplotype block structure underlying the ADH gene cluster in this Native American sample. Quality of the genotypes was assessed by examining the call rate, departures from Hardy-Weinberg equilibrium, and number of Mendelian inconsistencies observed within pedigrees. All SNPs had call rates greater than 95%. Tests of Hardy-Weinberg equilibrium were conducted using the software program Pedstats (Wigginton and Abecasis, 2005). All SNPs had genotypes consistent with Hardy-Weinberg equilibrium with p-values that were >0.01 with the exception of rs1693482, which yielded a p-value of 0.0014. Mendelian inconsistencies were identified using Pedstats (Wiggenton and Abecasis, 2005). A total of 58 Mendelian inconsistencies were observed among the 21098 called genotypes suggesting an error rate <.3%.
Table 1.
Locations and minor allele frequencies of genotyped SNPs.
| Minor Allele Frequencies | |||||||
|---|---|---|---|---|---|---|---|
| SNP | Chromosome Position |
Gene | SNP Type/Location |
Alleles1 | Indians | Caucasians2 | African- Americans2 |
| rs1230155 | 99989258 | ADH5 | intergenic | A/G | 0.200 | 0.345 | 0.318 |
| rs7683802 | 99995137 | ADH5 | intron 7 | T/G | 0.063 | 0.102 | 0.107 |
| rs4699699 | 99997178 | ADH5 | intron 6 | C/G | 0.06 | 0.108 | 0.071 |
| rs4699700 | 99998334 | ADH5 | intron 4 | A/G | 0.074 | 0.148 | 0.261 |
| rs1154412 | 100001246 | ADH5 | intron 4 | C/T | 0.135 | 0.19 | 0.031 |
| rs7683704 | 100004225 | ADH5 | intron 2 | C/T | 0.07 | 0.108 | 0.268 |
| rs1154401 | 100009737 | ADH5 | intron 1 | C/G | 0.213 | 0.355 | 0.364 |
| rs1154400 | 100010009 | ADH5 | 5' end | T/C | 0.145 | 0.328 | 0.341 |
| rs7667261 | 100011299 | ADH5 | 5' end | C/G | 0.015 | 0.044 | 0.212 |
| rs2602846* | 100025150 | ADH5/ADH4 | intergenic | A/T | 0.133 | 0.299 | 0.123 |
| rs2602866 | 100034996 | ADH5/ADH4 | intergenic | G/A | 0.13 | 0.295 | 0.135 |
| rs1042365* | 100045499 | ADH4 | exon 9 | C/A | 0.136 | 0.294 | 0.138 |
| rs1042364* | 100045573 | ADH4 | exon 9 | G/A | 0.096 | 0.294 | 0.138 |
| rs29001229 (DWSHpy188I)* |
100046822 | ADH4 | IVS8 | T/C | 0.136 | 0.295 | 0.135 |
| rs1126672* | 100047811 | ADH4 | exon 8 coding- synon |
C/T | 0.133 | 0.305 | 0.165 |
| rs1126671 | 100048413 | ADH4 | exon 7 | G/A | 0.208 | 0.319 | 0.223 |
| rs1126670 | 100052732 | ADH4 | exon 6 | T/G | 0.21 | 0.315 | 0.217 |
| rs7694646* | 100059731 | ADH4 | intron 4 | T/A | 0.152 | 0.294 | 0.135 |
| rs4699714* | 100060537 | ADH4 | intron 4 | A/G | 0.135 | 0.304 | 0.119 |
| rs4148886 | 100064648 | ADH4 | intron 1 | A/G | 0.464 | 0.242 | 0.49 |
| rs1800759* | 100065508 | ADH4 | 5'UTR | A/C3 | 0.428 | 0.581 | 0.233 |
| rs3762894* | 100066083 | ADH4 | 5'UTR | T/C | 0.108 | 0.166 | 0.225 |
| rs4699718* | 100067790 | ADH4/ADH6 | intergenic | G/A | 0.136 | 0.298 | 0.122 |
| rs1984362* | 100070972 | ADH4/ADH6 | intergenic | C/T | 0.094 | 0.294 | 0.094 |
| rs4147545 | 100128752 | ADH6 | intron 6 | G/A3 | 0.438 | 0.658 | 0.453 |
| rs3857224 | 100129684 | ADH6 | intron 6 | T/C3 | 0.469 | 0.667 | 0.453 |
| rs6833176 | 100131162 | ADH6 | intron 5 | C/G | 0.397 | 0.461 | 0.186 |
| rs9307238 | 100136181 | ADH6 | intron 2 | G/A | 0.409 | 0.488 | 0.359 |
| rs1230026 | 100185618 | ADH6/ADH1A | intergenic | A/C | 0.235 | 0.223 | 0.074 |
| rs1618572 | 100195120 | ADH6/ADH1A | intergenic | C/G | 0.241 | 0.224 | 0.056 |
| rs2866151* | 100198511 | ADH1A | intron 8 | A/T | 0.425 | 0.456 | 0.233 |
| rs3819197 | 100200508 | ADH1A | intron 8 | C/T | 0.819 | 0.749 | 0.725 |
| rs1229976 | 100202077 | ADH1A | intron 6 | T/C | 0.242 | 0.224 | 0.073 |
| rs1229970 | 100204379 | ADH1A | intron 5 | G/T | 0.227 | 0.221 | 0.056 |
| rs6828526 | 100205886 | ADH1A | exon 4 | G/A | 0.000 | 0.001 | 0.009 |
| rs1229967 | 100207577 | ADH1A | intron 3 | G/C | 0.241 | 0.223 | 0.055 |
| rs3805325 | 100211398 | ADH1A | intron 1 | T/C | 0.282 | 0.078 | 0.128 |
| rs4147531* | 100212196 | ADH1A | 5'UTR | C/T | 0.429 | 0.457 | 0.233 |
| rs1229966 | 100213432 | ADH1A | 5' end | T/C4 | 0.328 | 0.346 | 0.605 |
| rs1826909* | 100217742 | ADH1A/1B | intergenic | C/T | 0.146 | 0.373 | 0.171 |
| rs1042026 | 100228465 | ADH1B | 3' UTR | A/G | 0.171 | 0.306 | 0.132 |
| rs2066702* | 100229016 | ADH1B | exon 9 coding- nonsyn |
C/T | 0.041 | 0.004 | 0.165 |
| rs1789883 | 100236374 | ADH1B | intron 5 | G/A | 0.118 | 0.027 | 0.015 |
| rs2066701 | 100238412 | ADH1B | intron 3 | C/T | 0.173 | 0.304 | 0.122 |
| rs1229984* | 100239318 | ADH1B | coding-nonsyn | G/A | 0.028 | 0.034 | 0.019 |
| rs1229983 | 100240001 | ADH1B | exon 2 | T/C | 0.110 | 0.024 | 0.044 |
| rs1353621 | 100241574 | ADH1B | intron 1 | A/G | 0.142 | 0.370 | 0.123 |
| rs1159918* | 100243008 | ADH1B | 3'UTR | T/G3 | 0.292 | 0.652 | 0.355 |
| rs1229982* | 100243931 | ADH1B | 3'UTR | G/T | 0.273 | 0.22 | 0.443 |
| rs1614972 | 100258154 | ADH1C | intron 8 | C/T | 0.147 | 0.330 | 0.493 |
| rs35719513 (P351T) |
100260782 | ADH1C | exon 8 | C/A | 0.079 | 0.005 | 0 |
| rs698* | 100260788 | ADH1C | exon 8 coding- nonsyn |
A/G | 0.403 | 0.389 | 0.185 |
| rs1789903 | 100262040 | ADH1C | intron 6 | C/G | 0.410 | 0.389 | 0.169 |
| rs1693482* | 100263964 | ADH1C | exon 6 coding- nonsyn |
C/T | 0.391 | 0.383 | 0.178 |
| rs1693426 | 100266329 | ADH1C | intron 4 | A/G | 0.409 | 0.387 | 0.171 |
| rs1789915 | 100266370 | ADH1C | exon 4 | A/G | 0.197 | 0.261 | 0.144 |
| rs3133158 | 100270611 | ADH1C | intron 1 | C/G | 0.204 | 0.285 | 0.160 |
| rs1789924 | 100274285 | ADH1C | 5' end | C/T | 0.414 | 0.389 | 0.179 |
| rs1229849 | 100284684 | ADH1C/ADH7 | intergenic | T/A | 0.197 | 0.280 | 0.147 |
| rs283406 | 100298470 | ADH1C/ADH7 | intergenic | C/T | 0.033 | 0.079 | 0.082 |
| rs284794 | 100323028 | ADH1C/ADH7 | intergenic | A/T | 0.055 | 0.109 | 0.157 |
| rs284786* | 100333976 | ADH7 | 3'UTR | A/T | 0.294 | 0.272 | 0.459 |
| rs284779* | 100338260 | ADH7 | intron 7 | C/G | 0.392 | 0.458 | 0.170 |
| rs2584464 | 100339048 | ADH7 | intron 7 | A/G5 | 0.416 | 0.516 | 0.678 |
| rs971074 | 100341860 | ADH7 | exon 6 | G/A | 0.073 | 0.104 | 0.177 |
| rs1154468 | 100354256 | ADH7 | intron 1 | A/T | 0.488 | 0.328 | 0.161 |
| SBP2 | 100356465 | ADH7 | 5' end | A/G | 0.102 | 0.130 | 0.051 |
| rs1154476 | 100360726 | ADH7 | intergenic | G/A | 0.497 | 0.384 | 0.138 |
| rs894363 | 100376845 | ADH7 | intergenic | C/T | 0.497 | 0.379 | 0.165 |
Notes:
Indicates SNPs that were genotyped in the full sample,
Alleles indicated are from the + strand and are ordered major/minor based on prevalence estimates from the present sample,
Allele frequencies cited from Edenberg et al. (2006),
The designated 'minor' allele in the Native American sample was observed at a frequency >0.50 among the European-American sample,
The designated 'minor' allele in the Native American sample was observed at a frequency >0.50 among the African-American sample,
The designated 'minor' allele in the Native American sample was observed at a frequency >0.50 among the European-American and African-American samples
Statistical Analysis
Initial analyses were conducted using Haploview (Barrett et al., 2005) to evaluate the haplotype block structure of the ADH gene cluster in this Native American sample. Blocks were defined using the confidence interval approach described by Gabriel et al. (2002). Given that a proportion of the study sample reported a mixed ethnic background, within-family tests of association were conducted to reduce potential bias due to population stratification. Thus, tests of association were conducted using the PDT (Martin et al., 2001) as implemented in the UNPHASED software package (Dudbridge, 2003). The PDT analyzes all informative trios within an extended pedigree as well as discordant sibships with one affected and one unaffected sibling that possess discrepant genotypes at the marker to be analyzed. The 'averaged' PDT statistic, which gives equal weight to all families included in the analysis, was used to evaluate the evidence for association. Three phenotypes (i.e., DSM-III-R defined Alcohol Dependence, presence of withdrawal symptoms, and presence of 'severe' alcohol dependence symptoms) were tested for association. To correct for multiple testing across these three phenotypes and the 22 SNPs genotyped in the full sample, a critical p-value of 0.002 was selected to reduce the potential for Type I error while considering the correlations between SNP genotypes (average R2=.44 for adjacent SNP pairs).
Results
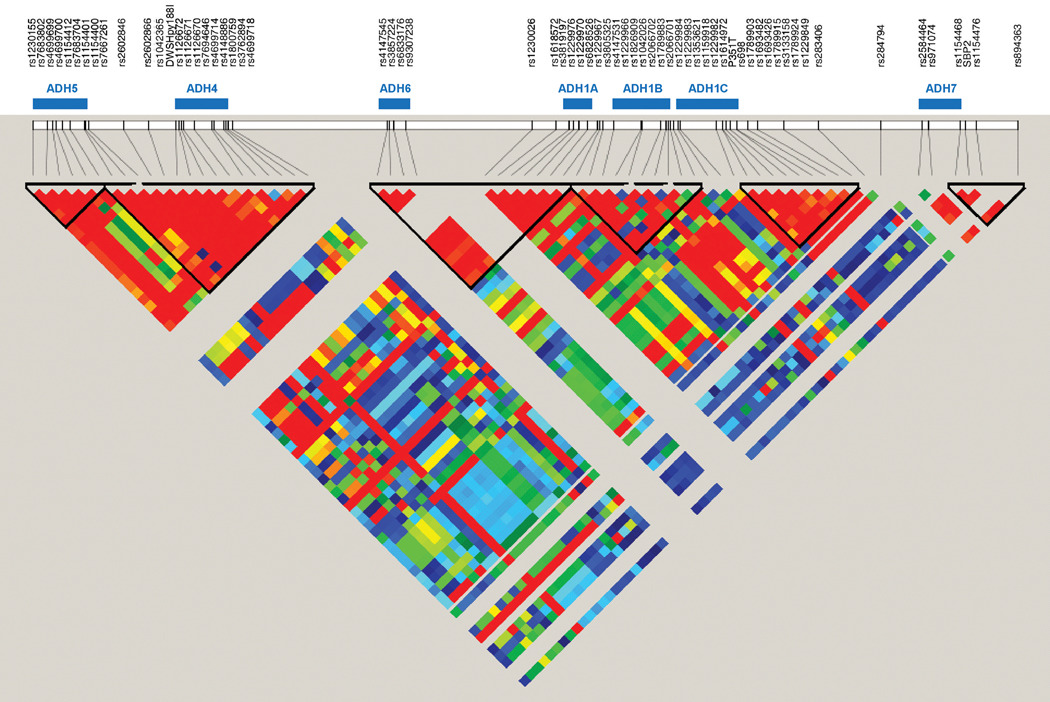
One hundred and three unrelated individuals were drawn from the larger sample to calculate linkage disequilibrium (LD) statistics and estimate the haplotype block structure of the region in the present sample using Haploview (Barrett et al., 2005). The degree of LD between SNPs and the haplotype block structure are shown in Figure 1. Evidence of LD was observed across the region with an average D' value across SNP pairs of 0.64. Nonetheless, there was stronger LD observed within than between genes with an average D' value across SNP pairs within genes of 0.91 and an average D' value across SNP pairs of adjacent genes of 0.70. Further, each gene appeared to be defined by a unique haplotype block in this population with the exception of ADH6 and ADH1A, which were fully contained within a single block. The only strong evidence of recombination within a gene was observed for ADH7 and to a lesser extent for ADH1B. For example, in ADH7 the SNPs at the 5' end show evidence of LD with one another, but those at the 3' end do not. These results are largely consistent with those from the European-American sample described by Edenberg et al (2006), which showed evidence for LD across the ADH gene cluster that was stronger within than between genes.
Figure 1.
Linkage disequilibrium (LD) between SNPs genotyped across the chromosome 4q ADH gene cluster. Gene locations are indicated by the blue bars. Absent or low levels of LD are indicated by the blue and green squares, respectively, whereas increasing levels of LD are indicated by the yellow, orange, and red squares in ascending order of LD.
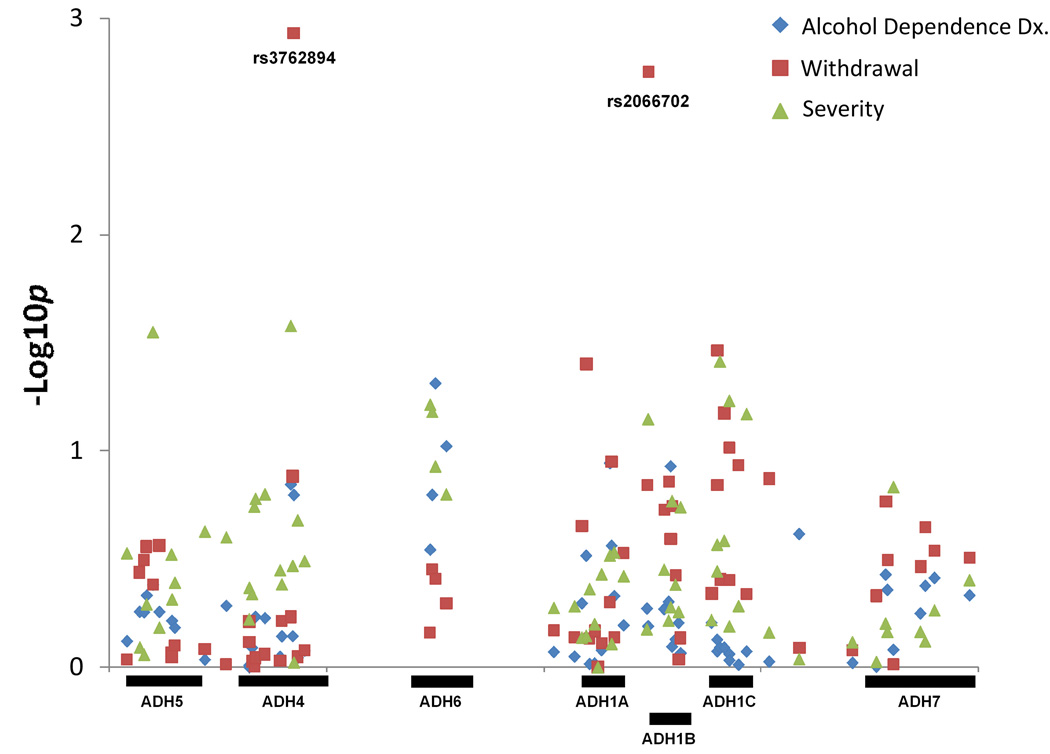
The pedigree disequilibrium test (PDT) was used to test for association between ADH gene cluster SNPs and 3 alcohol dependence related phenotypes: (1) DSM-III-R defined alcohol dependence, (2) the presence of alcohol withdrawal symptoms, and (3) the presence of 'severe' alcohol dependence symptoms (see Table 2 for complete results). For the alcohol dependence diagnosis and 'severe' alcohol dependence symptoms, several SNPs were nominally significant (p<0.05) but none of the SNPs yielded evidence for association at p<0.002. For the withdrawal symptoms phenotype, two SNPS showed significant evidence for association at p<0.002: rs3762894 located in the promoter region of ADH4 (chi-square=10.53, p=0.0012) and rs2066702 located in exon 9 of ADH1B (chi-square=9.77, p=0.0018). For both SNPs, transmission of the minor allele (rs3762894 - C allele; rs2066702 - T allele) was associated with reduced risk for withdrawal symptoms. The locations of these SNPs as well as the results for all tests of association are shown in the Manhattan plot displayed in Figure 2.
Table 2.
Pedigree Disequilibrium Tests of association for individual SNPs and three alcohol dependence phenotypes.
| p-values | ||||
|---|---|---|---|---|
| SNP | Gene | Alcohol Dependence Diagnosis |
Experienced Withdrawal Symptoms |
Experienced Severe Symptoms |
| rs1230155 | ADH5 | 0.7561 | 0.9254 | 0.2974 |
| rs7683802 | ADH5 | 0.5527 | 0.3642 | 0.8073 |
| rs4699699 | ADH5 | 0.5560 | 0.3206 | 0.8734 |
| rs4699700 | ADH5 | 0.4647 | 0.2767 | 0.5125 |
| rs1154412 | ADH5 | 0.4147 | 0.4171 | 0.0285 |
| rs7683704 | ADH5 | 0.5539 | 0.2733 | 0.6551 |
| rs1154401 | ADH5 | 0.8639 | 0.8597 | 0.3020 |
| rs1154400 | ADH5 | 0.6093 | 0.8938 | 0.4862 |
| rs7667261 | ADH5 | 0.6547 | 0.7970 | 0.4071 |
| rs2602846* | ADH5/ADH4 | 0.9198 | 0.8234 | 0.2365 |
| rs2602866 | ADH5/ADH4 | 0.5191 | 0.9668 | 0.2510 |
| rs1042365* | ADH4 | 0.9770 | 0.7640 | 0.5993 |
| rs1042364* | ADH4 | 0.9975 | 0.6163 | 0.4294 |
| rs29001229 (DWSHpy188I)* |
ADH4 | 0.8111 | 0.9402 | 0.4591 |
| rs1126672* | ADH4 | 0.9336 | 0.9973 | 0.1815 |
| rs1126671 | ADH4 | 0.5827 | 0.9056 | 0.1670 |
| rs1126670 | ADH4 | 0.5911 | 0.8728 | 0.1592 |
| rs7694646* | ADH4 | 0.8928 | 0.9365 | 0.3564 |
| rs4699714* | ADH4 | 0.7175 | 0.6134 | 0.4137 |
| rs4148886 | ADH4 | 0.1427 | 0.5866 | 0.0266 |
| rs1800759* | ADH4 | 0.7167 | 0.1312 | 0.3405 |
| rs3762894* | ADH4 | 0.1596 | 0.0012 | 0.9496 |
| rs4699718* | ADH4/ADH6 | 0.9005 | 0.8942 | 0.2098 |
| rs1984362* | ADH4/ADH6 | 0.8366 | 0.8358 | 0.3232 |
| rs4147545 | ADH6 | 0.2857 | 0.6914 | 0.0615 |
| rs3857224 | ADH6 | 0.1595 | 0.3529 | 0.0663 |
| rs6833176 | ADH6 | 0.0487 | 0.3893 | 0.1185 |
| rs9307238 | ADH6 | 0.0950 | 0.5078 | 0.1592 |
| rs1230026 | ADH6/ADH1A | 0.8477 | 0.6762 | 0.5316 |
| rs1618572 | ADH6/ADH1A | 0.8908 | 0.7292 | 0.5229 |
| rs2866151* | ADH1A | 0.5055 | 0.2234 | 0.7288 |
| rs3819197 | ADH1A | 0.3042 | 0.0396 | 0.7143 |
| rs1229976 | ADH1A | 0.9635 | 0.7384 | 0.4354 |
| rs1229970 | ADH1A | 0.9563 | 0.6809 | 0.6310 |
| rs6828526 | ADH1A | 1.0000 | 1.0000 | 1.0000 |
| rs1229967 | ADH1A | 0.8321 | 0.7754 | 0.3722 |
| rs3805325 | ADH1A | 0.1138 | 0.5002 | 0.3048 |
| rs4147531* | ADH1A | 0.2741 | 0.1124 | 0.7801 |
| rs1229966 | ADH1A | 0.4680 | 0.7293 | 0.2949 |
| rs1826909* | ADH1A/1B | 0.6383 | 0.2966 | 0.3797 |
| rs1042026 | ADH1B | 0.5340 | 0.1445 | 0.6680 |
| rs2066702* | ADH1B | 0.6446 | 0.0018 | 0.0717 |
| rs1789883 | ADH1B | 0.5377 | 0.1878 | 0.3544 |
| rs2066701 | ADH1B | 0.4972 | 0.1383 | 0.6085 |
| rs1229984* | ADH1B | 0.1177 | 0.2561 | 0.5283 |
| rs1229983 | ADH1B | 0.8007 | 0.1807 | 0.1709 |
| rs1353621 | ADH1B | 0.7413 | 0.3749 | 0.4154 |
| rs1159918* | ADH1B | 0.6215 | 0.9254 | 0.5548 |
| rs1229982* | ADH1B | 0.8587 | 0.7335 | 0.1824 |
| rs1614972 | ADH1C | 0.6216 | 0.4558 | 0.6042 |
| rs35719513 (P351T) |
ADH1C | 0.7438 | 0.1446 | 0.3611 |
| rs698* | ADH1C | 0.8423 | 0.0344 | 0.2718 |
| rs1789903 | ADH1C | 0.8085 | 0.3926 | 0.0389 |
| rs1693482* | ADH1C | 0.8101 | 0.0668 | 0.2606 |
| rs1693426 | ADH1C | 0.8665 | 0.3960 | 0.0590 |
| rs1789915 | ADH1C | 0.9266 | 0.0964 | 0.6475 |
| rs3133158 | ADH1C | 0.9713 | 0.1166 | 0.5221 |
| rs1789924 | ADH1C | 0.8435 | 0.4608 | 0.0679 |
| rs1229849 | ADH1C/ADH7 | 0.9401 | 0.1347 | 0.6898 |
| rs283406 | ADH1C/ADH7 | 0.2419 | 0.8143 | 0.9149 |
| rs284794 | ADH1C/ADH7 | 0.9504 | 0.8343 | 0.7660 |
| rs284786* | ADH7 | 0.9911 | 0.4685 | 0.9435 |
| rs284779* | ADH7 | 0.3723 | 0.1719 | 0.6271 |
| rs2584464 | ADH7 | 0.4379 | 0.3188 | 0.6852 |
| rs971074 | ADH7 | 0.8276 | 0.9676 | 0.1473 |
| rs1154468 | ADH7 | 0.5625 | 0.3428 | 0.6853 |
| SBP2 | ADH7 | 0.4194 | 0.2265 | 0.7587 |
| rs1154476 | ADH7 | 0.3853 | 0.2905 | 0.5450 |
| rs894363 | ADH7 | 0.4638 | 0.3110 | 0.3960 |
Notes:
Indicates SNPs that were genotyped in the full sample, italicized text indicates p-value<0.05, bold text indicates p-value<0.002.
Figure 2.
Manhattan plot of the association signals for the SNPs typed across the ADH gene cluster and the three analyzed phenotypes. The X-axis displays the location of the SNP with each gene indicated by a labeled black bar, and the y-axis displays the −log(p) value for each SNP.
A set of follow-up analyses were then conducted to test for association between haplotypes constructed for a set of six SNPs, rs1126672, rs3762894, rs2066702,rs1229984, rs698, and rs1693482, that yielded significant evidence for association in the present study or have a suggested functional impact on gene expression. A sliding window of 2 SNPs was used to construct haplotypes for all contiguous SNP pairs, which were then evaluated for evidence of association with the three described alcohol use phenotypes using the PDT. The strongest evidence for association across all phenotypes was observed for the analysis of rs3762894 and rs2066702 (chi-square=24.01, p=0.000006; see Table 3a for complete results). Analysis of the individual haplotypes suggested that transmission of haplotypes containing a minor allele at either SNP (C allele of rs3762894 or T allele of rs2066702) was associated with a reduced risk for experiencing withdrawal symptoms (Table 3b).
Table 3.
Haplotype analysis of putatively functional SNPs using a sliding window of 2 SNPs.
| a.) Summary of results for all sets of SNPs. | |||
|---|---|---|---|
| p-values | |||
| SNPs | Alcohol Dependence Diagnosis |
Experienced Withdrawal Symptoms |
Experienced Severe Symptoms |
| rs1126672,rs3762894 | 0.6408 | 0.01528 | 0.7022 |
| rs3762894,rs2066702 | 0.2499 | 0.000006126 | 0.3977 |
| rs2066702,rs1229984 | 0.2573 | .007018 | 0.2022 |
| rs1229984,rs698 | 0.8451 | 0.02986 | 0.2704 |
| rs698,rs1693482 | 0.04523 | 0.004394 | 0.009667 |
| b.) Association results for withdrawal symptoms and haplotypes constructed from rs3762894 and rs2066702. | |||||
|---|---|---|---|---|---|
| Haplotype | Z | p-value | Transmitted | Not Transmitted |
Frequency |
| C/C | −3.274 | 0.001061 | 12.63 | 20.23 | 0.103 |
| T/C | 4.085 | 0.00004403 | 192.4 | 180.2 | 0.8762 |
| T/T | −2.933 | 0.003359 | 1 | 5.598 | 0.02081 |
Global test: Chi-square = 24.01, p = 6.126e-006.
Discussion
The present study sought to evaluate the evidence for association between the 7 ADH genes located on chromosome 4q and alcohol dependence as well as phenotypes associated with long-term alcohol misuse. Two SNPs, rs2066702 located in exon 9 of ADH1B and rs3762894 located at the 5' end of ADH4, yielded significant evidence for an association with withdrawal symptoms. For both SNPs, the minor allele (C allele of rs3762894; T allele of rs2066702) showed a protective relation to the development of withdrawal symptoms. This result was further supported by an analysis of haplotypes constructed from these two SNPs that suggested haplotypes containing either of the minor alleles was protective against alcohol withdrawal relative to the ancestral haplotype (i.e., a haplotype consisting of the major allele at both SNPs). Only nominally significant evidence of an association with alcohol dependence or the severe use phenotype was found with the tested polymorphisms. Though previous studies conducted in this population reported evidence of linkage between the region and the severe use phenotype (Ehlers et al., 2004b) and associations between SNPs located in ADH1B and ADH1C and DSM-III-R defined alcohol dependence diagnoses (Wall et al., 2003), the present study included an expanded sample using a distinct analytic approach, which may have led to difficulties in reproducing the previous findings.
Both of the SNPs associated with withdrawal symptoms have shown previous evidence of association with alcohol dependence and related phenotypes. Specifically, rs2066702, which has been described in the literature as identifying the ADH1B*3 allele, has been associated with alcohol dependence in samples of African descent (Edenberg et al., 2006; Ehlers et al., 2001a, 2007; Luo et al., 2006) and an earlier study of this Native American population that used a subset of the participants described in the present study (Wall et al., 2003). Similarly, rs3762894 has been associated with alcohol dependence in several recent studies of ADH4 polymorphisms (Edenberg et al., 2006; MacGregor et al., 2009). Both SNPs have also been shown to affect the kinetic properties of ADH with the minor alleles of each SNP producing more active ADH isozymes than the major alleles (Birley et al., 2009; Edenberg, 2007; Thomasson et al., 1995). It has been suggested that these more active isozymes lead to a more rapid buildup of acetaldehyde, thus leading to a stronger response to alcohol and possibly more severe withdrawal symptoms. Nonetheless, there have been negative results reported for these SNPs (e.g., Kuo et al., 2008), thus posing the question of why the associations have been observed in some studies but not others.
Genetically complex disorders like alcohol dependence are likely influenced by a number of genes each exerting only a small effect on the broad clinical phenotype (Lander and Schork, 1994). These small effect sizes can complicate the search for susceptibility loci given that normal sampling variability will produce both positive and negative results when studies are insufficiently powered due to small sample sizes as is frequently the case. Nonetheless, such genes might be detected if they have a larger effect on a more narrowly defined phenotype. For example, withdrawal symptoms, the presence of which can indicate alcohol dependence with a "physiological component" as defined by DSM-IV (American Psychiatric Association 1994), appear to have particular clinical relevance and may identify an important subpopulation of alcohol dependent individuals with a more severe clinical course (Langenbucher et al., 2000; Schuckit et al., 1998). It is possible that genes involved in alcohol metabolism may play an important role in the etiology of the disorder within this subpopulation relative to individuals diagnosed with alcohol dependence without a "physiological component." The results of the present study as well as those of a previous linkage scan for alcohol dependence and related phenotypes (Ehlers et al., 2004b) are consistent with these conclusions in demonstrating that genetic linkage and association can be detected for withdrawal symptoms even when such associations are not observed for the broader alcohol dependence diagnosis.
The results of the present study also provide evidence that the observed associations between the ADH4 and ADH1B SNPs and alcohol dependence phenotypes previously reported in African, Asian, and Caucasian populations can be extended to Native American populations. This is particularly relevant for Native American populations given that the increased prevalence of alcohol dependence among Native Americans relative to Caucasians has unfortunately resulted in many negative stereotypes of Native Americans. Among these is the common belief that Native Americans may be more genetically susceptible to developing alcohol dependence due to unique differences in the metabolism of alcohol. The results of the present study suggest that in relation to the ADH genes located on chromosome 4q, the evidence for association is consistent with that observed in other ethnic groups, thus casting doubt on such theories.
The results of the present study should be interpreted in the context of several limitations. First, the findings may not generalize to other Native Americans or represent all Native Americans in this population. Second, the study gathered clinical data using retrospective methods; therefore, more information is needed using longitudinal techniques. Third, the association between ADH variants and alcohol-related phenotypes in this Native American population may not extend to other large population samples due to differences in genetic and environmental variables. Despite these limitations, this report represents an important step in an ongoing investigation to determine risk and protective factors associated with the development of substance use disorders in this high risk and understudied ethnic group.
In summary, the present study examined evidence for association between the cluster of ADH genes located on chromosome 4q and alcohol dependence and related phenotypes. Only nominal evidence for association between SNPs in these gene and alcohol dependence was observed, but the presence of withdrawal symptoms was significantly related to a SNP at the 5' end of ADH4 and a coding SNP in exon 9 of ADH1B with a known functional impact on ADH activity. Evidence of association between these two genes and alcohol misuse phenotypes has been reported in previous studies of ethnic groups including Caucasians, individuals of African descent, and Native Americans.
Acknowledgments
This research was supported by grants from the National Institutes of Health from the National Institute on Alcoholism and Alcohol Abuse (NIAAA) to Ian R. Gizer (T32 AA007573, PI - F. Crews) and to Howard J. Edenberg (AA006460) and from the NIAAA and the National Center on Minority Health and Health Disparities (NCMHD) (5R37 AA010201) to Cindy L. Ehlers.
Reference List
- Allgulander C, Nowak J, Rice JP. Psychopathology and treatment of 30,344 twins in Sweden. II. Heritability estimates of psychiatric diagnosis and treatment in 12,884 twin pairs. Acta Psychiatr Scand. 1991;83:12–15. doi: 10.1111/j.1600-0447.1991.tb05504.x. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnosis and statistical manual of mental disorders (DSM-III) Washington, DC: American Psychaitric Association; 1980. [Google Scholar]
- American Psychiatric Association, Task Force on DSM-IV. Diagnostic and statistical manual of mental disorders (DSM-IV) Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, Martin NG, Whitfield JB. ADH single nucleotide polymorphism associations with alcohol metabolism in vivo. Hum Mol Genet. 2009;18:1533–1542. doi: 10.1093/hmg/ddp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borras E, Coutelle C, Rosell A, Fernandez-Muixi F, Broch M, Crosas B, Hjelmqvist L, Lorenzo A, Gutierrez C, Santos M, Szczepanek M, Heilig M, Quattrocchi P, Farres J, Vidal F, Richart C, Mach T, Bogdal J, Jornvall H, Seitz HK, Couzigou P, Pares X. Genetic polymorphism of alcohol dehydrogenase in Europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatology. 2000;31:984–989. doi: 10.1053/he.2000.5978. [DOI] [PubMed] [Google Scholar]
- Bosron WF, Ehrig T, Li TK. Genetic factors in alcohol metabolism and alcoholism. Semin Liver Dis. 1993;13:126–135. doi: 10.1055/s-2007-1007344. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Heath AC, Reich T, Hesselbrock VM, Kramer JR, Nurnberger JI, Jr, Schuckit MA. Can we subtype alcoholism? A latent class analysis of data from relatives of alcoholics in a multicenter family study of alcoholism. Alcohol Clin Exp Res. 1996;20:1462–1471. doi: 10.1111/j.1530-0277.1996.tb01150.x. [DOI] [PubMed] [Google Scholar]
- Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–1175. doi: 10.1056/NEJMsb025007. [DOI] [PubMed] [Google Scholar]
- Carr LG, Xu Y, Ho WH, Edenberg HJ. Nucleotide sequence of the ADH2(3) gene encoding the human alcohol dehydrogenase beta 3 subunit. Alcohol Clin Exp Res. 1989;13:594–596. doi: 10.1111/j.1530-0277.1989.tb00383.x. [DOI] [PubMed] [Google Scholar]
- Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, Yin SJ. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65:795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Loh EW, Hsu YP, Chen CC, Yu JM, Cheng AT. Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br J Psychiatry. 1996;168:762–767. doi: 10.1192/bjp.168.6.762. [DOI] [PubMed] [Google Scholar]
- Chen YC, Peng GS, Wang MF, Tsao TP, Yin SJ. Polymorphism of ethanol-metabolism genes and alcoholism: correlation of allelic variations with the pharmacokinetic and pharmacodynamic consequences. Chem Biol Interact. 2009;178:2–7. doi: 10.1016/j.cbi.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Corbett J, Saccone NL, Foroud T, Goate A, Edenberg H, Nurnberger J, Porjesz B, Begleiter H, Reich T, Rice JP. A sex-adjusted and age-adjusted genome screen for nested alcohol dependence diagnoses. Psychiatr Genet. 2005;15:25–30. doi: 10.1097/00041444-200503000-00005. [DOI] [PubMed] [Google Scholar]
- Crabb DW. Ethanol oxidizing enzymes: roles in alcohol metabolism and alcoholic liver disease. Prog Liver Dis. 1995;13:151–172. [PubMed] [Google Scholar]
- Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Garcia-Andrade C, Wall TL, Sobel DF, Phillips E. Determinants of P3 amplitude and response to alcohol in Native American Mission Indians. Neuropsychopharmacology. 1998;18:282–292. doi: 10.1016/S0893-133X(97)00160-7. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Garcia-Andrade C, Wall TL, Cloutier D, Phillips E. Electroencephalographic responses to alcohol challenge in Native American Mission Indians. Biol Psychiatry. 1999;45:776–787. doi: 10.1016/s0006-3223(98)00113-9. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Garcia-Andrade C, Phillips E. EEG asymmetry: relationship to mood and risk for alcoholism in Mission Indian youth. Biol Psychiatry. 2001b;50:129–136. doi: 10.1016/s0006-3223(01)01132-5. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Harris L, Carr L. Association of the ADH2*3 allele with a negative family history of alcoholism in African American young adults. Alcohol Clin Exp Res. 2001a;25:1773–1777. [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004b;129B:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wall TL, Betancourt M, Gilder DA. The clinical course of alcoholism in 243 Mission Indians. Am J Psychiatry. 2004a;161:1204–1210. doi: 10.1176/appi.ajp.161.7.1204. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic scan for alcohol craving in Mission Indians. Psychiatr Genet. 2005;15:71–75. doi: 10.1097/00041444-200503000-00012. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Montane-Jaime K, Moore S, Shafe S, Joseph R, Carr LG. Association of the ADHIB*3 allele with alcohol-related phenotypes in Trinidad. Alcohol Clin Exp Res. 2007;31:216–220. doi: 10.1111/j.1530-0277.2006.00298.x. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Garcia-Andrade C, Wall TL, Ehlers CL. Alcohol expectancies in a Native American population. Alcohol Clin Exp Res. 1996;20:1438–1442. doi: 10.1111/j.1530-0277.1996.tb01146.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Andrade C, Wall TL, Ehlers CL. The firewater myth and response to alcohol in Mission Indians. Am J Psychiatry. 1997;154:983–988. doi: 10.1176/ajp.154.7.983. [DOI] [PubMed] [Google Scholar]
- Gilder DA, Gizer IR, Ehlers CL. Item response theory analysis of binge drinking and its relationship to lifetime alcohol use disorder symptom severity in an American Indian community sample. Alcohol Clin Exp Res. 2011;35(3):984–995. doi: 10.1111/j.1530-0277.2010.01429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilder DA, Wall TL, Ehlers CL. Psychiatric diagnoses among Mission Indian children with and without a parental history of alcohol dependence. J Stud Alcohol. 2002;63:18–23. [PubMed] [Google Scholar]
- Gilder DA, Wall TL, Ehlers CL. Comorbidity of select anxiety and affective disorders with alcohol dependence in Southwest California Indians. Alcohol Clin Exp Res. 2004;28:1805–1813. doi: 10.1097/01.alc.0000148116.27875.b0. [DOI] [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Hermansen L, Guze SB, Winokur G. Alcohol problems in adoptees raised apart from alcoholic biological parents. Arch Gen Psychiatry. 1973;28:238–243. doi: 10.1001/archpsyc.1973.01750320068011. [DOI] [PubMed] [Google Scholar]
- Guindalini C, Scivoletto S, Ferreira RG, Breen G, Zilberman M, Peluso MA, Zatz M. Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am J Psychiatry. 2005;162:1005–1007. doi: 10.1176/appi.ajp.162.5.1005. [DOI] [PubMed] [Google Scholar]
- Hall DA, Chambers GK, Lea RA. Haplotype analysis at the alcohol dehydrogenase gene region in New Zealand Maori. J Hum Genet. 2007;52:191–194. doi: 10.1007/s10038-006-0094-1. [DOI] [PubMed] [Google Scholar]
- Han Y, Gu S, Oota H, Osier MV, Pakstis AJ, Speed WC, Kidd JR, Kidd KK. Evidence of positive selection on a class I ADH locus. Am J Hum Genet. 2007;80:441–456. doi: 10.1086/512485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin D, Aharonovich E, Liu X, Mamman Z, Matseoane K, Carr LG, Li TK. Alcohol dependence symptoms and alcohol dehydrogenase 2 polymorphism: Israeli Ashkenazis, Sephardics, and recent Russian immigrants. Alcohol Clin Exp Res. 2002;26:1315–1321. doi: 10.1097/01.ALC.0000029597.07916.A9. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heckathorn DD. Respondent-driven sampling: a new approach to the study of hidden populations. Soc Probl. 1997;44:174–199. [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Hesselbrock VM, Segal B, Hesselbrock MN. Alcohol dependence among Alaska Natives entering alcoholism treatment: a gender comparison. J Stud Alcohol. 2000;61:150–156. doi: 10.15288/jsa.2000.61.150. [DOI] [PubMed] [Google Scholar]
- Higuchi S, Matsushita S, Murayama M, Takagi S, Hayashida M. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry. 1995;152:1219–1221. doi: 10.1176/ajp.152.8.1219. [DOI] [PubMed] [Google Scholar]
- Hurley TD, Edenberg HJ, Bosron WF. Expression and kinetic characterization of variants of human beta 1 beta 1 alcohol dehydrogenase containing substitutions at amino acid 47. J Biol Chem. 1990;265:16366–16372. [PubMed] [Google Scholar]
- Kalton G, Anderson DW. Sampling rare populations. J Roy Stat Soc. 1986;149:65–82. [Google Scholar]
- Kendler KS, Prescott CA, Neale MC, Pedersen NL. Temperance board registration for alcohol abuse in a national sample of Swedish male twins, born 1902 to 1949. Arch Gen Psychiatry. 1997;54:178–184. doi: 10.1001/archpsyc.1997.01830140090015. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Nishimura FT, Abe S, Fukunaga T, Tanii H, Saijoh K. Polymorphisms in the promoter region of the human class II alcohol dehydrogenase (ADH4) gene affect both transcriptional activity and ethanol metabolism in Japanese subjects. J Toxicol Sci. 2009;34:89–97. doi: 10.2131/jts.34.89. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, van Oordden EJ, Alexander J, Jiang C, Sullivan PF, Patterson DG, Walsh D, Kendler KS, Riley BP. Association of ADH and ALDH Genes With Alcohol Dependence in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) Sample. Alcohol Clin Exp Res. 2008;32:785–795. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Langenbucher J, Martin CS, Labouvie E, Sanjuan PM, Bavly L, Pollock NK. Toward the DSM-V: the Withdrawal-Gate Model versus the DSM-IV in the diagnosis of alcohol abuse and dependence. J Consult Clin Psychol. 2000;68:799–809. [PubMed] [Google Scholar]
- Li TK. Pharmacogenetics of responses to alcohol and genes that influence alcohol drinking. J Stud Alcohol. 2000;61:5–12. doi: 10.15288/jsa.2000.61.5. [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Luczak SE, Elvine-Kreis B, Shea SH, Carr LG, Wall TL. Genetic risk for alcoholism relates to level of response to alcohol in Asian-American men and women. J Stud Alcohol. 2002;63:74–82. [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005;15:755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Diplotype trend regression analysis of the ADH gene cluster and the ALDH2 gene: multiple significant associations with alcohol dependence. Am J Hum Genet. 2006;78:973–987. doi: 10.1086/504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Multiple ADH genes modulate risk for drug dependence in both African- and European-Americans. Hum Mol Genet. 2007;16:380–390. doi: 10.1093/hmg/ddl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Xue Y, Liu Y, Wang Z, Cui X, Li P, Fu S. Polymorphism study of seven SNPs at ADH genes in 15 Chinese populations. Hereditas. 2005;142:103–111. doi: 10.1111/j.1601-5223.2005.01910.x. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CS, Chung T, Kirisci L, Langenbucher JW. Item response theory analysis of diagnostic criteria for alcohol and cannabis use disorders in adolescents: implications for DSM-V. J Abnorm Psychol. 2006;115:807–814. doi: 10.1037/0021-843X.115.4.807. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Kaplan NL. Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Genet. 2001;68:1065–1067. doi: 10.1086/319525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DM, Pedersen SL, Lobos EA, Todd RD, Wall TL. ADH1B*3 and response to alcohol in African-Americans. Alcohol Clin Exp Res. 2010;34:1274–1281. doi: 10.1111/j.1530-0277.2010.01205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhib FB, Lin LS, Stueve A, Miller RL, Ford WL, Johnson WD, Smith PJ. A venue-based method for sampling hard-to-reach populations. Public Health Rep. 2001;116 Suppl 1:216–222. doi: 10.1093/phr/116.S1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan CJ, Robin RW, Osier MV, Sambuughin N, Goldfarb LG, Kittles RA, Hesselbrock D, Goldman D, Long JC. Allelic variation at alcohol metabolism genes (ADH1B, ADH1C, ALDH2) and alcohol dependence in an American Indian population. Hum Genet. 2003;113:325–336. doi: 10.1007/s00439-003-0971-z. [DOI] [PubMed] [Google Scholar]
- Nelson CB, Little RJ, Heath AC, Kessler RC. Patterns of DSM-III-R alcohol dependence symptom progression in a general population survey. Psychol Med. 1996;26:449–460. doi: 10.1017/s0033291700035534. [DOI] [PubMed] [Google Scholar]
- Neumark YD, Friedlander Y, Durst R, Leitersdorf E, Jaffe D, Ramchandani VA, O'Connor S, Carr LG, Li TK. Alcohol dehydrogenase polymorphisms influence alcohol-elimination rates in a male Jewish population. Alcohol Clin Exp Res. 2004;28:10–14. doi: 10.1097/01.ALC.0000108667.79219.4D. [DOI] [PubMed] [Google Scholar]
- Nishimura FT, Kimura Y, Abe S, Fukunaga T, Saijoh K. Effects of polymorphisms in untranslated regions of the class I alcohol dehydrogenase (ADH) genes on alcohol metabolism in Japanese subjects and transcriptional activity in HepG2 cells. Nihon Arukoru Yakubutsu Igakkai Zasshi. 2009;44:139–155. [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, Robinson VP, Neale MC, van den Oord EJ, Walsh D, Riley BP, Kendler KS. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Preuss UW, Ridinger M, Rujescu D, Samochowiec J, Fehr C, Wurst FM, Koller G, Bondy B, Wodarz N, Debniak T, Grzywacz A, Soyka M, Zill P. Association of ADH4 genetic variants with alcohol dependence risk and related phenotypes: results from a larger multicenter association study. Addict Biol. 2010;16:323–333. doi: 10.1111/j.1369-1600.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- Reich T, Cloninger CR, Van EP, Rice JP, Mullaney J. Secular trends in the familial transmission of alcoholism. Alcohol Clin Exp Res. 1988;12:458–464. doi: 10.1111/j.1530-0277.1988.tb00227.x. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Hum Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet B Neuropsychiatr Genet. 2000;96B:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Saha TD, Chou SP, Grant BF. Toward an alcohol use disorder continuum using item response theory: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Psychol Med. 2006;36:931–941. doi: 10.1017/S003329170600746X. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Daeppen JB, Eng M, Li TK, Hesselbrock VM, Nurnberger JI, Jr, Bucholz KK. Clinical relevance of the distinction between alcohol dependence with and without a physiological component. Am J Psychiatry. 1998;155:733–740. doi: 10.1176/ajp.155.6.733. [DOI] [PubMed] [Google Scholar]
- Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, Tian CH, Zhou CF, Zhou RL, Wang J, Zhao ZL, Xia GY. Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res. 1997;21:1272–1277. [PubMed] [Google Scholar]
- Sherva R, Rice JP, Neuman RJ, Rochberg N, Saccone NL, Bierut LJ. Associations and interactions between SNPs in the alcohol metabolizing genes and alcoholism phenotypes in European Americans. Alcohol Clin Exp Res. 2009;33:848–857. doi: 10.1111/j.1530-0277.2009.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak B, Frisch A, Maman Z, Aharonovich E, Alderson D, Carr LG, Weizman A, Hasin D. Effect of ADH1B genotype on alcohol consumption in young Israeli Jews. Alcohol Clin Exp Res. 2007;31:1297–1301. doi: 10.1111/j.1530-0277.2007.00438.x. [DOI] [PubMed] [Google Scholar]
- Takeshita T, Morimoto K, Mao X, Hashimoto T, Furuyama J. Characterization of the three genotypes of low Km aldehyde dehydrogenase in a Japanese population. Hum Genet. 1994;94:217–223. doi: 10.1007/BF00208273. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Beard JD, Li TK. ADH2 gene polymorphisms are determinants of alcohol pharmacokinetics. Alcohol Clin Exp Res. 1995;19:1494–1499. doi: 10.1111/j.1530-0277.1995.tb01013.x. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Crabb DW, Edenberg HJ, Li TK, Hwu HG, Chen CC, Yeh EK, Yin SJ. Low frequency of the ADH2*2 allele among Atayal natives of Taiwan with alcohol use disorders. Alcohol Clin Exp Res. 1994;18:640–643. doi: 10.1111/j.1530-0277.1994.tb00923.x. [DOI] [PubMed] [Google Scholar]
- van Beek JH, Willemsen G, de Moor MH, Hottenga JJ, Boomsma DI. Associations between ADH gene variants and alcohol phenotypes in Dutch adults. Twin Res Hum Genet. 2010;13:30–42. doi: 10.1375/twin.13.1.30. PM:20158305. [DOI] [PubMed] [Google Scholar]
- Wall TL, Thomasson HR, Schuckit MA, Ehlers CL. Subjective feelings of alcohol intoxication in Asians with genetic variations of ALDH2 alleles. Alcohol Clin Exp Res. 1992;16:991–995. doi: 10.1111/j.1530-0277.1992.tb01907.x. [DOI] [PubMed] [Google Scholar]
- Wall TL, Gallen CC, Ehlers CL. Effects of alcohol on the EEG in Asian men with genetic variations of ALDH2. Biol Psychiatry. 1993;34:91–99. doi: 10.1016/0006-3223(93)90261-b. [DOI] [PubMed] [Google Scholar]
- Wall TL, Garcia-Andrade C, Thomasson HR, Cole M, Ehlers CL. Alcohol elimination in Native American Mission Indians: an investigation of interindividual variation. Alcohol Clin Exp Res. 1996;20:1159–1164. doi: 10.1111/j.1530-0277.1996.tb01105.x. [DOI] [PubMed] [Google Scholar]
- Wall TL, Johnson ML, Horn SM, Carr LG, Smith TL, Schuckit MA. Evaluation of the self-rating of the effects of alcohol form in Asian Americans with aldehyde dehydrogenase polymorphisms. J Stud Alcohol. 1999;60:784–789. doi: 10.15288/jsa.1999.60.784. [DOI] [PubMed] [Google Scholar]
- Wall TL, Garcia-Andrade C, Wong V, Lau P, Ehlers CL. Parental history of alcoholism and problem behaviors in Native-American children and adolescents. Alcohol Clin Exp Res. 2000;24:30–34. [PubMed] [Google Scholar]
- Wall TL, Carr LG, Ehlers CL. Protective association of genetic variation in alcohol dehydrogenase with alcohol dependence in Native American Mission Indians. Am J Psychiatry. 2003;160:41–46. doi: 10.1176/appi.ajp.160.1.41. [DOI] [PubMed] [Google Scholar]
- Wall TL, Shea SH, Luczak SE, Cook TA, Carr LG. Genetic associations of alcohol dehydrogenase with alcohol use disorders and endophenotypes in white college students. J Abnorm Psychol. 2005;114:456–465. doi: 10.1037/0021-843X.114.3.456. [DOI] [PubMed] [Google Scholar]
- Whitfield JB. Meta-analysis of the effects of alcohol dehydrogenase genotype on alcohol dependence and alcoholic liver disease. Alcohol Alcohol. 1997;32:613–619. doi: 10.1093/oxfordjournals.alcalc.a008303. [DOI] [PubMed] [Google Scholar]
- Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van EP, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]