Abstract
Genome-wide association studies (GWAS) have been frequently conducted on general or isolated populations with related individuals. However, there is a lack of consensus on which strategy is most appropriate for analyzing dichotomous phenotypes in general pedigrees. Using simulation studies, we compared several strategies including generalized estimating equations (GEE) strategies with various working correlation structures, generalized linear mixed model (GLMM) and a variance component strategy (denoted LMEBIN) that treats dichotomous outcomes as continuous with special attentions to their performance with rare variants, rare diseases and small sample sizes. In our simulations, when the sample size is not small, for type I error, only GEE and LMEBIN maintain nominal type I error in most cases with exceptions for GEE with very rare disease and genetic variants. GEE and LMEBIN have similar statistical power and slightly outperform GLMM when the prevalence is low. In terms of computational efficiency, GEE with sandwich variance estimator outperforms GLMM and LMEBIN. We apply the strategies to GWAS of gout in the Framingham Heart Study. Based on our results, we would recommend using GEE ind-san in the GWAS for common variants and GEE ind-fij or LMEBIN for rare variants for GWAS of dichotomous outcomes with general pedigrees.
Keywords: genetic association, dichotomous phenotypes, familial relatedness
INTRODUCTION
A genome-wide association study (GWAS) is a powerful tool that is currently the most popular and successful way for identifying genetic loci associated with complex human traits and diseases. A variety of statistical approaches have been used in GWAS depending on sampling designs and types of outcomes. While there have been several publications on how to analyze continuous traits in general pedigrees [Kang et al., 2010; Zhang et al., 2010], strategies appropriate for dichotomous outcomes in general pedigrees were less studied. For this reason, we propose and study several statistical approaches that can be applied to GWAS of dichotomous outcomes with general pedigree data.
For continuous trait GWAS, [Zhang et al., 2010] proposed two linear mixed effects (LME) model based approaches that improves the computational efficiency of regular LME with large datasets. One is a compressed LME model approach that clusters individuals into groups based on kinship coefficients and reduce the effective sample size, and the other called population parameter previously determined (P3D) that eliminates the need to estimate population parameters (e.g. variance components) separately for every marker.
When the sample contains only unrelated individuals, logistic regression, a type of generalized linear models (GLM), can be used in association analyses of dichotomous outcomes (denote as GLM) But when the sample contains family data, GLM may increase the false positive rate. For continuous traits, a LME model [Almasy and Blangero 1998; Atkinson et al., 2009; Chen and Yang 2010]has been adopted to analyze GWAS of continuous traits to account for within-pedigree familial correlations using the relationship coefficient (twice the kinship coefficient) matrix. We consider an approach treating the dichotomous outcome as continuous and applying LME, similar to [Kang et al., 2010] (denoted as LMEBIN).
Two other methods, GLMM [Breslow and Clayton 1993] and GEE [Liang and Zeger 1986] are also considered. GLMM, a likelihood-based approach, synthesizes GLM and LME by including random effects in the mean model of GLM. GEE, a popular alternative to GLMM, models the marginal mean and covariance that incorporates a user-specified working correlation matrix using quasi-likelihood functions that do not require a full specification of the joint distribution. GEE is designed to be robust to misspecification of working correlation structure or deviation from specified outcome distribution [Liang and Zeger 1986].
In the present report, we use various simulation studies to compare GLMM, LMEBIN and GEE for dichotomous phenotypes by evaluating their type I error rates and statistical power in genetic association testing. The results are contrasted with GLM. In addition, for GEE we examine combinations of working correlation structure and variance estimator also by evaluating their type I error rates and statistical power. We consider independence (ind), exchangeable (exch), kinship coefficient (kin) and unstructured (unst) working correlation structures with sandwich variance estimator (san) and two jackknife-type variance estimators, one-step (j1s) and fully iterated (fij) jackknife variance estimators [Paik 1988; Lipsitz et al., 1994; Yan and Fan 2004]. Table I presents the abbreviations for different combinations of working correlation structure and variance estimator in the GEE analyses. As an example, the strategy of independence working correlation matrix with sandwich variance estimator in GEE is abbreviated as ind-san. In our simulations, kinship, geepack and lme4 R packages are used for LMEBIN, GEE and GLMM, respectively.
Table I.
Abbreviations for different combinations of working correlation structure and variance estimator used in GEE analyses.
| Variance estimator | Working correlation structure | |||
|---|---|---|---|---|
| independence | exchangeable | kinship | unstructured | |
| Sandwich | ind-san | exch-san | kin-san | unst |
| One-step jackknife | ind-j1s | exch-j1s | kin-j1s | - |
| Fully-iterated jackknife | ind-fij | exch-fij | kin-fij | - |
There are other existing approaches for testing genetic association for dichotomous traits with pedigree data. For multiplex case-control (MCC) design, data that consists of related cases and unrelated controls, [Slager and Schaid 2001] proposed a statistical test based on Armitage test for trend that uses a variance to account for family relationships and involves identical by descent (IBD) in computation. This approach can also be applied to data that consists of related cases and related controls and is extended to allow for modeling for covariates [Slager et al., 2003]. In addition, [Browning et al., 2005] proposed to use an individual weighting to account for the correlations between individuals due to IBD sharing. Unlike the methods we compared, the three methods used IBD sharing and were proposed for MCC or similar design for candidate-gene study. In a dense SNP map, high linkage disequilibrium (LD) is expected between nearby SNPs, which may lead to excess IBD sharing that should be handled when applied the methods to GWAS.
Lastly, as the number of SNPs being tested for association in a GWAS has grown rapidly from hundreds of thousands to approaching 10 million, it is also important for the applied statistical approaches to be time-efficient. Therefore, we also compare the computational efficiency of these approaches. We apply all strategies to analyze gout association in the Framingham Heart Study (FHS) GWAS of 550K SNPs with special attention to a rare variant that reached genome-wide significance (p-value< 5×10−8) [Yang et al., 2010].
METHODS
LMEBIN: The model for LMEBIN has the form,
| (1) |
where yij is the dichotomous outcome of the j-th individual in the i-th family, xij are the covariate values of the j-th individual in the i-th family, β is a vector of fixed effect parameters, are subject specific random effects correlated within a family, is the variance due to these effects assuming homogeneous variance among different individuals, and Ωi is the matrix of the coefficient of relationships or twice the coefficient of kinships of the i-th family, εij ~ N (0,σ2) is the error residual. LME is implemented in lmekin function of kinship R package. Despite the LME model is established for continuous dependent variables, we apply it to dichotomous outcomes. We used a modification of lmekin function and kinship R package of version 1.1.0–18 (http://cran.r-project.org/web/packages/kinship/). The estimates were obtained by maximum likelihood approach.
GLMM: Unlike LMEBIN that treats dichotomous outcomes as continuous, GLMM uses the logit link function g to relate the linear predictors of the j-th individual in the i-th family to the expected value of the response (μij).
| (2) |
In addition, instead of using relationship coefficient matrix as in LMEBIN, GLMM (implemented in lme4 R package of version 0.999375-32, http://cran.r-project.org/web/packages/lme4/) uses a covariance matrix that is determined by a parameter vector with much smaller dimension θ [Bates and DebRoy 2004]. The estimates were obtained by using restricted maximum likelihood approach.
GEE: GEE is a popular alternative to GLMM that models mean as (2) in GLMM and user-specified covariance structure (of the i-th family) as (3) below via quasi-likelihood, instead of specifying the exact likelihoods.
| (3) |
where φ is a common scale papameter, Ri (α) is the user-specified working correlation structure completely described by the parameter vector α, Ai is the diagonal matrix with entries aij = a (μij), a is a known variance function. We used geepack R package of version 1.0–17 (http://cran.r-project.org/web/packages/geepack/). The estimates were obtained by simultaneously solving estimating equations using Fisher scoring algorithm. The package allows user-specified fixed correlation structure that is appropriate when all pedigrees are of the same structure. In our first, second and fourth simulation studies, where 400 three-generation pedigrees of the same structure were generated, we used kinship coefficients matrix as the working correlation structure in addition to independent, exchangeable and unstructured correlation structures. However, in practice, this is not feasible since all pedigrees may not have the same structure. GEE implemented in R gee or geepack packages can use kinship coefficient matrix as working correlation matrix only when all families have exactly the same pedigree structure. Modification of these packages is needed in order to allow different families have different structures.
Jackknife variance estimator: Jackknife variance estimator is defined in what follows,
where N is the number of clusters (pedigrees in the current report), p is the number of parameters in the mean, β̂−i and β̂ are the estimates of β leaving out the i-th cluster and with all observations, respectively. In geepack R package, Fisher scoring method is used to find the roots of estimating equations. Fully iterated jackknife estimator (fij) uses β̂−i from full iteration of Fisher scoring, while one-step jackknife estimator (j1s) uses β̂−i from one step Fisher scoring.
Simulation Studies
Simulations are performed to evaluate the type I error and power of GEE strategies, LMEBIN and GLMM. The results are contrasted with logistic regression (denoted GLM here) that treats all individuals as independent. For GEE strategies, we consider independence, exchangeable, and kinship coefficients correlation structures with sandwich, one-step jackknife and fully iterated jackknife variance estimators.
For the first simulation study, four hundred three-generation pedigrees each with 10 members and identical structure (2 grand-parents, 4 parents and 4 offspring) are generated. Under additive genetic model, we first use SOLAR [Almasy and Blangero 1998] that uses a linear mixed effects model to simulate continuous phenotypes which follow a multivariate Normal distribution in a pedigree. A bi-allelic quantitative trait locus (QTL) that explains 1% of the phenotype variation is simulated and polygenic variation of 0.3 and 0.6 is considered. The SNP is simulated to be in linkage equilibrium (LE)/LD (r2 =0, 0.2, 0.5 and 0.8) with the QTL. In addition, the SNP and the QTL have the same MAF of 0.005, 0.01, 0.05, 0.1 and 0.3. We then dichotomize the simulated phenotypes with prevalence of 0.01, 0.05 and 0.2, and genotype relative risk of 1.3 under additive genetic model. Type I error is assessed with 10,000 replications and power is assessed empirically with 1,000 replications at a 0.05 significance level. That is, for each method and simulation scenario with inflated type I error, we used the 5% quantile of p-values from type I error simulations as the critical value. Under additive genetic model, GEE analyses are carried out with geepack R package, while GLMM analysis is carried out with lme4 R package, and LMEBIN is carried out with kinship R package by taking dichotomous trait as continuous. The GEE results with convergence problems were excluded for each considered working correlation structure.
To evaluate how the methods perform when the correlations of underlying continuous phenotypes among family members deviated from the relationship coefficients, the second simulation study further adds household effects to the simulated continuous phenotypes generated by SOLAR in the first simulation study. The additional household effects in each pedigree follow a multivariate Normal distribution with mean 0 and the covariance matrix (10×10) with each diagonal entry being 1 and each off-diagonal entry being 0.2. So, before dichotomization, in the within pedigree covariance matrix, now the total trait variance (diagonal) is 2 and the covariance of any two pedigree members (off-diagonal) is 0.2 greater than the relationship coefficient as in LMEBIN (and SOLAR). This unrealistic design is created mainly to see how LMEBIN would perform when the underlying within pedigree covariance matrix is changed.
To evaluate whether the methods are robust to small sample size, the third simulation study used exactly the same design as in the first simulation study with the only difference in the number of pedigrees, which is thirty in this case. We only focus on evaluating type I error rates in the simulation study.
RESULTS
First simulation study (400 three-generation pedigrees each with 10 members)
Complete type I error results evaluated at a 0.05 significance level for all methods are displayed in Supplementary Table 1. Type I error estimates of selected strategies and scenarios are presented in Figure 1. The scenarios are selected to demonstrate which strategies have inflated type I error under the scenarios. For GEE, the results from different working correlation structures have a similar pattern, so we only present results for combinations of independence (ind) correlation structure with sandwich (san), one-step jackknife (j1s) and fully iterated jackknife (fij) variance estimators, and for the combination of kinship (kin) coefficient correlation structure with sandwich variance estimator. When a condition’s prevalence is low (k=0.01 and 0.05, top panels of Figure 1), GEE ind-san and GEE kin-san have inflated type I errors for SNPs of low minor allele frequency (MAF) 0.005 and 0.01. The inflation is reduced by using jackknife variance estimators, but only GEE ind-fij and GEE exch-fij eliminates the inflation in most scenarios. GLMM has conservative type I errors, while GLM and LMEBIN have valid type I errors. The lower panels of Figure 1 show that GLMM has inflated type I error when the prevalence is higher (k=0.2) and that GLM has inflated type I error when the prevalence is not low (k>0.01); also, the type I error of GLM and GLMM increases with increasing prevalence as well as with increasing correlation among family members (polygenic variance vG). In contrast, all GEE approaches have valid type I errors across various prevalences (≥0.05) and polygenic variances, while GEE ind-fij and LMEBIN have valid type I error across all scenarios.
Figure 1.
Type I error estimates of selected strategies for selected scenarios evaluated at 0.05 significance level with 10,000 replicates. The title of each panel indicates which parameters are fixed in the scenario. MAF (p), prevalence (k) and polygenic variation (vG).
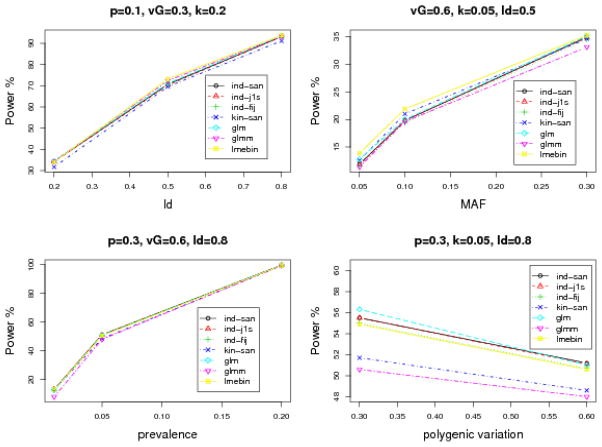
Power is estimated at a 0.05 significance level (Supplementary Table 2). To study the relationship between power estimates and simulation parameters - including linkage disequilibrium (LD) between the trait locus and the tested SNP, MAF, prevalence (k) and polygenic variation (vG) of the underlying continuous traits- we present GLM, GLMM, LMEBIN and GEE results in Figure 2. We observed that the statistical power increases when the LD between the SNP and the trait locus increases, when the MAF or the prevalence increases, and when the polygenic variation decreases. Power increases as the polygenic variation decreases because, when the polygenic variation (within pedigree correlation) decreases, each pedigree member contributes more information. In general, for GEE, the fully iterated jackknife variance estimator is slightly less powerful than the one-step jackknife variance estimator and the sandwich variance estimator, regardless of which working correlation structure is considered. The power is similar among different working correlation structures (median difference <1%; maximum difference 6.2%). When comparing across methods, LMEBIN and GEE have similar power, and they are slightly more powerful than GLMM and GLM (especially when prevalence is 0.01).
Figure 2.
The relationship between power evaluated at 0.05 significance level with 1,000 replicates and simulation parameters: LD (r2), MAF (p), prevalence (k) and polygenic variation (vG) of the simulated continuous traits. The title of each panel indicates which parameters are fixed in the scenario.
When using significance level of 0.001, the type I error and the power results are given in Supplement Tables 1-1 and 2-1, respectively. For type I error results GLMM has the least cases with inflated type I error rate. In general, most cases with conservative type I error in Supplementary Table 1 are no longer conservative except for GLMM and more cases with inflated type I error are observed for each approach except for GLMM. As for power, LMEBIN still has the best power in many cases.
The GEE results presented in Supplementary Tables 1, 1-1, 2 and 2-1 and Figures 1 and 2 exclude the results with convergence problems indicated in geepack output. The number of replicates with convergence problems for each GEE working correlation structure in simulations for assessing type I error are presented in Supplementary Table 3. The results show that the convergence problems in GEE tend to occur with low MAF, especially when the prevalence is also low. We further compare the median and the standard deviation of the regression coefficient estimates (Supplementary Table 4) and the standard error estimates from sandwich variance estimator (Supplementary Table 5) of SNP from results with and without convergence problems. In that setting, the regression coefficient estimates tend to be dramatically away from the null hypothesis 0 and the standard error estimates inflates dramatically except when using independence working correlation matrix, based on the results with convergence problems.
Second simulation study (including additional household effects in the 400 simulated pedigrees)
The type I error rates and statistical power results assessed at a 0.05 significance level are presented in Supplementary Tables 6 and 7, respectively. For type I error, the important findings as observed as well in the first simulation study are: 1) inflation is observed in GEE approaches when prevalence and MAF are low; 2) the inflation is eliminated in GEE ind-fij and GEE exch-fij; 3) inflation is observed in GLM and GLMM when prevalence is 0.2; and 4) LMEBIN has valid type I error in all scenarios. Power does not differ by more than 9.4% among all strategies across all scenarios (median difference 4.8%). The GEE results presented in Supplementary Tables 6 and 7 exclude the results with convergence problems. The number of replicates with convergence problems in GEE approaches for each working correlation structure in simulations for assessing type I error are presented in Supplementary Table 8, which shows similar pattern as in previous simulation study.
When using significance level of 0.001, the type I error and the power results are given in Supplement Tables 6-1 and 7-1, respectively. For type I error results, the results are similar as observed in the first simulation study (Supplementary Table 1-1). GLMM has the least cases with inflated type I error rate. In general, most cases with conservative type I error in Supplementary Table 6 are no longer conservative except for GLMM and more cases with inflated type I error are observed for each approach except for GLMM. As for power, kin-j1s has the best power in many cases in both Supplementary Tables 7 and 7-1.
An additional (the third) simulation study that uses FHS Offspring cohort consisting of about 3,000 individuals from 613 pedigrees (size 2-152, median size=3, Q3=5) is conducted to evaluate how the methods perform with pedigrees of unequal size. The results (Supplementary Tables 9, 9-1, 10, 10-1 and 11) are similar to the first study.
Fourth simulation study (30 three-generation pedigrees each with 10 members)
In this simulation study, we investigate robustness of the compared methods, particularly GEE with fij and LMEBIN, when the sample size is small, by evaluating their type I error rates. Due to such a small sample size, SNPs of low MAF (0.01 and 0.005) may become monomorphic and there may not be any affected individuals when prevalence is low (0.01) in simulations. In some scenarios, the small sample size also frequently caused numeric errors in lme4 R package (for GLMM) so that we are unable to obtain results for many replicates. Therefore, GLMM is not compared in this simulation study. In order to report more reliable results we focus on the scenarios with more than 5,000 replicates of results for each method. The type I error rates assessed at a 0.05 significance level are presented in Supplementary Table 12. Different from the first simulation study, GEE has inflated type I error rates in most scenarios and even though jackknife variance estimators reduce the inflation, the inflation may persist. In addition, LMEBIN has slightly inflated type I error rates in some scenarios more than observed in the first simulation study. In contrast, GLM is generally slightly conservative but has inflated type I error rates when prevalence is high and MAF is not low. When using 0.001 significance level (Supplementary Table 12-1), GLM has the least cases with inflated type I error rate. In general, GEE has inflated type I error almost in every scenario. LMEBIN has correct type I error only when MAF is 0.3. When the sample size further reduces to 20 or 10 pedigrees, similar results are observed (results not shown). The numbers of replicates with results, numbers with convergence problems in GEE, and numbers used to evaluate type I error rates are presented in Supplementary Tables 13, 14 and 15, respectively. Supplementary Tables 15 is obtained from Supplementary Tables 13 and 14.
Computational Efficiency
We measured the time taken by GEE ind-san, GEE ind-j1s, GLM, GLMM, and LMEBIN for analyzing 100 SNPs from FHS SNP Health Association Resource (SHARe) project using the FHS sample on a single Linux processor (2 × Dual-Core AMD Opteron(tm) Processor 2218 HE and total 12 GB RAM). Parallel computing was not used in this comparison. Table II reports the time (in seconds) used in analyzing the same 100 SNPs by GEE ind-san, GEE ind-j1s, GLM, GLMM and LMEBIN with sample size of 1,000, 2,000, 4,000 and 8,000 individuals from FHS sample. All the computations in Table II were done in the same R session. Table III reports the time (in seconds) used in analyzing the same 100, 200, 400 and 800 SNPs by GEE ind-san, GEE ind-j1s, GLM, GLMM and LMEBIN with 1,000 individuals from FHS sample. All the computations in Table III were done in the same R session. We did not measure time for GEE ind-fij, because it requires more iterations than GEE ind-j1s to reach convergence; thus, it would be the most time consuming. As expected, GLM has the best performance in computational efficiency, followed by GEE ind-san, GLMM, LMEBIN and GEE ind-j1s, and the computational time increases linearly with the number of analyzed SNPs. In a multi-CPU system that allows parallel computation, GWAF R package [Chen and Yang, 2010] provides a script for users to generate scripts to submit parallel jobs.
Table II.
The computation time (in seconds) used in analyzing the same 100 SNPs by GEE ind-san, GEE ind-j1s, GLM, GLMM and LMEBIN with sample size of the same 1,000, 2,000, 4,000 and 8,000 individuals from FHS sample.
| Sample size | GEE ind-san | GEE ind-j1s | GLM | GLMM | LMEBIN |
|---|---|---|---|---|---|
| 1,000 | 11 | 67 | 5 | 26 | 66 |
| 2,000 | 19 | 245 | 7 | 55 | 58 |
| 4,000 | 36 | 993 | 14 | 91 | 122 |
| 8,000 | 185 | 23671 | 28 | 131 | 570 |
Table III.
The computation time (in seconds) used in analyzing the same 100, 200, 400 and 800 SNPs by GEE ind-san, GEE ind-j1s, GLM, GLMM and LMEBIN with 1,000 individuals from the FHS sample.
| # of SNPs | GEE ind-san | GEE ind-j1s | GLM | GLMM | LMEBIN |
|---|---|---|---|---|---|
| 100 | 7 | 63 | 3 | 25 | 42 |
| 200 | 16 | 126 | 6 | 48 | 85 |
| 400 | 31 | 254 | 11 | 96 | 170 |
| 800 | 66 | 507 | 23 | 192 | 340 |
Application to FHS gout 550K GWAS
The sample size was 7,386 from 1,258 families and 197 (2.7%) had a history of gout [Yang et al., 2010]. For the GWAS of 550K SNPs, the genomic control inflation factor (λ) from GEE ind-san reduced from 1.17 to 1.04 after excluding 38,903 SNPs with convergence problems, and was1.04 from LEMBIN. Rs4753195 (MAF=0.042, call rate=99.8%) in MAML2 gene on chromosome 11 yielded a p-value of 4.47×10−8 by GEE ind-san. This SNP was not in the top hits of a recent meta-analysis of gout containing 28,283 individuals from 5 studies including FHS [Yang et al., 2010]. So we further applied GEE ind-j1s, GEE ind-fij, GLMM and LMEBIN to test the association between gout and rs4753195, which gave p-values 5.39×10−8, 6.65×10−8, 5.69×10−7 and 1.03×10−6 for GEE ind-j1s, GEE ind-fij, GLMM and LMEBIN, respectively. We also created 6.04×10 7 replicates of random SNP genotypes (MAF=0.042) under the null hypothesis using SOLAR and the same sample to evaluate the p-value from GEE ind-san empirically. From those replicates we obtained the empirical p-value= 8.61×10−7. In addition, we used the same MAF, prevalence, penetrance and the same sample to simulate 100 replicates of phenotypes conditional on rs4753195 to estimate power for GEE ind-san, LMEBIN and GLMM. We found GEE ind-san, LMEBIN and GLMM have 16%, 20% and 5% of power detecting the SNP with the genome-wide significance level of 5×10−8. With such low power, we think that the SNP is not necessarily a false positive. Therefore, we confirm that rs4753195 is not genome-wide significant but a suggestive hit by the approaches we applied.
DISCUSSION
In the present report, we compared several statistical strategies for GWAS of dichotomous outcomes with general pedigrees using three simulation studies. Based on the simulation results, when the sample size is not small, for type I error rates, we observed that LMEBIN has correct type I errors close to the 0.05 significance level, GEE strategies have inflated type I error rates when the MAF and the prevalence are both very low but the inflation can be reduced by jackknife type of variance estimators, and GLMM has inflated type I error rates when the prevalence is as high as 0.2. The inflated type I error rates for GEE in low MAF SNPs may be explained by the theoretic study of the property of GEE sandwich variance estimator by [Kauermann and Carroll 2001] that found the lower than expected coverage of confidence intervals for regression coefficients when the covariate design is unbalanced. As for statistical power, in general the differences in power among strategies are small and there is no single strategy that consistently outperforms the others. However, GEE and LMEBIN slightly outperform GLMM when the prevalence parameter is low. When using a more stringent significance level of 0.001 to investigate type I error rates, GLMM has the least inflation and is the only approach that has conservative type I error rates in more than one scenario. The reasons why GLMM does not maintain the correct type I error could be 1) Hauck-Donner effect [Hauck and Donner 1977] – in a binomial logit model, Wald statistic decreases to 0 when the distance between the regression coefficient estimate and the null value increases; and/or 2) As described in [Pinheiro and Bates 1995; Bates and DebRoy 2004], in the nonlinear mixed effects model, the evaluation of the likelihood function of the data may not have a closed-form, approximations to the likelihood may be used. The approximations may cause some bias. When the sample size is as small as 30 pedigrees each with 10 members, LMEBIN has the type I error rates closest to the 0.05 significance level with slight inflation observed in some scenarios, GLMM is not compared due to software error that terminates simulations in some scenarios, and GEE has more inflated type I error rates and the inflation can be reduced by jackknife variance estimators. In contrast, the type I error rate of GLM that ignores familial correlation tends to be conservative/inflated when the prevalence parameter is low/high. When evaluated at the 0.001 significance level, GEE tends to have inflated type I error rates, LMEBIN has correct type I error rate only when the MAF is not less than 0.1. In contrast, GLM has the least scenarios with inflated type I error rates and is the only approach that has conservative type I error rates.
To investigate why GLM’s conservativeness, we performed additional simulation study similar to the first one but taking out the familial correlations in both phenotypes and genotypes simulated under the null. The results are presented in Supplementary Tables 16 (at significance level 0.05), 16-1 (at significance level 0.001), and 17 (median and standard deviation of regression coefficient of SNP). Compare Supplementary Table 16 to Supplementary Table 1, GLM does not have inflation as expected, however, the conservativeness is still observed when the prevalence is low. The results should indicate that being conservative when the prevalence is low is a nature of GLM. When using significance level of 0.001, GLM becomes slightly inflated as appeared in Supplementary Tables 16-1 and 1-1. The conservativeness of GLM may be due to Hauck-Donner effect as shown in Supplementary Table 17 where in some cases the median of the regression coefficient estimate is away from 0 and the standard deviation of the regression coefficient is up to 6.6. In addition, when using likelihood ratio test (LRT), GLM (column glm.lrt) has inflation in some cases. Results from GEE ind-san, GEE ind-j1s, GEE ind-fij, GLMM-Wald test (column glmm) and GLMM-LRT (column glmm.lrt) were also presented.
Based on our simulations and computational efficiency comparisons, we would recommend the following strategy for GWAS of dichotomous outcomes with general pedigrees: using GEE ind-san in the GWAS for common variants and using GEE ind-fij or LMEBIN for rare variants (defined as a SNP with MAF 0.05 or less based on our first three simulation studies). In addition, it is known that GEE sandwich variance estimator is not robust when the number of clusters is small (n<30). Jackknife variance estimators have been suggested to replace sandwich variance estimator in this case [Lipsitz et al., 1994, Paik 1988]. When the sample size is small and the disease prevalence is low, LMEBIN or GLM may replace GEE. It is also suggested that GEE results with convergence problems should be excluded as demonstrated in the simulations (Supplementary Tables 4 and 5) and as shown in the application that the exclusion reduced the λ estimate. Even though LMEBIN is the only strategy with valid type I error in most cases and adequate statistical power, its regression coefficient cannot have the interpretation of odds ratio (unlike other strategies). This is a major drawback of LMEBIN and it precludes the fixed effects meta-analysis in potential collaboration with other studies. If a GWAS with small sample size uses GEE ind-san and has high genomic control parameter estimate after excluding results with convergence problems, ind-j1s can be used to reduce the systematic inflation. One reasonable modification to our suggested strategy is to replace GEE ind-san with GLM to SNPs of low MAF, because in general GLM has less inflated type I error for SNPs of low MAF than GEE ind-san, as shown in our simulations. In the application to FHS GWAS of gout, our analyses confirmed that rs4753195 is not genome-wide significant (p-value> 5×10−8) but a suggestive hit by the approaches we applied. When significant results are identified in GWAS in similar situations (low MAF and/or prevalence), we suggest validate the results with simulations as done in the application.
In the present study, we used various simulation studies to compare GEE, GLMM and LMEBIN by type I error rates, statistical power and computational efficiency for GWAS of dichotomous outcomes with general pedigree data. Our simulation results show that LMEBIN is the most robust approach to low MAF SNPs, low disease prevalence and small sample size but with less computational efficiency and un-interpretable effect estimates. Conversely, even though GEE is not as robust as LMEBIN, it is computationally efficient with interpretable results and the inflation in type I error rates can be reduced or eliminated by incorporating jackknife variance estimators or using GLM instead. In FHS, the GWAS pipeline for dichotomous outcomes uses the proposed strategy of GEE ind-san incorporating with GLM [Chen and Yang 2010] and has been successful. The present study reassures the validity of the proposed strategy and serves as a good reference that provides useful and practical information to investigators for their GWAS.
Supplementary Material
Acknowledgments
This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study (Contract No. N01-HC-25195), its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278) and NIH grants R01 NS017950-28 and R01-HL093328-01. A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center.
References
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson B, Therneau T, Zhao JH. kinship: mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees. 2009 http://cran.r-project.org/web/packages/kinship/
- Bates DM, DebRoy S. Linear mixed models and penalized least squares. J Multivariat Anal. 2004;91:1–17. [Google Scholar]
- Breslow NE, Clayton DG. Approximate Inference in Generalized Linear Mixed Models. J Ame Statist Assoc. 1993;88:9–25. [Google Scholar]
- Browning SR, Briley JD, Briley LP, Chandra G, Charnecki JH, Ehm MG, Johansson KA, Jones BJ, Karter AJ, Yarnall DP, Wagner MJ. Case-control single-marker and haplotypic association analysis of pedigree data. Genet Epidemiol. 2005;28:110–122. doi: 10.1002/gepi.20051. [DOI] [PubMed] [Google Scholar]
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck WW, Jr, Donner A. Wald’s Test as Applied to Hypotheses in Logit Analysis. J Amer Statist Assoc. 1977;72:851–853. [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S, Freimer NB, Sabattisabatti C, Eskineeskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauermann G, Carroll RJ. A note on the efficiency of sandwich covariance matrix estimation. J Amer Statist Assoc. 2001;96:1387–1396. [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Lipsitz SR, Dear BG, Zhao LP. Jackknife estimators of variance for parameter estimated from estimating equations with applications to clustered survival data. Biometrics. 1994;50:842–846. [PubMed] [Google Scholar]
- Paik MC. Repeated measurement analysis for nonnormal data in small samples. Commun Stat Simulat. 1988;17:1155–1171. [Google Scholar]
- Pinheiro JC, Bates DM. Approximations to the log-likelihood function in the nonlinear mixed-effects model. J Comput Graph Stat. 1995;4:12–35. [Google Scholar]
- Slager SL, Schaid DJ. Evaluation of Candidate Genes in Case-Control Studies: A Statistical Method to Account for Related Subjects. Am J Hum Genet. 2001;68:1457–1462. doi: 10.1086/320608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager SL, Schaid DJ, Wang L, Thibodeau SN. Candidate-gene association studies with pedigree data: Controlling for environmental covariates. Genet Epidemiol. 2003;24:273–283. doi: 10.1002/gepi.10228. [DOI] [PubMed] [Google Scholar]
- Yan J, Fine J. Estimating equations for association structures. Stat Med. 2004;23:859–874. doi: 10.1002/sim.1650. [DOI] [PubMed] [Google Scholar]
- Yang Q, Kottgen A, Dehghan A, Smith AV, Glazer NL, Chen MH, Chasman DI, Aspelund T, Eiriksdottir G, Harris TB, Launer L, Nalls M, Hernandez D, Arking DE, Boerwinkle E, Grove ML, Li M, Kao WL, Chonchol M, Haritunians T, Li G, Lumley T, Psaty BM, Shlipak M, Hwang SJ, Larson MG, O’Donnell CJ, Upadhyay A, van Duijn CM, Hofman A, Rivadeneira F, Stricker B, Uitterlinden AG, Pare G, Parker AN, Ridker PM, Siscovick DS, Gudnason V, Witteman JC, Fox CS, Coresh J. Multiple genetic loci influence serum urate and their relationship with gout and cardiovascular disease risk factors. Circ Cardiovasc Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.