Abstract
Background
Network-based approaches may leverage genome-wide association (GWA) analysis by testing for the aggregate association across several pathway members. We aimed to examine if networks of genes that represent experimentally determined protein-protein interactions are enriched in genes associated with risk of coronary heart disease (CHD).
Methods and Results
GWA analyses of ~700,000 SNPs in 899 incident CHD cases and 1,823 age- and sex-matched controls within the Nurses’ Health and the Health Professionals Follow-Up Studies were used to assign gene-wise p-values. A large database of protein-protein interactions (PPI) was used to assemble 8,300 unbiased protein complexes and corresponding gene-sets. Superimposed gene-wise p-values were used to rank gene-sets based on their enrichment in genes associated with CHD. After correcting for the number of complexes tested, one gene-set was overrepresented in CHD-associated genes (p-value=0.002). Centered on the beta-1-adrenergic receptor gene (ADRB1), this complex included 18 protein interaction partners that, so far, have not been identified as candidate loci for CHD. Five of the 19 genes in the top-complex are reported to be involved in abnormal cardiovascular system physiology based on knock-out mice (4-fold enrichment; p-value, Fisher’s exact test= 0.006). Ingenuity pathway analysis revealed that especially canonical pathways related to blood pressure regulation were significantly enriched in the genes from the top complex.
Conclusions
The integration of a GWA study with PPI data successfully identifies a set of candidate susceptibility genes for incident CHD that would have been missed in single-marker GWA analysis.
Keywords: Genetics of cardiovascular disease, acute myocardial infarction, epidemiology
Genome-wide association (GWA) studies provide a unique opportunity for the unbiased exploration of novel genetic variation of importance to phenotypic traits. The first series of GWA studies of coronary heart disease (CHD) and more broadly defined cardiovascular disease (CVD) phenotypes elucidated DNA sequence variations at the 9p21.3 locus as a robustly replicated risk-conferring region,1, 2,3 but through a series of larger GWA study consortia about 10 susceptibility loci have been reported.4–6 The recent publication of results from the multi-ethnic Coronary Artery Disease (C4D) Genetics Consortium,7 the first Han Chinese GWA study,8 and the CARDIoGRAM consortium with more than 20,000 coronary artery disease cases,9 yielded an additional 18 new loci. However, the complexity of the phenotype,10 small effect sizes, and between-study differences may complicate the identification of many true associations in meta-analyses that necessarily assumes homogeneity across the individual studies. Most GWA studies to date have focused on the identification of the strongest single-locus associations, but the identification of combined effects of many weakly associated variants is especially appealing for complex diseases, such as CHD, that is likely not caused by single variants or by a single biological pathway. Thus, another suggested approach for reducing the noise inherent in moderately powered high-density data collected within internally homogenous populations, is the integration of additional biological data on pathway organization through the use of a protein-protein interaction (PPI) database.11–16 By enabling tests of sets of single nucleotide polymorphisms (SNPs) within physically interacting gene products (direct or indirect), PPI data can augment GWA analysis since a set of SNPs, each with a moderate, but genuine association, in aggregate may have improved statistical significance. Although several databases provide gene-sets that resemble well-known canonical pathways, high-confidence PPI data may to a larger degree mimic the unbiased nature of GWA studies due to its increased coverage and detail of even non-canonical pathways.11, 17 Initial approaches have proven useful to suggest novel genes and gene-networks involved in other complex phenotypes such as obesity,18 type 2 diabetes,13 breast and pancreatic cancer,19 multiple sclerosis,20 and Crohn’s disease,21 that were not identified in the traditional GWA analysis. The completeness of such integrative analysis relies strongly on the gene-sets tested. We aimed to examine if networks of genes that represent experimentally determined protein-protein interactions are enriched in genes associated with risk of incident CHD. To leverage our GWA analysis of CHD within two homogenous American prospective cohorts including 899 incident cases collected through more than 10 years of follow-up, we used our PPI database InWeb,14 which covers ~13,000 human proteins and 173,500 high-confidence experimentally-derived protein-protein interactions based on 11 publicly available PPI-databases.
Methods
Study population
The Nurses’ Health Study (NHS) enrolled 121,701 female nurses aged 30 to 55 who returned a mailed questionnaire in 1976 regarding lifestyle and medical history. The Health Professionals Follow-up Study (HPFS) enrolled 51,529 males aged 40 to 75 who returned a similar questionnaire in 1986. Participants of both cohorts have received follow-up questionnaires biennially to record newly diagnosed illnesses. Detailed descriptions of the study cohorts have been published previously.22, 23
Blood collection and DNA extraction in nested case-control study
Between 1989 and 1990, a blood sample was requested from all active participants in NHS and collected from 32,826 women. Similarly, blood samples were requested between 1993 and 1995 and obtained from 18,225 HPFS participants. For details on storage of blood samples, please see the online supplement.
In both cohorts, nested case-control studies were designed using incident CHD, with non-fatal myocardial infarction (MI) and fatal CHD as the outcome. Diagnosis of MI was confirmed on the basis of the criteria of the World Health Organization (symptoms plus either diagnostic electrocardiographic changes or elevated levels of cardiac enzymes). Deaths were identified from state vital records and the National Death Index or reported by the participant’s next of kin or the postal system. Fatal CHD was confirmed by an examination of hospital or autopsy records, by the listing of CHD as the cause of death on the death certificate, if CHD was the underlying and most plausible cause, and if evidence of previous CHD was available. Among participants who provided blood samples and who were free of diagnosed cardiovascular disease or cancer at blood draw, we identified 474 women and 454 men with incident CHD between blood draw and June, 2004. Using risk-set sampling,24 controls were selected randomly and matched in a 1:2 ratio on age, smoking, and month of blood return, among participants who were free of cardiovascular disease at the time CHD was diagnosed in the case. In this study design, a control for an early case may be included again if the person develops CHD during follow-up, thus after counting such converters only once (as cases), the total number of samples sent for genotyping were 1354 HPFS samples and 1521 NHS samples.
The present study was approved by the institutional review boards at Brigham and Women’s Hospital and Harvard School of Public Health.
Genotyping and Quality Control
Details on the protocol for DNA extraction has been included in the online supplement. Genotyping was done using the Affymetrix Genome-Wide Human 6.0 array and the Birdseed calling algorithm.25 Genotypic data for a total of 1,330 HPFS samples (98%) passed laboratory technical quality control criteria and missing call rate <0.05. Likewise, 96% of the NHS samples were successfully genotyped. A subset of 312 NHS samples were not genotyped together with the remaining CHD case-control set as they overlapped with previous GWA studies of breast cancer (Illumina 550) and type 2 diabetes (Affymetrix 6.0). These samples were processed and subjected to quality control as part of the earlier GWAS (leaving n=272 samples with available data) and SNPs also present on the Affymetrix 6.0 platform were subsequently merged with the cleaned CHD data. Details on methods for data cleaning and assesment of population structure in the datasets are included in the online supplement. Due to very few samples with substantial evidence of non-European genetic ancestry, these samples were excluded from subsequent analysis (n=24). SNPs that were monomorphic, had a missing call rate ≥2%, a HWE p-value <1×10-4, or a MAF <0.02 were excluded, leaving a total of 724,881 in HPFS and SNPs that passed quality control in HPFS and 721,316 in NHS for analysis of called genotypes. Imputation of ~2.5 million SNPs was performed using MACH software (v1.0.16) with HapMap CEU phased II data (Release 22) as the reference panel.
Genome-wide association analysis of coronary heart disease
To analyze the association between each SNP (coded as counts of minor alleles) and risk of CHD, we ran logistic regression analysis using PLINK software.26 We adjusted for matching factors used in the design of the nested case-control study (age and smoking) and the top three eigenvectors. We also analyzed the MACH dosage files of the imputed SNPs (with MAF ≥0.05) in logistic regression models (adjusting for same covariates as above) using the ProbABEL package from the ABEL set of programs.27 Fixed-effects meta-analysis was performed to combine the study-specific β-estimates using the METAL package.28
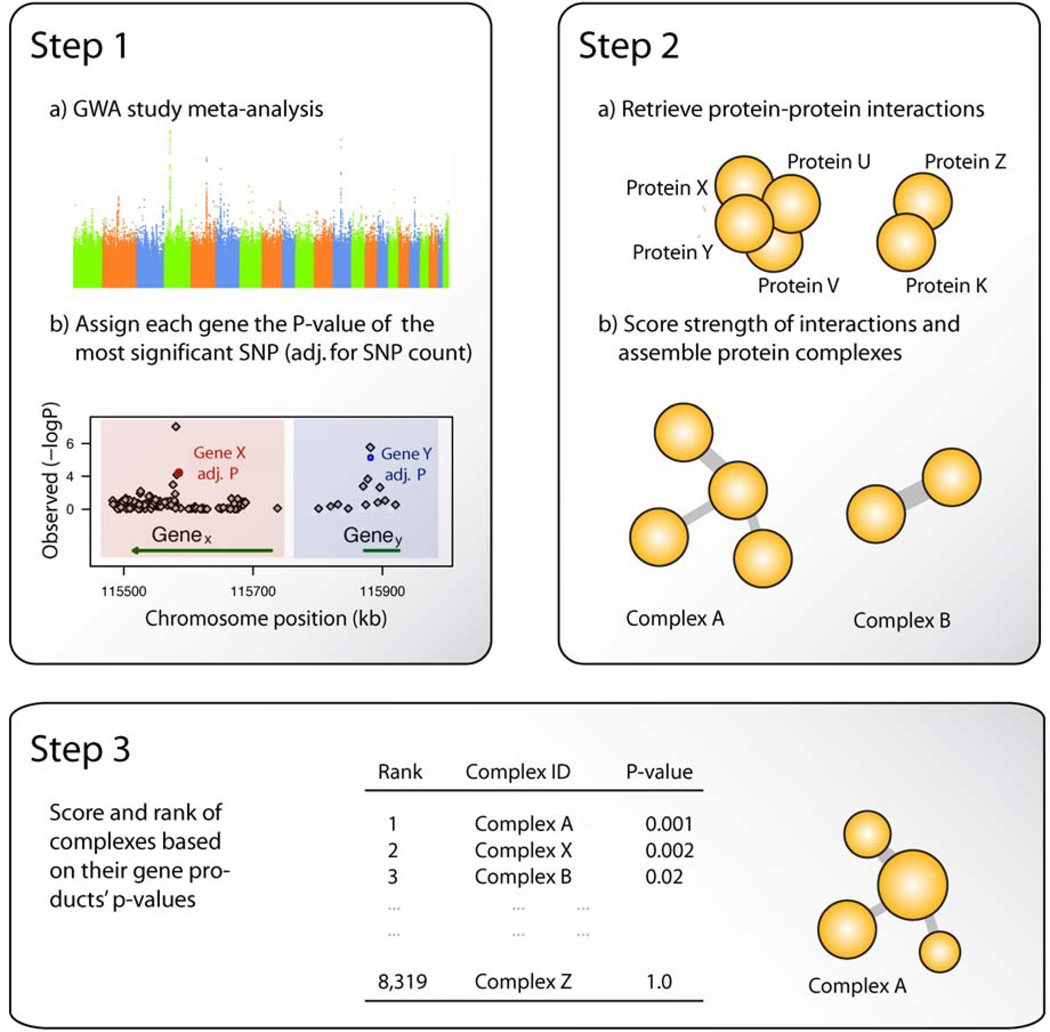
Systems biology-based approaches that integrate data on protein interactions are necessarily restricted to the protein-coding part of the genome. We mapped all GWA SNPs that passed quality control to 21,800 protein-coding genes (423,450 mapped SNPs, ~57% of all SNPs on the Affymetrix 6.0 arrays) (Figure 1a). This process is gene-centric such that SNPs that are not within genes or their 70 kb upstream and 20 kb downstream flanking regions were discarded. SNPs were allowed to map to more than one gene. Each gene was assigned a p-value based on the SNP with the lowest GWA p-value within the gene transcript(s) and its flanking regions. Subsequently, the Šidàk correction was applied to adjust the p-value for each gene by the number of effective tests (uncorrelated number of SNPs within each gene and its flaking regions, as per Galwey 2007).16, 29
Figure 1.
Conceptual framework for the integration of GWA data with protein-protein interaction data. The approach consists of three overall steps. First, GWA meta-analysis, SNPs are mapped to genes, genes are scored based on its most significant SNP, and the gene scores are adjusted by the number of independent SNPs mapped to the given gene. Second, protein complexes are assembled based on experimentally derived protein-protein interactions. Finally, the gene-sets underlying the protein complexes are scored based on their genes’ p-values and their protein-protein interaction confidence scores.
Protein-protein Interactions and CHD-specific protein complexes
Protein-protein interactions comprise both transient interactions (e.g. phosphorylation events) and stable interactions (e.g. the cytoskeleton). Our comprehensive, experimentally derived database of protein-protein interactions InWeb (version 2.9) covers ~13,000 human proteins and 350,029 protein-protein interactions of which 173,500 can be regarded as high-confidence interactions (as described below).14 The database is updated on a monthly basis with interactions retrieved from all major experimental PPI databases (details available in online supplement). Strengths of the InWeb database include the relative high coverage (4-fold increase in number of interaction compared the Human Protein Reference Database, HPRD)30 and a quantitative assessment of confidence in the reported interactions. The (continuous) confidence score (ranging from 0 [low support] to 1 [strong support]) is assigned by taking into account a) the number and quality of the publications reporting each of the interactions and b) the number of shared interaction partners of two interacting proteins.14 The assembly of 8,531 gene-sets was accomplished by iteratively assigning each protein in the database and its first-order interaction partners to a protein complex (Figure 1, Step 1b). As such a construction of protein complexes results in a relatively large number of overlapping complexes, complexes that were more than 80% similar (similarity of gene-sets assessed by the Jaccard Index) were merged. After superimposing the gene-wise p-values from the GWA analysis onto the network, we used a modified version of an approach published by Ideker et al. to iteratively assess whether any of the gene-sets that were derived from the protein complexes were enriched in CHD-associated genes.31 Given a gene-set of size k, this was accomplished by (1) converting all k gene p-values to z-scores using the inverse normal cumulative distribution function, (2) weighting them with the interaction confidence score of the protein-protein interaction with the central hub protein (a step that was not part of the original algorithm), (3) calculating a gene-set score by summing the weighted z-scores, and then (4) subtracting the sum of an average gene-set of size k (calculated based on 100,000 randomized gene-set scores), and dividing by the standard deviation of an average sub-network of size k. Formally, step 1 can be formulated as Zi = F−1(1 − pi),i ∈ {1,…k}, steps 2–3 as , and step 4 as , where pi denotes the p-value of gene i, zi denotes the z-score of gene i, F−1 denotes the inverse normal cumulative distribution function, the confidence score for the protein-protein interaction between gene product i and the central hub gene product, Sgene-set denotes the score of the gene-set after steps 1–4, μk and σk denote the mean and standard deviation of 100,000 randomized gene-set scores, and Zgene-set denotes the final gene-set z-score. Using this methodology, all gene-sets were ranked based on their computed z-scores (Figure 1c). Because SNPs were allowed to map to several overlapping genes, some gene-sets may be assigned artificially inflated scores if they comprise genes that overlap on a given chromosome and are scored based on the same low SNP p-value. To avoid this potential bias we discarded one of the genes in any overlapping gene pair in a given complex (genes were considered to overlap if their transcripts were closer than 200kb to each other). This approach is conservative as it avoids inflated complex scores, but in some cases may reduce significance of truly associated complexes that comprise co-localizing gene-products with independent associations. In our present analysis, the top complex remained the same with or without discarding overlapping genes (and for different exclusion thresholds). We assessed the significance of our observed top scoring complex by comparing its score with a background distribution of 100 scores generated under the null hypothesis that the complex is not associated with CHD case control status. The background distribution was estimated on the basis of 100 permutations of our GWA meta-analysis (randomizing the case control status) and re-computations of the gene scoring- and complex scoring step for each permutation. An ideal scenario would include up to 1 million permutations but the aggregate computing times for the GWA analysis, the gene scoring step, and the complex enrichment analysis did not allow for this.
After identification of the top-ranking complex we searched the literature to see if the genes were known as human CVD candidate genes. To assess over-representation of known CVD susceptibility genes we used a list of 123 genes reported by Samani et al. and updated it with GWA findings in the NIH Catalog of Genome-Wide Association Studies (Suppl. Table 1).3, 32 We also tested for overrepresentation of a list of 889 genes found to affect the cardiovascular system physiology (MP:0001544) in knockout mice (Mouse Genome Informatics database; www.informatics.jax.org, Jackson Laboratory, Bare Harbor Maine) (of which 837 were among the gene products in our PPI database). To ensure that the observation that genes from our top complex were overrepresented in the mouse cardiovascular physiology gene-set was not due to chance, we compared the observed enrichment score to a background distribution of 10,000 scores computed based on randomly sampled protein complexes. Each of the random complexes matched the observed complex in size, and each gene-product was sampled with a probability equal to its observed prevalence in the total set of protein complexes. In addition to the enrichment analysis of known human and mice CHD risk genes, we used the Ingenuity Pathway Analysis software tool (IPA, version 9.0, Ingenuity Systems Inc. 2011) to systematically test the complex genes for pathway enrichment.
Results
Characteristics of incident cases and matching controls in the two cohorts are presented in Table 1. The women in the NHS were slightly younger, more likely to smoke, and more likely to report a diagnosis of hypertension or diabetes. GWA analysis of each cohort separately and in meta-analysis did not reveal any markers that exceeded the genome-wide significant threshold (Supplement, fig 1).
Table 1.
Baseline characteristics of women and men in whom coronary heart disease developed during follow-up and matched controls in the Nurses’ Health Study (NHS) and the Health Professionals Follow-Up Study (HPFS).*
| HPFS | NHS | |||
|---|---|---|---|---|
| Characteristic | Cases | Controls | Cases | Controls |
| N | 425 | 878 | 464 | 945 |
| Age, years | 64.5 (8.6) | 64.2 (8.5) | 60.2 (6.3) | 59.8 (6.3) |
| Women, % | 0% | 0% | 100% | 100% |
| Hypertension, † % | 37.2% | 29.0% | 50.2% | 27.3% |
| Diabetes, † % | 9.0% | 3.8% | 14.4% | 6.24% |
| Current smoker, % | 9.7% | 8.7% | 27.8% | 26.1% |
| Total cholesterol, mg/dL | 5.5 (1.0) | 5.2 (1.0) | 6.1 (1.0) | 5.9 (1.0) |
| HDL cholesterol, mg/dL | 1.1 (0.3) | 1.2 (0.3) | 1.3 (0.4) | 1.6 (0.4) |
| Triglyceride, mg/dL | 1.8 (1.5) | 1.5 (2.2) | 1.6 (1.0) | 1.3 (0.7) |
| BMI, kg/m2 | 26.0 (3.2) | 25.6 (3.3) | 26.0 (6.6) | 24.5 (5.8) |
Age and smoking were matching factors. Values are means and standard deviation of continuous covariates (except triglyceride levels which is reported as median and IQR) or percentages. Triglyceride levels were log-transformed before analysis and only reported in fasting participants (HPFS= 65%, NHS= 79%).
Self-reported diagnosis before blood draw.
Based on the InWeb database, a total of 8,351 protein complexes were assembled based on large-scale proteomics data from human and model organisms. We restrained our analyses to high-confidence protein-protein interactions only, including a subset that we recently validated experimentally in human heart tissue.45 The resulting protein complexes were tested for enrichment in CHD-associated genes by using the gene-wise p-values from the GWA analysis to create z-scores and ranking the complexes (gene-sets) by their combined z-scores, adjusted for the size of each gene-set, and weighted by the confidence of the interactions between the peripheral gene-products and the central protein of the complex. After correcting for the number of complexes tested, one gene-set was significantly overrepresented in CHD-associated genes from our GWA meta-analysis (p-value=0.002). The gene complex was centered on the known candidate gene for the beta-1-adrenergic receptor (ADRB1) (fig. 2). To ensure that the top complex was not merely significantly enriched in genes with low p-values but indeed significantly associated with CHD case control status, we permuted the phenotype-genotype association in the GWA analysis 100 times and re-computed the complex score at each iteration. We found that the score for the observed ADRB1 complex was superior to any of the scores for the randomized complexes. In Figure 2, the additional 18 genes that were part of the complex of interacting proteins are scaled according to their gene-wise p-values. As shown in more detail in Table 2, the genes; membrane-associated guanylate kinase inverted 1 (MAGI1), the protein kinase cAMP-dependent catalytic alpha (PRKACA), and the Golgi associated PDZ and coiled-coil motif containing (GOPC) were nominally significant after correcting for the number of independent SNPs in each gene, whereas the remainder showed weaker, or no association. In the combined test of a gene-set, all known interaction partners are included regardless of their GWA signal and the strength of the association for the complex relies on the sum of all gene-wise p-values of the interacting genes. Our results did not change when we based our analysis on the imputed GWA data rather than the hard-call genotypes.
Figure 2.
Top-ranking protein complex from the genome-wide analysis of coronary heart disease in the Nurses’ Health and the Health Professionals Follow-Up Studies. The gene products (nodes) are scaled in size according to their significance (larger indicates smaller p-value). Edges between the nodes denote experimentally-derived protein-protein interactions. Red nodes denote genes in the complex with corrected gene-wise p-values < 0.05.
Full gene names available in online supplement.
Table 2.
Genes and primary SNPs in the top-ranking protein complex based on the GWA meta-analysis of risk of CHD in the Nurses’ Health and Health Professionals Follow-Up Studies.
| Gene | Gene p-value* |
Top SNP | MAF | OR | Top SNP, raw p-value |
# SNPs in gene |
# independent SNPs in gene |
|---|---|---|---|---|---|---|---|
| MAGI1 | 7.8E-04 | rs7620106 | 0.40 | 1.30 | 9.1E-06 | 251 | 86 |
| PRKACA | 0.004 | rs40282 | 0.46 | 1.20 | 0.002 | 2 | 2 |
| GOPC | 0.028 | rs12664183 | 0.28 | 1.23 | 0.001 | 100 | 27 |
| ADRB1 | 0.041 | rs17653278 | 0.06 | 0.70 | 0.003 | 41 | 12 |
| MAGI3 | 0.073 | rs4839312 | 0.26 | 1.21 | 0.005 | 62 | 14 |
| MAGI2 | 0.086 | rs2065198 | 0.46 | 1.22 | 0.001 | 579 | 149 |
| GRB2 | 0.107 | rs7223674 | 0.05 | 0.72 | 0.014 | 32 | 8 |
| DLGAP2 | 0.143 | rs7836020 | 0.45 | 1.18 | 0.005 | 100 | 33 |
| ARRB1 | 0.217 | rs2279129 | 0.08 | 0.75 | 0.013 | 34 | 19 |
| DLG4 | 0.251 | rs5412 | 0.16 | 1.15 | 0.069 | 7 | 4 |
| GNAL | 0.277 | rs2848465 | 0.22 | 0.83 | 0.009 | 85 | 36 |
| GIPC1 | 0.304 | rs4926215 | 0.47 | 0.89 | 0.042 | 15 | 8 |
| DLG1 | 0.335 | rs7616531 | 0.26 | 1.17 | 0.020 | 56 | 20 |
| GPRASP1 | 0.348 | rs17340189 | 0.11 | 1.15 | 0.090 | 6 | 5 |
| ADRA2A | 0.355 | rs7908645 | 0.34 | 1.13 | 0.056 | 15 | 8 |
| SH3GL3 | 0.441 | rs8025427 | 0.42 | 1.15 | 0.018 | 68 | 31 |
| GNAS | 0.508 | rs1022697 | 0.43 | 1.13 | 0.032 | 50 | 21 |
| SH3GL2 | 0.562 | rs10810813 | 0.16 | 0.83 | 0.019 | 162 | 43 |
| PDE4D | 0.677 | rs17799450 | 0.08 | 1.34 | 0.015 | 312 | 74 |
adjusted for the number of independent SNP within loci (see last column, independent SNPs in gene). Full gene names available in online supplement.
Next, we assessed whether the ADRB1 complex was enriched in known human or mice CVD risk genes. No significant overlap with the list of 123 susceptibility genes reported by Samani et al. and the genetic loci identified in GWA studies of CVD was observed (p-value=0.1). 3, 32 To test for enrichment in CHD-specific evidence from mouse studies, we searched for the genes in the top-complex in an a priori defined set of genes causing abnormal cardiovascular physiology in knockout mice. Among a total of 889 genes reported for that phenotype, 837 human homologs were among the 12,793 genes included in our analysis, and 5 were part of the 19 genes in the ADRB1 complex; representing a 4-fold enrichment (p-value, Fisher’s exact test =0.006). The five genes also found in mice knockout gene-sets, were ADRB1, ADRA2A, ARRB1, PDE4D, and GRB2 of which all except PDE4D were reported to play a role in the regulation of blood pressure, cardiac function, and hypertrophy. Because proteins that are known to interact physically are more likely to have similar functional annotation,33 possible chance-correlations resulting in a gene with a low p-value could potentially result in a falsely associated complex if the falsely associated gene’s annotation resembles the phenotype of interest. To test for this possible bias, we subjected the mouse gene-set enrichment analysis to 10,000 random complexes sampled from the PPI network and found that only 13 out of the 10,000 randomized enrichment scores were lower than our observed score (p-value=0.001).
We used Ingenuity Pathway Analysis to examine whether the annotations of the genes in the ADRB1 complex were enriched for any particular phenotype. Between 10 and 12 of the 19 genes were reported in cardiovascular, neurological, endocrine, and immunological disorders (Table 3). Moreover, several cardiovascular related pathways were enriched in genes from the complex. The top canonical pathway was cardiac hypertrophy signaling. To better ensure that the observed enrichment was not due to chance, we sampled 100 random gene-sets comprising 19 genes each, and performed IPA analysis based on each set. Only one random gene-set exhibited enrichment in cardiovascular disease genes as strong as the observed enrichment for the ADRB1 complex gene-set. Thus, we conclude that our top complex was significantly enriched in genes associated with cardiovascular disease (p-value< 0.05). None of the random gene-sets were significantly enriched in the cardiac hypertrophy canonical pathway, suggesting that the ADRB1 complex gene set was significantly enriched in genes from this pathway too. We confined this IPA permutation analysis to 100 iterations as the software does not allow automation and all runs were done manually.
Table 3.
Diseases and Disorders, and canonical pathways enriched in genes from top complex, identified by Ingenuity Pathway Analysis
| IPA Disease/Disorder | P-value for enrichment | # genes |
|---|---|---|
| Respiratory Disease | 2.34E-07 - 5.00E-02 | 3 |
| Cardiovascular Disease | 1.12E-05 - 4.31E-02 | 12 |
| Neurological Disease | 2.81E-05 - 3.51E-02 | 12 |
| Endocrine System Disorders | 3.63E-05 - 2.94E-02 | 10 |
| Immunological Disease | 3.63E-05 - 1.56E-02 | 11 |
| IPA canonical pathway | P-value for enrichment | Ratio (# genes in top complex/total # genes in pathway) |
| Cardiac Hypertrophy Signaling | 1.62E-07 | 0.024 (6/246) |
| G Beta Gamma Signaling | 3.28E-06 | 0.034 (4/117) |
| cAMP-mediated Signaling | 5.75E-06 | 0.023 (5/216) |
| PTEN Signaling | 6.83E-06 | 0.033 (4/123) |
| Cardiac b-adrenergic Signaling | 1.43E-05 | 0.026 (4/151) |
Discussion
We conducted a protein network-based GWA analysis to leverage our moderately powered GWA study of CHD. Using GWA data from two individually homogeneous studies, we integrated the gene-wise p-values with a large database of protein-protein interactions. By exploiting the complementary nature of genetic variation and biochemical data, we successfully identified a gene complex of 19 candidate genes that may play a role in the etiology of incident CHD. Subsequent pathway enrichment analysis indicated that the top complex was significantly enriched in (a) genes from the canonical cardiac hypertrophy signaling pathway (the highest ranking pathway in the IPA analysis), (b) genes annotated with cardiovascular disease (the second most enriched trait in the IPA analysis), and (c) mouse genes annotated in the cardiovascular system physiology. Our results provide preliminary evidence that known CHD-related genes coalesce onto distinct protein complexes. Most of the genes in the top complex had relatively small effect sizes, making them unlikely findings in traditional single-locus GWA analyses of CHD.
To our knowledge, our study of incident CHD is the first attempt at integration of data on the human interactome with GWA data in relation to incident CHD. As shown in the enrichment analyses, the top complex comprises several genes that previously have been annotated to cardiovascular disease and, in particular, the cardiac hypertrophy signaling pathways. Except for ADRB1, these known genes were not nominally significant by themselves but leveraged due to their interaction with genes that comprised SNPs, which exhibited association with CHD in our GWA study. In addition, the top complex was significantly enriched in genes from the Mouse Genetics Initiative database that were annotated in the ‘cardiovascular system physiology’. The genes ADRB1, GRB2, ADRA2A were found to overlap between all three a priori defined gene-sets (overview provided in Table 4). It is well-known that the β1-adrenergic receptor plays an important role in the regulation of cardiac contractility. In candidate genetic studies, ADRB1 SNPs have been associated with blood pressure34 and risk of future CHD, which might be particularly true for individuals with elevated blood pressure.35 Studies on the adrenergic pathway genes, including ADRA2A, that encodes the α2A-adrenergic receptor, have not shown consistent associations. However, recently a polymorphism in ADRA2A that caused overexpression of the protein, was shown to strongly reduce insulin secretion from pancreatic cells and be associated with an elevated risk of type 2 diabetes.36 The GRB2 gene encodes the growth factor receptor-bound protein 2. So far, information on this genetic locus links it to an important role in lymphocytes and growth cells, but no human genetic epidemiologic studies have investigated this locus in relation to cardiometabolic disorders.
Table 4.
Overview of genes* in the identified top complex and their implication in the IPA cardiovascular disease set (CVD), the cardiac hypertrophy signaling pathway (Hypertrophy) and the mouse knock out models of abnormal cardiovascular physiology (MGI).
| Gene | SNP | CVD | Hypertrophy | MGI |
|---|---|---|---|---|
| MAGI1 | rs7620106 | yes | No | no |
| PRKACA | rs40282 | no | Yes | no |
| GOPC | rs12664183 | no | No | no |
| ADRB1 | rs17653278 | yes | yes | yes |
| MAGI3 | rs4839312 | yes | no | no |
| MAGI2 | rs2065198 | yes | no | no |
| GRB2 | rs7223674 | yes | yes | yes |
| DLGAP2 | rs7836020 | yes | no | no |
| ARRB1 | rs2279129 | no | no | yes |
| DLG4 | rs5412 | no | no | no |
| GNAL | rs2848465 | yes | no | no |
| GIPC1 | rs4926215 | no | yes | no |
| DLG1 | rs7616531 | no | no | no |
| GPRASP1 | rs17340189 | no | no | no |
| ADRA2A | rs7908645 | yes | yes | yes |
| SH3GL3 | rs8025427 | yes | no | no |
| GNAS | rs1022697 | yes | yes | no |
| SH3GL2 | rs10810813 | yes | no | no |
| PDE4D | rs17799450 | yes | no | yes |
Full gene names available in online supplement.
Alternative approaches for augmenting GWA data by testing significance beyond single locus associations include pathway-based approaches, such as methods that search the protein interactome for dense subnetworks enriched in GWA signal19, 20 and methods that assess pre-defined gene-sets for enrichment in GWA signal,16, 17, 37 The former class of methods are inspired by early work of Ideker,31, 38 and employ an heuristic search algorithm to identify subnetworks that are enriched in gene-products that in aggregate associate with the phenotype. The advantage of these methods is that they do not assume any a priori delineation of pathways. However, the main drawback is that they rely on user-specified parameters that control the size of the subnetworks identified by the algorithm. In addition, none of them incorporate information on the confidence of the experimentally derived protein-protein interactions. While our approach resembles the recently presented dmGWAS approach,19 only ours incorporated a score on confidence in the reported interactions. Another strength of our approach is that it is based on a PPI database that, despite its high coverage (our analysis includes twice as many interactions as those used in the dmGWAS method), solely includes high-confidence experimentally derived interactions. While InWeb does not rely on predicted protein-protein interactions, which are more prone to false-positive interactions, it still entails approximately 173,500 interactions from a total of 11 databases. Our integration-based approach has strengths, but limitations as well. One of the inherent limitations is that it only covers roughly 60% of all SNPs present on genotyping platforms. Consequently, SNPs within distal enhancer regions are discarded, as are other long-range regulatory relationships. However, systematic tissue- and condition-specific expression quantitative trait loci analyses are increasingly contributing to the development of more refined SNP to gene mapping schemes. Among other limitations, we had a relative small sample size in our GWA study of incident CHD and were limited to Caucasians. However, the application of the novel PPI approach still allowed us to uncover gene sets that were not otherwise identified. Replication in another prospective study setting is important to verify and demonstrate the significance of the ADRB1 complex in incident cardiovascular disease. Genome-wide data in a cohort with prospectively collected CHD cases would be preferable. Likely only larger GWA consortia will have sufficient number of incident cases to accomplish this. Other approaches to follow-up on our proof-of-principle approach and findings include investigations of rare variants through targeted re-sequencing or expression profiling across CHD-relevant tissues from appropriate cases and controls.
In conclusion, our approach suggests that integration of other layers of biological evidence with a moderately powered GWA study of CHD in two homogenous study populations can yield potentially interesting sets of candidate genes that would be missed in traditional statistical GWA analyses. We identified one gene-set, centered on ADRB1, that was overrepresented in CHD-associated genes in our GWA study and also enriched in genes involved in the cardiovascular disease phenotype and particularly blood pressure regulation pathways. Our novel approach highlighted 19 genes that warrant further association and functional studies in terms of risk of CHD and blood pressure.
New approaches for the analysis of genome-wide association (GWA) data that allow for integration with complementary data are needed for phenotypes that do not support large-scale recruitment schemes or meta-analysis. Most GWA studies to date have focused on the identification of the strongest single-locus associations, but the identification of combined effects of many weakly associated variants is especially appealing for complex diseases, such as CHD, that is likely not caused by single variants or by a single biological pathway. We used a network-based analysis based on protein-protein interaction data and GWA results for coronary heart disease (CHD) in 899 CHD cases and 1,823 controls from the Nurses’ Health and the Health Professionals Follow-up studies. P-values for each gene were assigned according to the smallest p-values for SNPs within 70kb upstream and 20kb downstream of the gene, and corrected for the effective number of independent SNPs for each gene. Networks of genes that represent direct protein-protein interactions were examined to identify protein complexes with evidence for association in aggregate. After correcting for the number of complexes tested, a significant association was observed between CHD and genes that encode proteins that interact with the beta-1-adrenergic receptor gene (ADRB1). This complex included 18 protein interaction partners that, so far, have not been identified as candidate loci for CHD. This comprehensive approach highlights that network-based analysis have the potential to reveal additional novel genes of interest to a phenotype beyond those discovered in GWA studies of common variants.
Supplementary Material
Acknowledgments
Funding Sources:
This study was supported by HL34594, CA87969, HL35464, and CA55075 from the National Institutes of Health, Bethesda, MD, with additional support for genotyping from Merck Research Laboratories, North Wales, PA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosures:
Merck Research Laboratories supported the genotyping of the Nurses’ Health and Health Professionals Follow-Up Studies though an unrestricted grant.
References
- 1.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 2.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O'Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Ardissino D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Mannucci PM, Schwartz SM, Siscovick DS, Yee J, Friedlander Y, Elosua R, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Kathiresan S, Meigs JB, Williams G, Nathan DM, MacRae CA, O'Donnell CJ, Salomaa V, Havulinna AS, Peltonen L, Melander O, Berglund G, Voight BF, Kathiresan S, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Musunuru K, Daly MJ, Purcell S, Voight BF, Purcell S, Nemesh J, Korn JM, McCarroll SA, Schwartz SM, Yee J, Kathiresan S, Lucas G, Subirana I, Elosua R, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Samani NJ, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall AS, Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Schunkert H, Samani NJ, Erdmann J, Ouwehand W, Hengstenberg C, Deloukas P, Scholz M, Cambien F, Reilly MP, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein SE, Rader DJ, Scheffold T, Berger K, Stoll M, Huge A, Girelli D, Martinelli N, Olivieri O, Corrocher R, Morgan T, Spertus JA, McKeown P, Patterson CC, Schunkert H, Erdmann E, Linsel-Nitschke P, Lieb W, Ziegler A, Konig IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Engert JC, Do R, Xie C, Anand S, Kathiresan S, Ardissino D, Mannucci PM, Siscovick D, O'Donnell CJ, Samani NJ, Melander O, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Altshuler D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Tregouet DA, Cambien F, Bruse P, Aherrahrou Z, Wagner AK, Stark K, Schwartz SM, Salomaa V, Elosua R, Melander O, Voight BF, O'Donnell CJ, Peltonen L, Siscovick DS, Altshuler D, Merlini PA, Peyvandi F, Bernardinelli L, Ardissino D, Schillert A, Blankenberg S, Zeller T, Wild P, Schwarz DF, Tiret L, Perret C, Schreiber S, El Mokhtari NE, Schafer A, Marz W, Renner W, Bugert P, Kluter H, Schrezenmeir J, Rubin D, Ball SG, Balmforth AJ, Wichmann HE, Meitinger T, Fischer M, Meisinger C, Baumert J, Peters A, Ouwehand WH, Deloukas P, Thompson JR, Ziegler A, Samani NJ, Schunkert H. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peden JF, Hopewell JC, Saleheen D, Chambers JC, Hager J, Soranzo N, Collins R, Danesh J, Elliott P, Farrall M, Stirrups K, Zhang W, Hamsten A, Parish S, Lathrop M, Watkins HC, Clarke R, Deloukas P, Kooner JS, Goel A, Ongen H, Strawbridge RJ, Heath S, Malarstig A, Helgadottir A, Ohrvik J, Murtaza M, Potter S, Hunt SE, Delepine M, Jalilzadeh S, Axelsson T, Syvanen AC, Gwilliam R, Bumpstead S, Gray E, Edkins S, Folkersen L, Kyriakou T, Franco-Cereceda A, Gabrielsen A, Seedorf U, Eriksson P, Offer A, Bowman L, Sleight P, Armitage J, Peto R, Abecasis G, Ahmed N, Caulfield M, Donnelly P, Froguel P, Kooner AS, McCarthy MI, Samani NJ, Scott J, Sehmi J, Silveira A, Hellenius ML, van 't Hooft FM, Olsson G, Rust S, Assmann G, Barlera S, Tognoni G, Franzosi MG, Linksted P, Green FR, Rasheed A, Zaidi M, Shah N, Samuel M, Mallick NH, Azhar M, Zaman KS, Samad A, Ishaq M, Gardezi AR, Memon FU, Frossard PM, Spector T, Peltonen L, Nieminen MS, Sinisalo J, Salomaa V, Ripatti S, Bennett D, Leander K, Gigante B, de FU, Pietri S, Gori F, Marchioli R, Sivapalaratnam S, Kastelein JJ, Trip MD, Theodoraki EV, Dedoussis GV, Engert JC, Yusuf S, Anand SS. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 8.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, Cheng X, Li C, Yang R, Wang CC, Wu G, Lu QL, Bai Y, Huang YF, Yin D, Yang Q, Wang XJ, Dai DP, Zhang RF, Wan J, Ren JH, Li SS, Zhao YY, Fu FF, Huang Y, Li QX, Shi SW, Lin N, Pan ZW, Li Y, Yu B, Wu YX, Ke YH, Lei J, Wang N, Luo CY, Ji LY, Gao LJ, Li L, Liu H, Huang EW, Cui J, Jia N, Ren X, Li H, Ke T, Zhang XQ, Liu JY, Liu MG, Xia H, Yang B, Shi LS, Xia YL, Tu X, Wang QK. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 9.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de FU, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitsios GD, Dahabreh IJ, Trikalinos TA, Schmid CH, Huggins GS, Kent DM. Heterogeneity of the phenotypic definition of coronary artery disease and its impact on genetic association studies. Circ Cardiovasc Genet. 2011;4:58–67. doi: 10.1161/CIRCGENETICS.110.957738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbers CC, van Eijk KR, Franke L, Mulder F, van der Schouw YT, Wijmenga C, Onland-Moret NC. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet Epidemiol. 2009;33:419–431. doi: 10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- 12.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry JR, McCarthy MI, Hattersley AT, Zeggini E, Weedon MN, Frayling TM. Interrogating type 2 diabetes genome-wide association data using a biological pathway-based approach. Diabetes. 2009;58:1463–1467. doi: 10.2337/db08-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, Hinsby AM, Tumer Z, Pociot F, Tommerup N, Moreau Y, Brunak S. A human phenome interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25:309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
- 15.Brorsson C, Hansen NT, Lage K, Bergholdt R, Brunak S, Pociot F. Identification of T1D susceptibility genes within the MHC region by combining protein interaction networks and SNP genotyping data. Diabetes Obes Metab. 2009;11 Suppl 1:60–66. doi: 10.1111/j.1463-1326.2008.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pers TH, Hansen NT, Lage K, Koefoed P, Dworzynski P, Miller ML, Flint TJ, Mellerup E, Dam H, Andreassen OA, Djurovic S, Melle I, Borglum AD, Werge T, Purcell S, Ferreira MA, Kouskoumvekaki I, Workman CT, Hansen T, Mors O, Brunak S. Meta-analysis of heterogeneous data sources for genome-scale identification of risk genes in complex phenotypes. Genet Epidemiol. 2011;35:318–332. doi: 10.1002/gepi.20580. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am J Hum Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu YJ, Guo YF, Zhang LS, Pei YF, Yu N, Yu P, Papasian CJ, Deng HW. Biological pathway-based genome-wide association analysis identified the vasoactive intestinal peptide (VIP) pathway important for obesity. Obesity. 2010;18:2339–2346. doi: 10.1038/oby.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, Wu W, Uitdehaag BM, Kappos L, Polman CH, Matthews PM, Hauser SL, Gibson RA, Oksenberg JR, Barnes MR. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18:2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C, Wilson DC, Walters T, Kim C, Frackelton EC, Lionetti P, Barabino A, Van LJ, Guthery S, Denson L, Piccoli D, Li M, Dubinsky M, Silverberg M, Griffiths A, Grant SF, Satsangi J, Baldassano R, Hakonarson H. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colditz GA, Manson JE, Hankinson SE. The Nurses' Health Study: 20-year contribution to the understanding of health among women. J Womens Health. 1997;6:49–62. doi: 10.1089/jwh.1997.6.49. [DOI] [PubMed] [Google Scholar]
- 23.Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC. Reproducibility and validity of a expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol. 1992;135:1114–1126. doi: 10.1093/oxfordjournals.aje.a116211. [DOI] [PubMed] [Google Scholar]
- 24.Prentice RL, Breslow NE. Retrospective studies and failure time models. Biometrika. 1978;65:153–158. [Google Scholar]
- 25.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, Gabriel SB, Purcell S, Daly MJ, Altshuler D. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aulchenko YS, Struchalin MV, Van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, vey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galwey NW. A new measure of the effective number of tests, a practical tool for comparing families of non-independent significance tests. Genet Epidemiol. 2009;33:559–568. doi: 10.1002/gepi.20408. [DOI] [PubMed] [Google Scholar]
- 30.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC, Yatish AJ, Kavitha MP, Menezes M, Choudhury DR, Suresh S, Ghosh N, Saravana R, Chandran S, Krishna S, Joy M, Anand SK, Madavan V, Joseph A, Wong GW, Schiemann WP, Constantinescu SN, Huang L, Khosravi-Far R, Steen H, Tewari M, Ghaffari S, Blobe GC, Dang CV, Garcia JG, Pevsner J, Jensen ON, Roepstorff P, Deshpande KS, Chinnaiyan AM, Hamosh A, Chakravarti A, Pandey A. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18 Suppl 1:S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 32.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 34.Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, Rose L, Rice K, Verwoert GC, Launer LJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, Caulfield M, Van Duijn CM, Ridker PM, Munroe PB, Levy D. Association of Hypertension Drug Target Genes With Blood Pressure and Hypertension in 86 588 Individuals. Hypertension. 2011;57:903–910. doi: 10.1161/HYPERTENSIONAHA.110.158667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leineweber K, Heusch G. Beta 1- and beta 2-adrenoceptor polymorphisms and cardiovascular diseases. Br J Pharmacol. 2009;158:61–69. doi: 10.1111/j.1476-5381.2009.00187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosengren AH, Jokubka R, Tojjar D, Granhall C, Hansson O, Li DQ, Nagaraj V, Reinbothe TM, Tuncel J, Eliasson L, Groop L, Rorsman P, Salehi A, Lyssenko V, Luthman H, Renstrom E. Overexpression of alpha2A-adrenergic receptors contributes to type 2 diabetes. Science. 2010;327:217–220. doi: 10.1126/science.1176827. [DOI] [PubMed] [Google Scholar]
- 37.Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6(8) doi: 10.1371/journal.pgen.1001058. pii: e1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.