The ability to monitor phosphorylation events can provide valuable information pertaining to signal transduction regulation and for developing effective therapeutics targeted at aberrant kinase activities.[1] Kinase activity can be optically reported using sensors based on short peptides or domains resembling amino acids near the phosphorylation site of a substrate protein. Of these, peptide-based reporters exhibit large fluorescence changes from small molecule fluorophores chemically attached to the peptide sensor,[2] but introducing these reporters into cells is challenging. Protein-based reporters contain genetically appended fluorescent proteins and have revealed novel spatiotemporal information regarding kinases in living cells despite their moderate signal changes.[3] Nonetheless, the subcellular location, trafficking and lifetime of a full-length substrate protein cannot be faithfully replicated by comparatively simplified peptide or domain sensor elements.[4] Moreover, many kinases derive substrate specificity using distal residues in addition to those proximal to the phosphorylation site, and can have multiple substrate proteins.[5] Therefore, current kinase reporters provide limited information on the phosphorylation state of a particular substrate protein.
Here we present a method to optically report the phosphorylation status of a specific full-length substrate protein, signal transducer and activator of transcription 3 (STAT3), which plays a leading role in many oncogenic and developmental pathways.[6] A small molecule fluorophore in the format of an unnatural amino acid (Uaa) was genetically introduced into STAT3 to sense its phosphorylation state. A large fluorescence change was observed when the STAT3 probe was phosphorylated by Src kinase in vitro and when it was incubated with endogenously activated STAT3 from mammalian nuclear extracts. This method enables optical investigation of protein phosphorylation on the substrate level with high specificity.
Our strategy is to genetically incorporate a fluorescent Uaa into the target protein at a site close to the residue subject to phosphorylation (Figure 1a). The negatively charged phosphate group may alter local polarity or pH, properties to which the fluorophore of the Uaa is designed to be sensitive. By using a full-length substrate protein, one can incorporate the fluorescent Uaa at any site close to the phosphorylated residue in the tertiary structure, providing more flexibility in choosing the optimal sensor location than peptide-based methods. The closely positioned phosphorylated residue and the fluorescent Uaa can be within the same target protein or in different proteins if the target protein is oligomeric or part of a complex.
Figure 1.
Reporting the phosphorylation status of a substrate protein using genetically encoded Uaas. a) Schematic illustration. The broken line indicates that the phosphorylated residue and the fluorescent Uaa can be on the same or separate proteins. b) Crystal structure of the STAT3β homodimer depicting two monomers (pink and cyan) binding DNA (grey) (PDB 1BG1). c) Region framed in (b) illustrating the location of pTyr705 (red) in relation to Trp564 (green). d) Structure of L-(7-hydroxycoumarin-4-yl) ethylglycine (7HC).
Upon phosphorylation on Tyr705, STAT3 dimerizes through the reciprocal binding of phosphotyrosine (pTyr) into the SH2 domain of an opposing monomer (Figure 1b). The dimer subsequently translocates into the nucleus as an activated transcription factor. We reasoned that introduction of the negatively charged pTyr705 into the SH2 domain would alter the pH within the binding pocket and that a pH-sensitive fluorophore should report such a change. A good candidate is 7-hydroxycoumarin (quantum yield=0.63), whose fluorescence intensity and excitation wavelength are pH-dependent with a pKa of ~7.8.[7] Based on the crystal structure of the DNA-bound STAT3β homodimer (Figure 1b),[8] we selected Trp564 for mutation to L-(7-hydroxycoumarin-4-yl) ethylglycine (7HC, Figure 1d). Trp564 is located within the second layer of the SH2 binding pocket close to the pTyr of the opposing monomer, but distant from Tyr705 of the same monomer and outside of the DNA binding domain (Figure 1c). Trp is also similar in size to 7HC. Collectively, these properties should minimize any potential interference from introducing 7HC.
7HC was genetically incorporated into the STAT3β isoform in E. coli using a reported orthogonal tRNA/aminoacyl-tRNA synthetase pair[9] to suppress the 564TAG codon in our optimized expression system (see Supporting Information). To verify 7HC incorporation, cell lysates were analyzed by Western blot using an antibody against STAT3 (Figure 2a). Full-length STAT3β was observed only when 7HC was added to the growth medium. Wild type (wt) and 7HC-containing STAT3β proteins (7HC-STAT3β) were purified with Ni-NTA chromatography. A single bright blue fluorescent band was observed only for purified 7HC-STAT3β on SDS-PAGE (Figure 2b). Incorporation of 7HC into STAT3β at the TAG site was confirmed using MS (Figure S1).
Figure 2.
7HC-STAT3β, similar to wt STAT3β, can be phosphorylated and binds a consensus DNA sequence. a) Western blot analysis of E. coli lysates from cells expressing STAT3β(564TAG) and the 7HC-specific tRNA/synthetase pair. b) Photograph of SDS-PAGE analysis of wt STAT3β and 7HC-STAT3β(564TAG) expressed in the presence and absence of 7HC. The gel was exposed to 365 nm UV light. (c) Western blot of protein samples incubated with and without Src kinase. Probing with a STAT3-specific antibody ensured that comparable amounts of STAT3 were loaded. (d) EMSA using 32P labelled hSIE DNA probe. Probe incubated with wt STAT3β and 7HC-STAT3β was upshifted indicating that both proteins bind the hSIE probe. Specific competition was seen with excess unlabeled probe yielding dissociation constants for wt STAT3β (Kd = 6.3 ± 0.6 nM, n = 4) and 7HC-STAT3β (Kd = 6.8 ± 1.6 nM, n = 4).
To determine if Trp564 mutation to 7HC affects STAT3 function, purified 7HC-STAT3β protein was tested in vitro for its ability to be phosphorylated by the nonreceptor tyrosine kinase Src and its ability to bind the high-affinity sis-inducible element (hSIE) consensus DNA sequence in an electrophoretic mobility shift assay (EMSA). After incubation with Src, samples were separated by SDS-PAGE and transferred onto a blot, which was probed with a STAT3 antibody specific to pY705 (Figure 2c). A clear band at the same molecular weight was seen for both wt STAT3β and 7HC-STAT3β only when phosphorylated. In the EMSA, the 32P-labelled DNA probe shifted to the same position for both phosphorylated wt STAT3β and 7HC-STAT3β (Figure 2d). When excess non-radiolabelled hSIE probe was introduced into EMSA binding reactions, specific competition was seen for both 7HC-STAT3β and wt STAT3β. The relative affinities for hSIE, quantified by Kd, were almost identical. These results indicate that 7HC-STAT3β can be phoshorylated and bind a consensus DNA sequence with similar affinity to wt STAT3β, suggesting that substitution of Trp564 with 7HC does not alter STAT3 function.
We next tested if the 7HC could sense and report the phosphorylation of STAT3β using fluorometry (Figure 3a). Before phosphorylation, 7HC-STAT3β exhibited very weak fluorescence with a single emission peak at 448 nm. After incubation with Src kinase, the fluorescence intensity of 7HC-STAT3β increased markedly. A 13 (13 ± 4.3, n = 6) fold increase was detected for 20 nM 7HC-STAT3β, indicating that the reporter is highly sensitive. In addition, a second emission peak emerged at 416 nm. When calf intestinal phosphatase (CIP) was added, the fluorescence intensity dropped back to the level of unphosphorylated 7HC-STAT3β, indicating that the fluorescence change is reversible and dependent on phosphorylation status.
Figure 3.
7HC-STAT3β reversibly reports the phosphorylation status of STAT3. a) Fluorescence emission of 7HC-STAT3β before and after phosphorylation by Src kinase followed by dephosphorylation by CIP. b) Fluorescence emission of 7HC-STAT3β mutants Y705F and R609Q before and after phosphorylation by Src kinase. c) Western blots for 7HC-STAT3β and mutants using an antibody against phosphorylated STAT3. Equivalent amounts of STAT3 protein were loaded in each lane.
To confirm that the observed fluorescence change in 7HC-STAT3β was specifically due to phosphorylation of Tyr705, we introduced into 7HC-STAT3β a Y705F mutation, which abolishes STAT3 phosphorylation at Tyr705 [10] (Figure 3c). This mutant had the same fluorescence emission spectrum as 7HC-STAT3β, but showed no fluorescence change upon incubation with Src (Figure 3b). We made another mutation, R609Q, which prevents binding of pTyr705 into the SH2 domain.[11] The 7HC-STAT3β (R609Q) mutant could still be phosphorylated by Src kinase (Figure 3c), yet exhibited no fluorescence change (Figure 3b). Collectively, these results indicate that the observed fluorescence change can be attributed to the phosphorylation of Tyr705 that subsequently binds to the SH2 pocket containing 7HC.
To understand the sensing mechanism, we measured the fluorescence spectra of 7HC at different pH in aqueous buffer (Figure 4a). Consistent with 7-hydroxycoumarin,[7] 7HC showed an excitation peak at 325 nm at low pH corresponding to the neutral phenol form, and at 365 nm at high pH corresponding to the anionic phenolate form. Consistent with the pH-induced shift of 7HC, the excitation peak for 7HC-STAT3β shifted from 325 nm to 365 nm upon phosphorylation (Figure 4c). In addition, when excited at a wavelength longer than the isosbestic point (335 nm), the emission intensity of 7HC increased with the pH (Figure 4b) due to higher concentration of the phenolate species at the ground state. Using a similar excitation wavelength to 7HC, the fluorescence intensity of 7HC-STAT3β increased after phosphorylation, further implying a local pH increase. Both the shifted excitation peak and increased emission intensity of 7HC-STAT3β consistently suggest that the pH within 7HC microenvironment increased upon phosphorylation. This pH increase results in deprotonation of phenolic 7HC in 7HC-STAT3β to the phenolate form, which may occur due to an altered local hydrogen-bonding network induced by the incoming phosphate group. Moreover, crystal structures of the unphosphorylated and phosphorylated STAT3 protein show almost no conformational change after phosphorylation of Tyr705,[12] suggesting that a conformational change upon pTyr705 binding to the SH2 domain is not responsible for the observed 7HC fluorescence change.
Figure 4.
7HC in the 7HC-STAT3β protein experiences a pH change upon phosphorylation. a) Fluorescence excitation spectra of 7HC in aqueous buffer with emission recorded at 450 nm. b) Fluorescence emission spectra of 7HC in aqueous buffer with excitation at 363 nm. c) Fluorescence excitation spectra of 7HC-STAT3β with emission recorded at 450 nm before and after phosphorylation by Src kinase.
Another unique spectroscopic feature of 7HC-STAT3β is the appearance of an emission peak at 416 nm after phosphorylation, providing a characteristic readout that has not been reported in other proteins containing 7HC.[9] This emission peak corresponds to the excited state of the neutral phenol form of 7HC.[13] When 7-hydroxycoumarin is excited in aqueous solution above pH 2, only a single emission peak at 456 nm, corresponding to the excited phenolate species, is observed regardless of which ground species is excited.[7] We observed the same for 7HC in aqueous buffer (Figure 4b). This is due to rapid deprotonation of the neutral phenol form of 7-hydroxycoumarin at the excited state, which occurs within the lifetime of the singlet excited state in aqueous solution.[7] When 7-hydroxycoumarin is excited in H2O mixed with solvents that are less efficient proton acceptors, as the mole fraction of H2O decreases, the emission peak corresponding to the excited neutral phenol form of 7-hydroxycoumarin increases.[14] This emission thus indicates a decreasing accessibility of the fluorophore to H2O. In the 7HC-STAT3β protein, a single emission peak corresponding to the phenolate form was observed before phosphorylation (Figure 3a and 3b), signifying 7HC water accessibility and very quick excited state deprotonation. The additional 416 nm emission peak corresponding to the neutral phenol form of 7HC emerged only after phosphorylation (Figure 3a). This indicates that deprotonation of the phenol form at the excited state was no longer rapid and that 7HC became shielded from water, possibly due to pTyr705 and its neighboring residues filling the SH2 pocket.
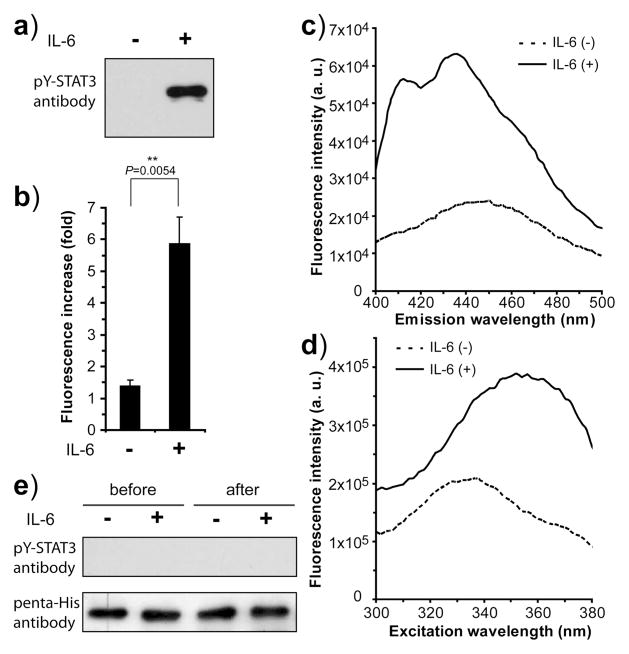
To test if 7HC-STAT3β can report the phosphorylation status of STAT3 proteins in mammalian cellular media, we incubated 7HC-STAT3β with nuclear extracts from human hepatoma HepG2 cells. A potent physiological activator of STAT3 is the cytokine interleukin-6 (IL-6). Upon IL-6 binding to its cytokine receptor, STAT3 is phosphorylated at Tyr705 by the receptor-associated and activated Janus kinase, and then translocates into the nucleus. Consistent with a previous report,[15] we detected a high level of phosphorylated STAT3 in the nucleus of HepG2 cells only when treated with IL-6 (Figure 5a). We incubated the same amount of 7HC-STAT3β with these nuclear extracts and found the fluorescence intensity increased only slightly (1.4 fold) for those from uninduced cells but significantly (5.9 fold) for those treated with IL-6 (Figure 5b). This indicates that 7HC-STAT3β can indeed optically report the phosphorylation status of endogenous STAT3.
Figure 5.
7HC-STAT3β reports the phosphorylation status of endogenous STAT3 from HepG2 cells. a) Western blot showing that STAT3 was phosphorylated in the nucleus of HepG2 cells only when activated by IL-6. b) Fluorescence increase of 7HC-STAT3β upon incubation with nuclear extracts. The values (± s.e.m.) were: IL-6 (−) 1.4 ± 0.2 and IL-6 (+) 5.9 ± 0.8, n = 3 from 3 independent batches of cells. The IL-6 activated nuclear fraction was statistically different from the uninduced sample (Student’s t-test, two-tailed, unpaired). c) Fluorescence emission spectra and d) excitation spectra of 7HC-STAT3β after incubation with the nuclear extracts of HepG2 cells with (+) and without (−) IL-6 induction. e) Western blot showing that 7HC-STAT3β was not phosphorylated by cell lysates. 7HC-STAT3β mixed with cell lysates before (t = 0) and after (t = 2 hr) incubation were analyzed. 7HC-STAT3β was N-terminally truncated and thus ran at a different position from endogenous STAT3. The blot was also probed with the penta-His antibody to detect the C-terminal His6 tag appended on 7HC-STAT3β.
To understand the observed difference, we analyzed the excitation and emission spectra of the nuclear extract samples after incubation with 7HC-STAT3β (Figure 5c and d). The nuclear fraction of IL-6 induced cells showed the red-shifted excitation peak and double emission peaks characteristic of 7HC-STAT3β phosphorylated by Src as seen in Figure 4c and Figure 3a, whereas uninduced nuclear fractions showed the same excitation and emission spectra as unphosphorylated 7HC-STAT3β. These results indicate that only in the nuclear fraction of IL-6 induced cells did binding of 7HC-STAT3β to pTyr705 occur. Three possibilities can lead to such binding: 1) 7HC-STAT3β is phosphorylated by endogenous kinases in the nuclear extract, after which it forms a homodimer or a heterodimer with endogenous phosphorylated STAT3; 2) another phospho-protein binds the SH2 domain of 7HC-STAT3β; 3) unphosphorylated 7HC-STAT3β forms a heterodimer with phosphorylated endogenous STAT3. To examine the first possibility, an anti-phosphotyrosine STAT3 antibody was used to probe 7HC-STAT3β incubated in the nuclear lysate samples. Phosphorylation of 7HC-STAT3β was not detected in samples with or without IL-6 (Figure 5e). No other phospho-proteins other than STAT1 have been reported to bind the SH2 domain of pSTAT3, but we are nevertheless undertaking crosslinking experiments to determine whether additional proteins could compete with STAT3 in forming homodimers. In addition, it is known that STAT3α and STAT3β isoforms can form homodimers and heterodimers with each other.[16] We thus favor the conclusion that after being added to the nuclear fraction of IL-6 induced cells, 7HC-STAT3β is not phosphorylated but forms a heterodimer with endogenous phosphorylated STAT3 protein, resulting in the expected fluorescence intensity increase, characteristic double emission peaks, and excitation peak shift.
In summary, we developed a fluorescence reporter for the phosphorylation status of STAT3 by genetically incorporating the fluorescent Uaa 7HC into a selected site in STAT3β. As Trp564 is conserved in all 7 mammalian STAT proteins,[8] this method should be transferable to detect the phosphorylation of other STATs, which will be valuable to untangle the function of different STATs and various STAT isoforms selectively. A similar strategy could be applied to other SH2 domain-containing proteins, which participate in a variety of signal transduction pathways. A reporter based on the full-length substrate protein represents cellular characteristics of the target protein with high fidelity, and can be used to report kinase as well as phosphatase activity with high specificity. Toward the goal of expanding this method into mammalian cells, we are now evolving an orthogonal tRNA-synthetase pair that will enable the genetic incorporation of 7HC into proteins in live mammalian cells.
Experimental Section
Methods and experimental details for plasmid construction, protein expression and purification, phosphorylation reactions, Western blot, EMSA, fluorometry, nuclear extract experiments, and mass spectrometry are described in the Supporting Information.
Supplementary Material
Acknowledgments
We thank Dr. Tony Hunter for helpful discussions. L.W. thanks the support from the Salk Innovation grant, March of Dimes Foundation (5-FY08-110), CIRM (RN1-00577-1), NCI (P30CA014195) and NIH (1DP2OD004744).
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org.
Contributor Information
Vanessa K. Lacey, Jack H. Skirball Center for Chemical Biology & Proteomics The Salk Institute for Biological Studies 10010 N. Torrey Pines Road, La Jolla, California 92037, U.S.A
Angela R. Parrish, Jack H. Skirball Center for Chemical Biology & Proteomics The Salk Institute for Biological Studies 10010 N. Torrey Pines Road, La Jolla, California 92037, U.S.A
Shuliang Han, College of Chemistry, Peking University, Beijing 100871, China.
Dr. Zhouxin Shen, Section of Cell and Development Biology, University of California at San Diego, La Jolla, California 92037, U.S.A
Prof. Steven P. Briggs, Section of Cell and Development Biology, University of California at San Diego, La Jolla, California 92037, U.S.A
Prof. Yuguo Ma, College of Chemistry, Peking University, Beijing 100871, China
Prof. Lei Wang, Email: lwang@salk.edu, Jack H. Skirball Center for Chemical Biology & Proteomics The Salk Institute for Biological Studies 10010 N. Torrey Pines Road, La Jolla, California 92037, U.S.A
References
- 1.Knight ZA, Lin H, Shokat KM. Nat Rev Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shults MD, Imperiali B. J Am Chem Soc. 2003;125:14248–14249. doi: 10.1021/ja0380502. [DOI] [PubMed] [Google Scholar]; Shults MD, Janes KA, Lauffenburger DA, Imperiali B. Nat Methods. 2005;2:277–283. doi: 10.1038/nmeth747. [DOI] [PubMed] [Google Scholar]; Yeh RH, Yan X, Cammer M, Bresnick AR, Lawrence DS. J Biol Chem. 2002;277:11527–11532. doi: 10.1074/jbc.M111300200. [DOI] [PubMed] [Google Scholar]; Sharma V, Agnes RS, Lawrence DS. J Am Chem Soc. 2007;129:2742–2743. doi: 10.1021/ja068280r. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rothman DM, Shults MD, Imperiali B. Trends Cell Biol. 2005;15:502–510. doi: 10.1016/j.tcb.2005.07.003. [DOI] [PubMed] [Google Scholar]; Sharma V, Wang Q, Lawrence DS. Biochim Biophys Acta. 2008;1784:94–99. doi: 10.1016/j.bbapap.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Ma Y, Taylor SS, Tsien RY. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]; Violin JD, Zhang J, Tsien RY, Newton AC. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ting AY, Kain KH, Klemke RL, Tsien RY. Proc Natl Acad Sci U S A. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]; Humphries KM, Pennypacker JK, Taylor SS. J Biol Chem. 2007;282:22072–22079. doi: 10.1074/jbc.M702582200. [DOI] [PubMed] [Google Scholar]; Ouyang M, Huang H, Shaner NC, Remacle AG, Shiryaev SA, Strongin AY, Tsien RY, Wang Y. Cancer Res. 2010;70:2204–2212. doi: 10.1158/0008-5472.CAN-09-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lippincott-Schwartz J, Snapp E, Kenworthy A. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]; Zhang J, Campbell RE, Ting AY, Tsien RY. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]; Miyawaki A. Dev Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]; Zhang J, Allen MD. Mol Biosyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 4.Remenyi A, Good MC, Lim WA. Curr Opin Struct Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 6.Yu H, Pardoll D, Jove R. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fink DW, Koehler WR. Anal Chem. 1970;42:990–993. [Google Scholar]
- 8.Becker S, Groner B, Muller CW. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Xie J, Schultz PG. J Am Chem Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 10.Kretzschmar AK, Dinger MC, Henze C, Brocke-Heidrich K, Horn F. Biochem J. 2004;377:289–297. doi: 10.1042/BJ20030708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer BJ, Jackson PK, Van Etten RA, Baltimore D. Mol Cell Biol. 1992;12:609–618. doi: 10.1128/mcb.12.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Z, Mao X, Mertens C, Krishnaraj R, Qin J, Mandal PK, Romanowski MJ, McMurray JS, Chen X. Biochem Biophys Res Commun. 2008;374:1–5. doi: 10.1016/j.bbrc.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 13.Moriya T. Bull Chem Soc Jpn. 1983;56:6–14. [Google Scholar]
- 14.Zinsli PE. J Photochem. 1974;3:55–69. [Google Scholar]
- 15.Lutticken C, Wegenka UM, Yuan J, Buschmann J, Schindler C, Ziemiecki A, Harpur AG, Wilks AF, Yasukawa K, Taga T, et al. Science. 1994;263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 16.Park OK, Schaefer LK, Wang W, Schaefer TS. J Biol Chem. 2000;275:32244–32249. doi: 10.1074/jbc.M005082200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.