Abstract
Proteoglycans are complex glycoconjugates that regulate critical biological pathways in all higher organisms. Bikunin, the simplest proteoglycan having a single glycosaminoglycan chain, is a serine protease inhibitor used to treat acute pancreatitis. Unlike the template driven synthesis of nucleic acids and proteins, Golgi synthesized glycosaminoglycans are not believed to have predictable or deterministic sequence. Bikunin peptidoglycosaminoglycans were prepared and fractionated to obtain a collection of size similar and charge similar chains. Fourier transform mass spectral analysis identified a small number of parent molecular-ions corresponding to mono-compositional peptidoglycosaminoglycans. Fragmentation using collision induced dissociation surprisingly afforded a single sequence for each mono-compositional parent-ion, unequivocally demonstrating the presence of a defined sequence. The common biosynthetic pathway for all proteoglycans suggests that even more structurally complex proteoglycans, such as heparan sulfate, may have defined sequences, requiring a readjustment of our understanding of information storage in complex glycans.
The human genome, proteome, and glycome, the latter of which is the major component of the metabolome, have come under increased scrutiny in the development of novel therapeutic strategies, particularly the next generation of biopharmaceuticals.1 Solving the chemical structure of glycan components of biomolecules is also critically important in answering fundamental questions in basic biology relevant to the translational science of medicine. The concept of whether glycans have a predictable or deterministic sequence is currently disputed.2,3 While the biosynthetic mechanism, resulting in the introduction of sequence into nucleic acids and proteins, is well understood and involves template driven biosynthesis, there is no comparable understanding of how sequence might be introduced into glycans within the endoplasmic reticulum and Golgi. Although enzymes can recognize remote sequences, imparting domains, the organized biosynthesis of the multiple domains within a full GAG chain may be beyond enzyme specificity. Despite all that is known about glycan biosynthesis, our understanding is still insufficient to infer sequence or even to suggest that glycans possess sequence. Indeed, it is valid to even question why a glycan would need a definable sequence. Many glycans, such as glycogen and starch,4 responsible for storing energy in plants and animals, or alginate and cellulose,5 responsible for providing structure to aquatic and terrestrial plants, have relatively simple physiological roles. In such cases, the specific arrangement of branches, linear sequence, and chain length may not be required, as multiple structures might fill these biological roles.
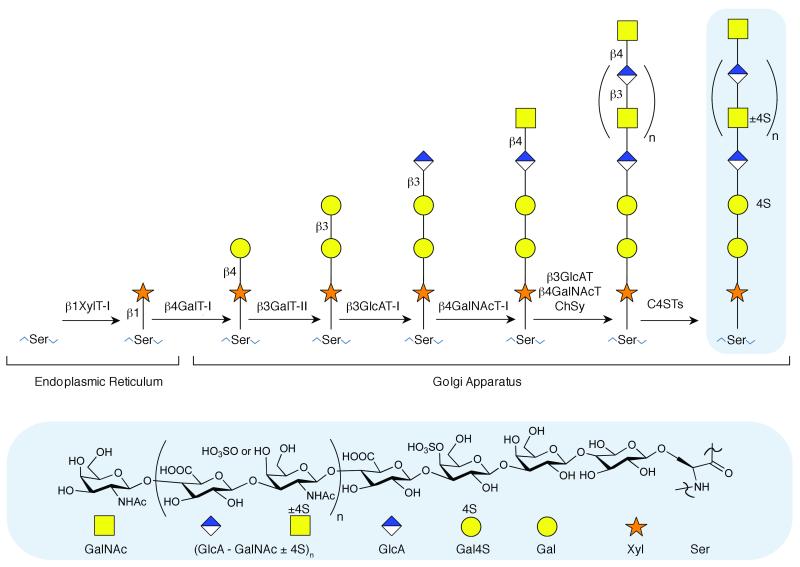
Some glycans, however, play a much more complex or even a dominant role in biology. Proteoglycans (PGs), for example, are glycoconjugates that contain glycosaminoglycan (GAG) side chains that are intricately modified during biosynthesis in the endoplasmic reticulum and Golgi (Figure 1).6-8 There are four families of PGs, hyaluronan, keratan sulfate, chondroitin sulfate/dermatan sulfate, and heparan sulfate/heparin. With the exception of hyaluronan, these consist of one or more linear, sulfated, polysaccharide chains (GAGs) covalently linked to a core protein.2 The GAG chains of these PGs are involved in complex biological roles such as in signal transduction (i.e., assembling and activating growth factor – growth factor receptor complexes)9 and in regulating pathways (i.e., the coagulation and complement cascades).10 These complex roles suggest sequence-dependent or sequence-specific mechanisms. Despite the suggestion that these glycans carry out sequence-dependent roles, only a handful of specific GAG sequences have a well-understood structure-activity relationship; the prototypical example, the heparin antithrombin binding pentasaccharide sequence is critically important for heparin’s anticoagulant activity.11,12
Figure 1.
Biosynthetic pathway for chondroitin sulfate A GAG, on a serine (Ser) residue of the core protein, beginning in the endoplasmic reticulum and concluding in the Golgi apparatus. The biosynthetic enzymes are: β1XylT-I, β-xylosyl transferase I; β4GalT-I, β-4-galactosyl transferase I; β3GalT-II, β-3-galactosyl transferase II; β3GlcAT-I, β-3-glucuronosyl transferase I; β4GalNAcT-I, β-4-N-acetyl galactosaminyl transferase I; β3GlcAT, β-3-glucuronosyl transferase; β4GalNAcT, β-4-N-acetyl galactosaminyl transferase; ChSy, chondroitin synthases; and C4STs, galactosyl 4-O-sulfo transferase and N-acetyl galactosaminyl-4-O-sulfotransferase.
If PGs have defined sequence, determined during their biosynthesis, then one might ask why has it been so difficult to determine GAG chain sequence. There are several reasons for the slow progress of this field, including: a lack of sufficient quantities of pure PGs; the multiple GAG chains often present on a single core protein may have multiple sequences; there are difficulties in purifying a single GAG chain for sequencing; there are difficulties in determining GAG sequence. In an effort to sequence a PG, we have carefully selected the bikunin PG as our target. Bikunin, a member of the kunin family of serine protease inhibitors,13-15 is a therapeutically relevant PG that is used as a drug for the treatment of acute pancreatitis in Japan.16 As a result, bikunin is available at a high level of purity in multimilligram quantities. Bikunin has the simplest chemical structure of any PG with a single site for O-linked modification by a GAG chain, located at Ser(10) in its 16 kDa core protein.17-20 While the protein component of bikunin is well characterized,21,22 its GAG chain structure is heterogeneous and has received less attention, due to the technical difficulties associated with GAG analysis. Although the bikunin GAG chain is quite short, it is very heterogeneous in size and composition, containing 27-39 saccharide residues and having a molecular mass (MR) ranging from 5,505-7,102 Da.23 In addition, enzymatic analysis shows that the bikunin GAG chains contain a single uronic acid stereochemistry (glucuronic acid), sulfo groups at only the 4-position of its galactosamine residue, and no N-sulfo group/N-acetyl group variability, common in the GAG chains of the more structurally complex heparan sulfate/heparin PG family.
Several critical issues, including GAG chain release from the core protein, GAG chain recovery and purification, and an appropriate sequencing strategy, must be addressed to successfully sequence bikunin. O-glycosidically-linked GAG chains are most commonly released from core protein through β-elimination,24 but this method utilizes harsh conditions that can result in GAG chain modification, complicating sequencing. Proteolysis, a mild method to recover peptidoglycosaminoglycan (pG) from a PG, also poses risks. If proteolysis is incomplete the mixture complexity can be increased by variability in the length of residual core peptide.23 Next, the recovery and purification of GAG or pG should require as few steps as possible so as not to lose sample or bias the mixture, while having sufficient resolution to obtain a single or a few individual chain lengths and compositions for sequencing. Finally, the analytical approach used for sequencing should be rapid, definitive, and ideally use widely available technology.
Methods for GAG sequencing reported to date have relied on various bottom-up approaches, e.g. the reassembly of fragments of GAGs, like pieces of a puzzle, into motifs or domains.25-29 Short sequences of GAG oligosaccharides have been read using a combination of enzymes and chemical chain scisson.30-33 Tandem MS of oligosaccharides has led to an understanding of the importance of charge-state in providing sequence-meaningful productions.27-29, 34-36
The present work demonstrates a new top-down glycomics approach that takes advantage of mass spectrometry to completely characterize the GAG/pG chains of a PG. Previous studies on bikunin have shown both ends of its GAG chain to contain ordered domains, reducing the number of possible sequences.23,37,38 In the current study, bikunin pG is recovered from the therapeutic urinary bikunin PG, fractionated by continuous elution preparative polyacrylamide gel electrophoresis (PAGE) to obtain a number of simple mixtures of size-similar and charge-similar pGs that are sequenced in a top-down approach using Fourier transform (FT) and FT ion cyclotron resonance (ICR) mass spectrometry (MS) with collision induced dissociation (CID) fragmentation.
RESULTS
Preparation of pG and determination of linkage region
Complete digestion of the bikunin PG core protein was accomplished by exhaustive treatment with actinase E. The complete chondroitin sulfate lyase ABC digestion of the resulting bikunin pG afforded several products, a single product corresponding to the linkage region at the reducing end, the Δ4,5 unsaturated disaccharides and tetrasaccharides coming from the middle of the chain, and a saturated trisaccharide from the chain’s non-reducing end. Centrifugal spin membrane (nominal molecular weight cut-off (MWCO) 10 kDa) separation afforded a retentate containing a single reducing end hexasaccharide, O-glycosidically linked to a serine residue (m/z 628.1118). Low collision energy (25 eV) activation applied to this abundant reducing end ion using FT-MS produced fragmentation with no sulfo group loss and identified the linkage region as ΔUA-GalNAc4S-GlcA-Gal4S-Gal-Xyl-Ser (Supplementary Results, Supplementary Figure 1). Thus, actinase E proteolysis of bikunin PG afforded a relatively simple mixture of pG chains.
The weight percentage of the pG prepared from bikunin PG after dialysis was ~ 30%. Polydisperse bikunin pG mixture had a number average molecular mass (MN) 7.0 kDa, and a weight average molecular mass (MW) 7.7 kDa based on PAGE using fractionated CS-A standards (Supplementary Figure 2).
FT-ICR-MS for pG component identifications
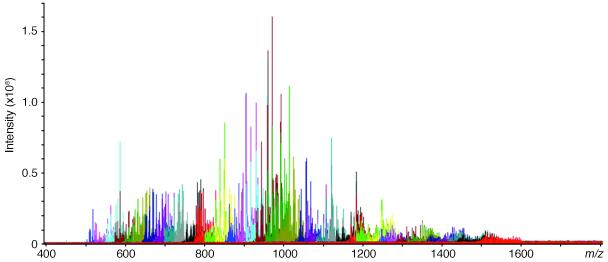
Previously, attempts to characterize the complex mixture of molecular compositions derived from bikunin at the MS level have provided minimal information due to heterogeneity in extent of sulfation, degree of polymerization, and extensive Na/H exchange. Windows of 50 m/z were selected with a quadrupole mass filter, introduced into the FT-ICR mass analyzer, and combined to generate a composite mass spectrum and provide sufficient signal for MS analysis.23 This technique results in an improved spectrum for a mixture of intact bikunin pG chains (Figure 2), although acquisition without quadrupole windowing can also be achieved due to advances in purification39,40 and reduction of Na/H heterogeneity using formic acid (0.1 vol. %) in the ESI solvent.41 Analysis of the quadrupole-windowed MS results in 47 molecular compositions as listed in Supplementary Table 1.
Figure 2.
FT-ICR-MS analysis of polydisperse and heterogeneous bikunin pG. “Quad-windowed” mass spectra are depicted with individual narrow m/z acquisitions in unique colors.
Evaluation of pG fractionated by continuous elution PAGE
Continuous elution PAGE was next used to separate bikunin into size-similar and charge-similar fractions. Bikunin pG fractions of 1.4 ml eluting from preparative electrophoresis were labeled as f1-f200, beginning with the ion front that coincided with phenol red dye. Initial examination by mini-slab PAGE with alcian blue followed by silver staining, showed that pG was first detected in f45. Fractions (≥ f50), containing sufficient amounts of pG for MS analysis, were then analyzed on 15% mini-slab PAGE together with molecular weight standards. pGs having MR ranging from 5.37-9.77 kDa were assigned based by electrophoresis on f50-f117 (Supplementary Figure 2, Supplementary Table 2). Uronic acid determination showed 13-52 μg bikunin pG/fraction, which corresponded to ~1.4-7.3 nmol bikunin pG/fraction (Supplementary Figure 3, Supplementary Table 3). The major pG (dp 35-5-Ser), represented ~ 2 mole % of the total.
Accurate mass measurements of bikunin pG fractions
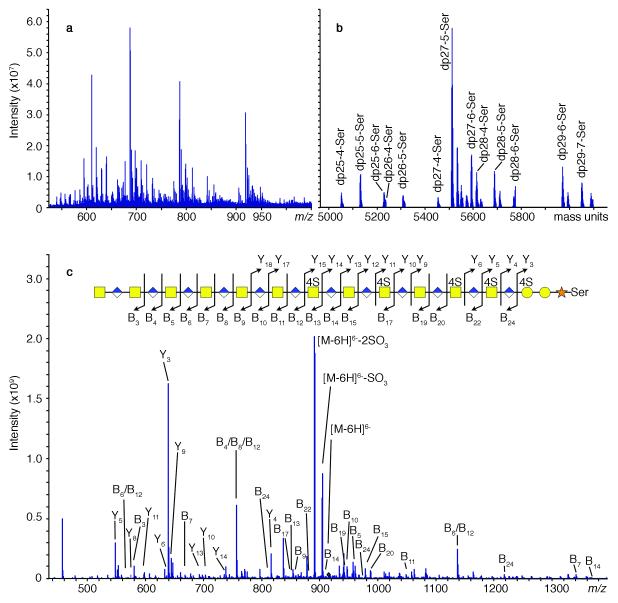
FT-MS identified a wide range of intact sized pGs, degree of polymerization (dp)23 to dp55, all of which were O-linked to one serine residue and included from 4-9 sulfo groups per chain (Supplementary Tables 4-14). Nearly the same range was seen on two different mass spectrometers in adjacent fractions. Chains having both even and odd number of saccharide units were observed. Odd-numbered chains were much more abundant based on relative parent-ion abundance. Compositional assignments were made with high confidence based on monoisotopic peaks and the isotopic envelope patterns. Different adjacent fractions, such as those corresponding to 5.80 kDa MR by PAGE and 5.87 kDa MR by PAGE, were analyzed by FT-MS and FT-ICR-MS (Figure 3a), deconvoluted manually (Figure 3b), and compared to statistically derived isotope patterns. The fraction corresponding to 5.80 kDa showed 18 components ranging from dp23-4-Ser to dp30-6-Ser (Supplementary Table 5) while the fraction corresponding to 5.87 kDa showed 20 components ranging from dp23-4-Ser to dp30-7-Ser. Much overlap was observed between adjacent fractions. Identified components of bikunin pG are listed (Supplementary Tables 4-14) and their spectra are shown (Supplementary Figures 4-13). Single components were identified with one to up to seven charge states, ranging from z = 5-14, dependent on the chain size of the pG analyzed.
Figure 3.
FT-ICR-MS analysis of a bikunin pG fraction. a. FT-ICR negative-ion mass spectrum of 5.80 kDa MR fraction by PAGE with 18 isobars and 63 parent-ions; b. Deconvolution of spectrum a; c. CID-FT-ICR-MS/MS spectra of parent-ion m/z = 917.38 (z = 6) and annotated fragment-ions providing sequence with dp27-5-Ser fragmentation pattern assigned from spectrum.
CID-FT & CID-FT-ICR-MS/MS analysis for sequencing pG ions
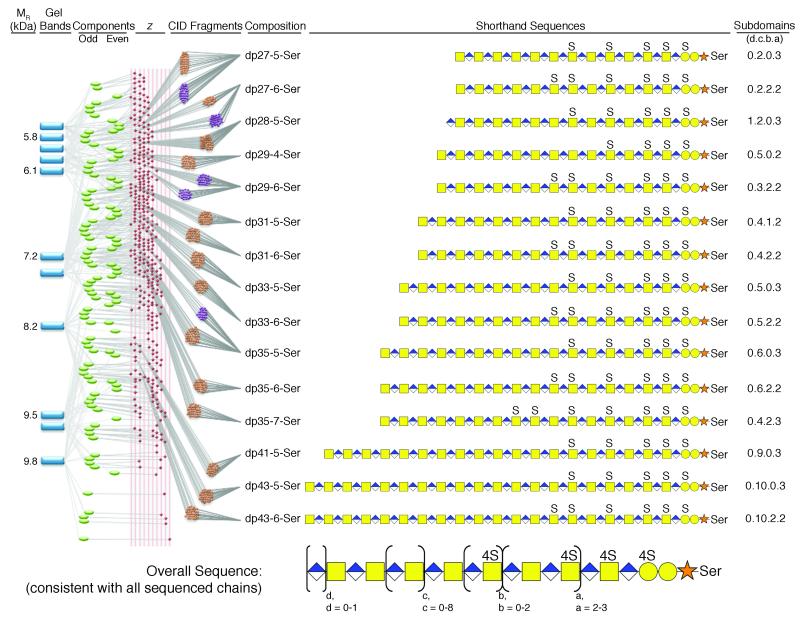
CID experiments performed on FT-MS allowed unambiguous sequencing of dp27-5-Ser, dp27-6-Ser, dp29-6-Ser, dp35-5-Ser. CID data from FT-ICR-MS (Figure 3c) afforded improved sensitivity and coverage for unambiguous sequencing of dp27-5-Ser, dp27-6-Ser, dp28-5-Ser, dp29-4-Ser, dp31-5-Ser, dp31-6-Ser, dp33-5-Ser, dp33-6-Ser, dp35-5-Ser, dp35-6-Ser, dp35-7-Ser, dp43-5-Ser, and dp43-6-Ser. With 19-37 fragments per CID spectrum, we could sequence 15 odd chains having 4-, 5-, 6-, and 7-sulfo groups ranging in chain length from 25-43 saccharide units, and an even chain of dp28-5-Ser (Supplementary Figures 14-33, Supplementary Tables 15-35). An example of a pG of single composition and having a single charge state sequenced on both types of FT mass spectrometers is dp27-5-Ser (Figure 3c, Supplementary Figure 14). The range of charge states used for CID experiments, which produced full sequence information, was z = 6-9. GAG chains with the same composition were observed in different fractions and different charge states, yielding redundant sequence data. On larger and more highly sulfated GAG chains it was difficult to obtain meaningful fragmentation data, especially when relying on the LTQ-orbitrap-FT-MS. Sulfo group placement on pGs with different GAG chain lengths having the same number of sulfo groups afforded exactly the same sulfo group placement for all sequenced chains with 4 through 7 sulfo groups. All 5-sulfo group-containing bikunin chains had the same sulfo group pattern, as did all 6-sulfo group-containing chains. Plasma bikunin pG42 was also examined using CID-FT-ICR-MS/MS and it showed the same sequence motif suggesting that the observed bikunin sequence was not altered or modified during filtration by the kidney or through the action of catabolic enzymes such as β-glucuronidases or sulfatases (Supplementary Figure 33, Supplementary Table 35). A decreasing number of possible sequences for bikunin GAG chains was calculated with increasing experimental constraints (Supplementary Table 36) and the sequence of bikunin GAG was determined (Figure 4, Supplementary Table 37).
Figure 4.
Bikunin sequencing flow chart (From left to right). The MR (kDa) determined based on PAGE of fractions (blue rectangles represent gel bands) of bikunin in pG prepared by continuous elution PAGE is shown. The deconvoluted MS obtained using FT-ICR-MS affords the mass of 3-5 odd and even components (green ovals) observed in each bikunin pG fraction is shown. Each MS spectrum showed multiple change states (z-values) shown as red diamonds from which parent-ions were selected for MS/MS giving CID fragments by analysis on FT (purple circles) or FT-ICR (brown circles). The composition is designated dp and sulfo group number (i.e., dp 27-5-Ser is 27 saccharide units with 5-sulfo groups O-glycosidically linked to a Ser residue). A shorthand sequence for each chain is shown with a, b, c and d subdomain repeats indicated by numbers (i.e., 0.2.0.3 for d = 0, c = 2, b = 0, a = 3). The overall sequence of bikunin CS-A pG shown at the bottom is consistent with all determined sequences.
DISCUSSION
Bikunin is a physiologically and pharmacologically important PG that is found both in human plasma and urine.13-16 From a structural standpoint, bikunin is the simplest PG consisting of a 143-amino acid core protein substituted at Asn(45) with an N-linked complex bianntenary glycan and substituted at Ser(10) with a single 23-55 saccharide unit O-linked chondroitin 4-sulfate chain.17-20 Based only on the size and composition of this GAG chain there are 210 billion sequence possibilities (Supplementary Table 36). Previous studies from our laboratory23,42 and others37,38 have found fixed structural motifs at both the reducing and non-reducing end the bikunin GAG chains greatly decreasing the expected number of sequences to 43 million (Supplementary Table 36). In the current study we undertook to completely sequence a single bikunin GAG chain using tandem mass spectrometry to establish whether there were motifs present within the center of the GAG chains that might further decrease the large number of expected bikunin sequences.
High-resolution separation was required to prepare a simple mixture containing a single prominent bikunin GAG chain for sequencing. Preliminary studies39,40,42 suggested that continuous elution preparative PAGE, offering the highest resolving power currently available, might permit the purification and recovery of the multi-microgram quantities of sample enriched in one to several GAG chains having a single composition (a fixed chain length and fixed number of sulfo groups). PAGE fractionation can be performed on the PG, pG or GAG. We selected to focus on pG as it offered advantage over fractionating the PG, where the core protein might reduce the dominant physical-chemical feature of the GAG chain and over fractionating the GAG chain, where harsh β-elimination conditions required for its preparation ran the risk of introducing structurally complicating artifacts.
Next, we selected the optimal approach for GAG chain sequencing. Our previous efforts had taken a combination of bottom-up and top-down glycomics approach relying on the application of polysaccharide lyases to fragment chains and use the oligosaccharide sequences to arrive at an overall chain sequence.23,26 There are limitations to this approach: not all the requisite enzymes are available to determine sequence;26 the available enzymes do not exhibit strict specificity;26,43 most of the available enzymes are endolytic making their use in direct sequencing difficult;43,44 and the exolytic polysaccharide lyases are not purely exolytic and are capable of jumping resistant sites making their use in sequencing difficult.43 ESI-MS offers some potential advantages for GAG sequencing but also has limitations including: full-length or highly sulfated GAG chains are difficult to analyze;2 additional spectral heterogeneity beyond composition (chain length and number of sulfo groups) are often introduced by Na/H exchange necessitating special care in ESI solvent selection and sample preparation;41 facile loss of sulfo groups complicates sequencing by giving fragmentation that is not sequence informative;45,46 and MS often has insufficient resolution and sensitivity for determination of small amounts of sample or samples consisting of even simple mixtures.23 We settled on an approach using an optimized solvent containing dilute formic acid to remove cations, simplifying spectra and forming negative-ions with high charge states (z-values), and thus, suppressing sulfo group loss.35,41 Direct infusion of simple mixtures containing bikunin pG compositions (~1 nmole) on FT-MS showed 18-34 compositions all with high z-values, suggesting CID might give sequence informative fragmentation. These high z-values could have been anticipated based on the modest number of sulfo groups present and the relatively large number of carboxyl groups associated with the polysaccharide analyte. In the past more highly sulfated and shorter oligosaccharides have afforded lower z-values with the CID spectra showing substantial loss of sulfo groups.35 Parent-ions were selected and the CID of each showed a surprisingly simple spectrum suggesting that a single composition represented a single sequence. Comparison of the sequences of individual parent-ions afforded a simple pattern or sequence motif (Figure 3b & 3c). Thus, based on our sampling of bikunin pG we anticipate that each of the ~150 compositions has a single sequence. Results from CID-FT-ICR-MS/MS provided enhanced mass accuracy, resolving power, and sensitivity (requiring ~1 pmole of bikunin pG PAGE fraction). CID-FT-ICR-MS/MS and CID-FT-MS/MS of the same precursor-ion were different but gave identical sequence. This manuscript is the first report of the concurrent use of two types of FT-MS instruments and it is surprising that the FT-MS provided excellent data that was thought to be only obtainable by sophisticated FT-ICR-MS instruments, suggesting that this sequencing approach will be generally available to the glycobiology community on more widely available spectrometers.
Bikunin pG analysis identified odd-numbered chains in higher abundance, probably due to their formation naturally occurring during biosynthesis. We initially hypothesized that the prominence of odd chains, GalNAc terminated, resulted from the removal of GlcA from the non-reducing end by β-d-glucuronidase, however, this is unlikely since plasma bikunin shows identical even and odd chain distribution as urinary bikunin.42 A previous study, suggesting that the non-reducing end was monosulfated based on recovery of a saturated monosulfated trisaccharide following enzymatic digestion,23 apparently missed the major saturated nonsulfated trisaccharide formed from the non-reducing end of the odd-numbered CS chain.
The biosynthetic assembly of bikunin’s CS chain occurs within the Golgi complex and bikunin is secreted by hepatocytes. In CS biosynthesis, sugar residues are sequentially attached by specific glycosyl transferases to first form the linkage region (Figure 1); then GAG elongation occurs with independent and alternating addition of GalNAc and GlcA, and sulfonation occurs during elongation. The kinetics of the biosynthetic events may perhaps also have a role in defining domain structure and catabolic or anabolic processing may play a role in stopping biosynthesis. It appears that there is some means whereby defined glycan domain structures are introduced, despite the apparent absence of an instructive template. Two ordered domains were identified in the bikunin GAG sequence, a sulfated domain near the reducing end, and a nonsulfated domain at the non-reducing end. In the sulfated domain, 12 residues adjacent to the tetrasaccharide linkage region showed a single sequence motif. Five residues, in which a motif is present, follow this domain but with variable sulfo group occupancy. The nonsulfated domain at the non-reducing end varied in length from ~6-22 residues.
One biological function of the nonsulfated domain is the binding of protein heavy chains (HC1, HC2) through ester linkages. HC1 and HC2 covalently-linked to bikunin bind inter-α-inhibitor protein to form a serine proteinase inhibitor complex. Plasma and urine levels of free and complexed bikunin are related to its anti-inflammatory activity. The domain structure of the bikunin GAG may be critical for biological switching of covalently-bound HC1 and HC2 from bikunin CS to hyaluronan.15,17,18,47
Chain length/molecular mass itself does not seem to be the major limitation in the application of FT-MS for sequencing GAGs. We have analyzed single chains of up to 80-saccharide units.40 Instead, increased molecular mass and sulfation level leads to more compositions and greater challenges in fractionation. In continuous elution PAGE of bikunin pG we observe 10-30 compositions in each band. When we select a single composition for sequencing by MS/MS, it might represent ~10 mole % of the mixture. Moreover, as the number of sulfo groups increases it becomes difficult to obtain molecular ions in which the charge state is greater than the number of sulfo groups, resulting in a reduced number of sequence informative fragments. Improved fractionation and FT-ICR-MS/MS methods may allow the application of these methods for more variable and highly sulfated GAGs in the future.
To put these results in perspective, one must consider how improbable is it that a PG has single sequence motif when there are so many possible sequence possibilities (Supplementary Table 36) despite the fact that there is no known biosynthetic mechanism that could explain how this sequence is installed. The common biosynthetic pathway for all PG families suggests that other defined sequences may be present in even the more structurally complex PGs, such as heparan sulfate. These findings clearly require a readjustment of our understanding of sequence and information storage in glycans having complex structure.
METHODS
Preparation and linkage region analysis of bikunin pG
Pharmaceutical grade bikunin PG (Mochida Pharmaceuticals) was purified from excipients and buffer salts by dialysis against distilled water using 30 kDa molecular weight centrifugal devices (Millipore) and then lyophilized. The PG (7 mg) was then proteolyzed by a 5% (w/w) actinase E (EC (Enzyme Commission) # 3.4.24.4, Kaken Biochemicals) digestion at pH 7.5 in 50 mM Tris-HCl/sodium acetate (Sigma). The enzymatic reaction proceeded at 45°C for 18 h and then isolated from the digestion mixture by strong-anion exchange (SAX) spin column chromatography (High-Capacity Maxi-Q, Sartorius). Anion exchange spin columns were pre-equilibrated with 50 mM sodium chloride, and after the digestion mixture was brought up to 50 mM, the samples were bound by 500 × g. Washed twice with 50 mM sodium chloride, the pG eluted with 1.5 M NaCl, desalted using YM-10 kDa MWCO centrifugal filter (Millipore) with deionized water washes and then lyophilized. The actinase E digestion and purification were repeated. Approximately 50 μg of bikunin pG digested completely using 250 mU chondroitin sulfate lyase ABC from Proteus vulgaris (EC 4.2.2.4, Seikagaku America) at 37°C for 18 h and it was then purified by a 10 kDa MWCO spin column filter to isolate reducing ends.
Fractionation of bikunin pG by continuous elution PAGE
Electrophoresis resolving buffer, electrode running buffer, and total acrylamide (T) monomer solutions, including resolving and stacking gel solutions, were prepared as previously.39 A gel of 10 cm column height using 15% T monomer resolving solution was allowed to polymerize overnight with 40 μl TEMED and 200 μl 10% (w/v) ammonium persulfate (APS), cast in a 37 mm diameter column (Bio-Rad). Above the polymerized resolving gel column, 4 ml of 5% T monomer stacking gel was cast. An aliquot of 2 mg of purified bikunin pG was loaded in a solution of 10 μg/ml (w/v) phenol red and 25% (w/v) sucrose. Electrophoresis was performed for 8 h at a constant power of 12 W, a peristaltic pump (Econo-pump, Bio-Rad) set to 0.7 ml/min and fraction collector (Model 2110, Bio-Rad) set to 2 min in conjunction accumulated separating fractions from the Model 491 Prep cell (Bio-Rad). Buffer salts from electrophoresis for each fraction were removed by strong anion exchange (Medium-Capacity Mini-Q, Sartorius) and thoroughly desalted by LC-grade water washes with Microcon YM-10 centrifugal filters (Millipore). Carbazole assay48 quantified the amount of bikunin pG per fraction based on a standard curve of chondroitin sulfate A (Celsus). The extent of separation was visualized by 15% T monomer solution using native mini-slab PAGE stained with Alcian blue, and molecular mass distribution properties (MR, MN, and MW) of the fractionated and unfractionated pGs were estimated on PAGE densitometry against identified bikunin standards using UN-SCANIT (Silk Scientific).40
ESI-LTQ-orbitrap-FT-MS analysis of bikunin pG
All pGs were analyzed in the negative-ion mode by electrospray ionization FT-MS on a Thermo Scientific LTQ orbitrap XL FT mass spectrometer with a standard, factory-installed ion source (Thermo Scientific). External calibration of mass spectra produced a mass accuracy of <3 ppm. Samples were dissolved in 50% aqueous methanol with 0.1% formic acid and delivered by an Agilent 1200 nano-LC pump at a flow rate of 20 μl/min. The purified bikunin pGs at volumes between 0.5 μl and 5 μl were directly infused through an Agilent 1200 autosampler. Mass spectra were acquired at a resolution between 30,000 and 60,000, and the charge deconvolution was performed manually with electronic spreadsheets. Acquisition parameters used to prevent in-source fragmentation included spray voltage 3-4.2 kV, capillary voltage −15 V, tube lens voltage −100 V, capillary temperature 250°C, sheath flow rate 25, and auxiliary gas flow rate 5. For collision-induced dissociation MS analysis of the linkage region pGs and intact pGs, precursor ions were isolated and activated in the linear ion trap for 800 μs before injection into the orbitrap for mass analysis. Parent-ions were fragmented by low-energy collision (25 eV to 65 eV) with a 3 m/z mass window. Acquired data were averaged to produce a single mass spectrum per fraction sample in MS and MS/MS. Fragments and pGs were identified manually and using Glycoworkbench.49 CID spectra are annotated using the Domon and Costello nomenclature with sulfo group loss shown by roman numeral.50
Nano-ESI-FT-ICR-MS analysis of bikunin pG
FT-ICR-MS experiments were performed in negative-ion mode with a 9.4 T Bruker Apex Ultra QeFTMS (Bruker) fitted with an Apollo II dual source. The sample solutions were infused at a rate of 5-10 μl/hour and ionized by nanoelectrospray using a pulled fused silica tip model FS360-75-15-D-20 (New Objective). Solutions of each bikunin pG fraction were introduced at a concentration of ~0.1 mg/ml in 50:50:0.1 methanol:H2O:FA (Sigma) to minimize Na/H heterogeneity.41
During mixture analysis for urinary bikunin pG, a quadrupole mass filter window of 50 m/z was advanced across the mass range of interest (400-2000 m/z).23 For each window, a mass spectrum was acquired and consisted of 24 signal-averaged acquisitions. For each mass spectrum, 1M points were acquired, padded with one zero fill, and apodized using a sinebell window. Independent windowed spectra were then combined to generate a composite mass spectrum. Mass spectra for purified fractions were acquired without the use of the quadrupole mass filter. For each mass spectrum, 24 acquisitions were signal averaged, consisting of 1M points, padded with one zero fill, and apodized using a sinebell window.
For tandem mass spectrometry experiments, mass-selected precursor ions were activated by CID in the hexapole collision cell of the Apex instrument with collision energy of 16-28 eV. The 36-48 acquisitions were signal averaged for each tandem mass spectrum during the analysis of urinary pG chains. Due to limited material during analysis of plasma bikunin pG chains, 148 acquisitions were signal averaged. External calibration of mass spectra produced mass accuracy of 1-2 ppm. Internal calibration was also performed using confidently assigned glycosidic bond cleavage products as internal calibrants, providing mass accuracy of <1 ppm. Product-ion assignment and annotation were identical to that used in FTMS.49, 50
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dmitri Zagorevski for his expertise in the proteomics core at RPI and the National Institutes of Health for support in the form of grant # GM38060.
Footnotes
AUTHOR CONTRIBUTIONS
M.L. and F.E.L. contributed experiments and data interpretation. T.N.L. contributed the fractions for analysis and assisted in writing. T.T. contributed the bikunin and assisted in writing. R.J.L. and I.J.A. contributed experimental planning, result interpretation and wrote the paper.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors have declared no conflict of interests.
REFERENCES
- 1.Feero WG, Guttmacher AE, Collins FS. Genomic medicine--an updated primer. N Engl J Med. 2010;362:2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 2.Ly M, Laremore TN, Linhardt RJ. Proteoglycomics: recent progress and future challenges. Omics. 2010;14:389–399. doi: 10.1089/omi.2009.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kreuger J, Spillmann D, Li J, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball S, et al. From glycogen to amylopectin: a model for the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- 5.Perez S, Mazeau K. Conformations, structures, and morphologies of celluloses. In: Dumitiu S, editor. Polysaccharides: Structural diversity and functional versatility. 2nd edn. Marcel Dekker; New York: 1998. pp. 41–68. [Google Scholar]
- 6.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 7.Silbert JE, Sugumaran G. Biosynthesis of chondroitin/dermatan sulfate. IUBMB Life. 2002;54:177–186. doi: 10.1080/15216540214923. [DOI] [PubMed] [Google Scholar]
- 8.Nairn AV, et al. Glycomics of proteoglycan biosynthesis in murine embryonic stem cell differentiation. J Proteome Res. 2007;6:4374–4387. doi: 10.1021/pr070446f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couchman JR. Transmembrane signaling proteoglycans. Annu Rev Cell Dev Biol. 2010;26:89–114. doi: 10.1146/annurev-cellbio-100109-104126. [DOI] [PubMed] [Google Scholar]
- 10.Linhardt RJ, Toida T. Role of glycosaminoglycans in cellular communication. Acc Chem Res. 2004;37:431–438. doi: 10.1021/ar030138x. [DOI] [PubMed] [Google Scholar]
- 11.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of 3-O- and 6-O-9sulfated glucosamine residues in the heparin-induced conformational change in antithrombin III. Biochemistry. 1987;26:6454–6461. doi: 10.1021/bi00394a024. [DOI] [PubMed] [Google Scholar]
- 12.Petitou M, van Boeckel CA. A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next? Angew Chem Int Ed Engl. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 13.Fries E, Blom AM. Bikunin--not just a plasma proteinase inhibitor. Int J Biochem Cell Biol. 2000;32:125–137. doi: 10.1016/s1357-2725(99)00125-9. [DOI] [PubMed] [Google Scholar]
- 14.Fries E, Kaczmarczyk A. Inter-alpha-inhibitor, hyaluronan and inflammation. Acta Biochim Pol. 2003;50:735–742. [PubMed] [Google Scholar]
- 15.Zhuo L, Salustri A, Kimata K. A physiological function of serum proteoglycan bikunin: the chondroitin sulfate moiety plays a central role. Glycoconj J. 2002;19:241–247. doi: 10.1023/A:1025331929373. [DOI] [PubMed] [Google Scholar]
- 16.Michalski C, et al. Preparation and properties of a therapeutic inter-alpha-trypsin inhibitor concentrate from human plasma. Vox Sang. 1994;67:329–336. doi: 10.1111/j.1423-0410.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 17.Enghild JJ, et al. Chondroitin 4-sulfate covalently cross-links the chains of the human blood protein pre-alpha-inhibitor. J Biol Chem. 1991;266:747–751. [PubMed] [Google Scholar]
- 18.Morelle W, et al. Chondroitin sulphate covalently cross-links the three polypeptide chains of inter-alpha-trypsin inhibitor. Eur J Biochem. 1994;221:881–888. doi: 10.1111/j.1432-1033.1994.tb18803.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhuo L, Hascall VC, Kimata K. Inter-alpha-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J Biol Chem. 2004;279:38079–38082. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]
- 20.Enghild JJ, et al. Organization of the inter-alpha-inhibitor heavy chains on the chondroitin sulfate originating from Ser(10) of bikunin: posttranslational modification of IalphaI-derived bikunin. Biochemisty. 1999;38:11804–11813. doi: 10.1021/bi9908540. [DOI] [PubMed] [Google Scholar]
- 21.Josic D, et al. Proteomic characterization of inter-alpha inhibitor proteins from human plasma. Proteomics. 2006;6:2874–2885. doi: 10.1002/pmic.200500563. [DOI] [PubMed] [Google Scholar]
- 22.Delaria KA, et al. Characterization of placental bikunin, a novel human serine protease inhibitor. J Biol Chem. 1997;272:12209–12214. doi: 10.1074/jbc.272.18.12209. [DOI] [PubMed] [Google Scholar]
- 23.Chi L, et al. Structural analysis of bikunin glycosaminoglycan. J Am Chem Soc. 2008;130:2617–2625. doi: 10.1021/ja0778500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conrad HE. Beta-elimination for release of O-linked glycosaminoglycans from proteoglycans. Curr Protoc Mol Biol. 2001;17:15.1–15.3. doi: 10.1002/0471142727.mb1715as31. [DOI] [PubMed] [Google Scholar]
- 25.Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Sequencing complex polysaccharides. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- 26.Laremore TN, et al. Domain structure elucidation of human decorin glycosaminoglycans. Biochem J. 2010;431:199–205. doi: 10.1042/BJ20100788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolff JJ, Amster IJ, Chi L, Linhardt RJ. Electron detachment dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2007;18:234–244. doi: 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolff JJ, Laremore TN, Aslam H, Linhardt RJ, Amster IJ. Electron-induced dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2008;19:1449–1458. doi: 10.1016/j.jasms.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff JJ, et al. Negative electron transfer dissociation of glycosaminoglycans. Anal Chem. 2010;82:3460–3466. doi: 10.1021/ac100554a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turnbull JE, Hopwood JJ, Gallagher JT. A strategy for rapid sequencing of heparan sulfate and heparin saccharides. Proc Natl Acad Sci USA. 1999;96:2698–2703. doi: 10.1073/pnas.96.6.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merry CLR, Lyon M, Deakin JA, Hopwood JJ, Gallagher JT. Highly sensitive sequencing of the sulfated domains of heparan sulfate. J Biol Chem. 1999;274:18455–18462. doi: 10.1074/jbc.274.26.18455. [DOI] [PubMed] [Google Scholar]
- 32.Turnbull JE, Gallager JT. Sequence analysis of heparan sulphate indicates defined location of N-sulphated glucosamine and iduronate 2-sulphate residues proximal to the protein linkage region. Biochem J. 1991;277:297–303. doi: 10.1042/bj2770297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Desai UR, Han X-J, Toida T, Linhardt RJ. Strategy for the sequence analysis of heparin. Glycobiology. 1995;5:765–774. doi: 10.1093/glycob/5.8.765. [DOI] [PubMed] [Google Scholar]
- 34.Zaia J, Li XQ, Chan SY, Costello CE. Tandem mass spectrometric strategies for determination of sulfation positions and uronic acid epimerization in chondroitin sulfate oligosaccharides. J Am Soc Mass Spectrom. 2003;14:1270–1281. doi: 10.1016/S1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 35.McClellan JE, Costello CE, O’Connor PB, Zaia J. Influence of charge state on product ion mass spectra and the determination of 4S/6S sulfation sequence of chondroitin sulfate oligosaccharides. Anal Chem. 2002;74:3760–3771. doi: 10.1021/ac025506+. [DOI] [PubMed] [Google Scholar]
- 36.Hitchcock AM, Yates KE, Costello CE, Zaia J. Comparative glycomics of connective tissue glycosaminoglycans. Proteomics. 2008;8:1384–1397. doi: 10.1002/pmic.200700787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyoda H, Kobayashi S, Sakamoto S, Toida T, Imanari T. Structural analysis of a low-sulfated chondroitin sulfate chain in human urinary trypsin inhibitor. Biol Pharm Bull. 1993;16:945–947. doi: 10.1248/bpb.16.945. [DOI] [PubMed] [Google Scholar]
- 38.Yamada S, et al. The sulphated carbohydrate-protein linkage region isolated from chondroitin 4-sulphate chains of inter-alpha-trypsin inhibitor in human plasma. Glycobiology. 1995;5:335–341. doi: 10.1093/glycob/5.3.335. [DOI] [PubMed] [Google Scholar]
- 39.Laremore TN, Ly M, Solakyildirim K, Zagorevski DV, Linhardt RJ. High-resolution preparative separation of glycosaminoglycan oligosaccharides by polyacrylamide gel electrophoresis. Anal Biochem. 2010;401:236–241. doi: 10.1016/j.ab.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ly M, et al. Analysis of E. coli K5 capsular polysaccharide heparosan. Anal Bioanal Chem. 2010;399:737–745. doi: 10.1007/s00216-010-3679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Influence of charge state and sodium cationization on the electron detachment dissociation and infrared multiphoton dissociation of glycosaminoglycan oligosaccharides. J Am Soc Mass Spectrom. 2008;19:790–798. doi: 10.1016/j.jasms.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laremore TN, Leach FE, III, Amster IJ, Linhardt RJ. Electrospray ionization Fourier transform mass spectrometric analysis of intact bikunin glycosaminoglycan from normal human plasma. Int J Mass Spectrom. doi: 10.1016/j.ijms.2010.09.020. doi: 10.1016/j.ijms.2010.09.020 (in press, corrected proof, available online 29 September 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ly M, Rensselaer Polytechnic Institute . Doctoral dissertation, Department of Chemistry and Chemical Biology. Troy: 2011. [Google Scholar]
- 44.Gu K, Liu J, Pervin A, Linhardt RJ. Comparison of the activity of two chondroitin AC lyases on dermatan sulfate. Carbohydr Res. 1993;244:369–377. doi: 10.1016/0008-6215(83)85014-9. [DOI] [PubMed] [Google Scholar]
- 45.Zaia J. Principles of mass spectrometry of glycosaminoglycans. J Biomacromol Mass Spectrom. 2005;1:3–36. [Google Scholar]
- 46.Gunay NS, Tadano-Aritomi K, Toida T, Ishizuka I, Linhardt RJ. Evaluation of counterions for electrospray ionization mass spectral analysis of a highly sulfated carbohydrate, sucrose octasulfate. Anal Chem. 2003;75:3226–3231. doi: 10.1021/ac034053l. [DOI] [PubMed] [Google Scholar]
- 47.Capon C, Mizon C, Lemoine J, Rodié-Talbère P, Mizon J. In acute inflammation, the chondroitin-4 sulphate carried by bikunin is not only longer; it is also undersulphated. Biochemie. 2003;85:101–107. doi: 10.1016/s0300-9084(03)00066-x. [DOI] [PubMed] [Google Scholar]
- 48.Bitter T, Muir HM. A modified uronic acid carbazole reaction. Anal Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 49.Ceroni A, et al. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 2008;7:1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 50.Domon B, Costello C. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.