Abstract
Obesity is a serious health problem worldwide associated with an increased risk of life-threatening diseases such as type 2 diabetes, atherosclerosis, and certain types of cancer. Fundamental for the development of novel therapeutics for obesity and its associated metabolic syndromes is an understanding of the regulation of fat cell development. Recent computational and experimental studies have shown that microRNAs (miRNAs) play a role in metabolic tissue development, lipid metabolism and glucose homeostasis. In addition, many miRNAs are dysregulated in metabolic tissues from obese animals and human, which potentially contributes to the pathogenesis of obesity associated complications. In this review, we will summarize the current state of understanding of the roles of miRNAs in metabolic tissues under normal development and obese conditions, and discuss the potential use of miNRAs as therapeutic targets.
Keywords: adipogenesis, adipose tissue, biomarker, diabetes, glucose homeostasis, insulin resistance, lipid metabolism, liver, microRNAs, obesity, pancreas, therapeutics
1. Introduction
Gene expression in human is precisely controlled in a cell, temporal, and condition specific manner. Therefore, completely understanding the regulatory mechanisms of gene expression is important in genomic medicine. One of the major discoveries in the last decade is microRNAs (miRNAs), which constitute an abundant and evolutionarily conserved class of post-transcriptional regulators of gene expression1–3. miRNAs are small endogenous noncoding RNAs that base pair to sites within target mRNAs, triggering either a block in translation or mRNA degradation or both4, 5. The expression of miRNAs is often tissue-specific or developmental-specific1, 2. The post-transcriptional programs controlled by miRNAs affect diverse biological processes, including development, cell differentiation, apoptosis, immune responses, metabolism and many diseases including various cancers, cardiovascular disease, viral infection and neurodegenerative diseases6–14. As example, signature miRNA expression patterns differentiate many types of related cancers better than more classical analyses of mRNA profiles or protein markers, and allow better prognosis of disease progression15.
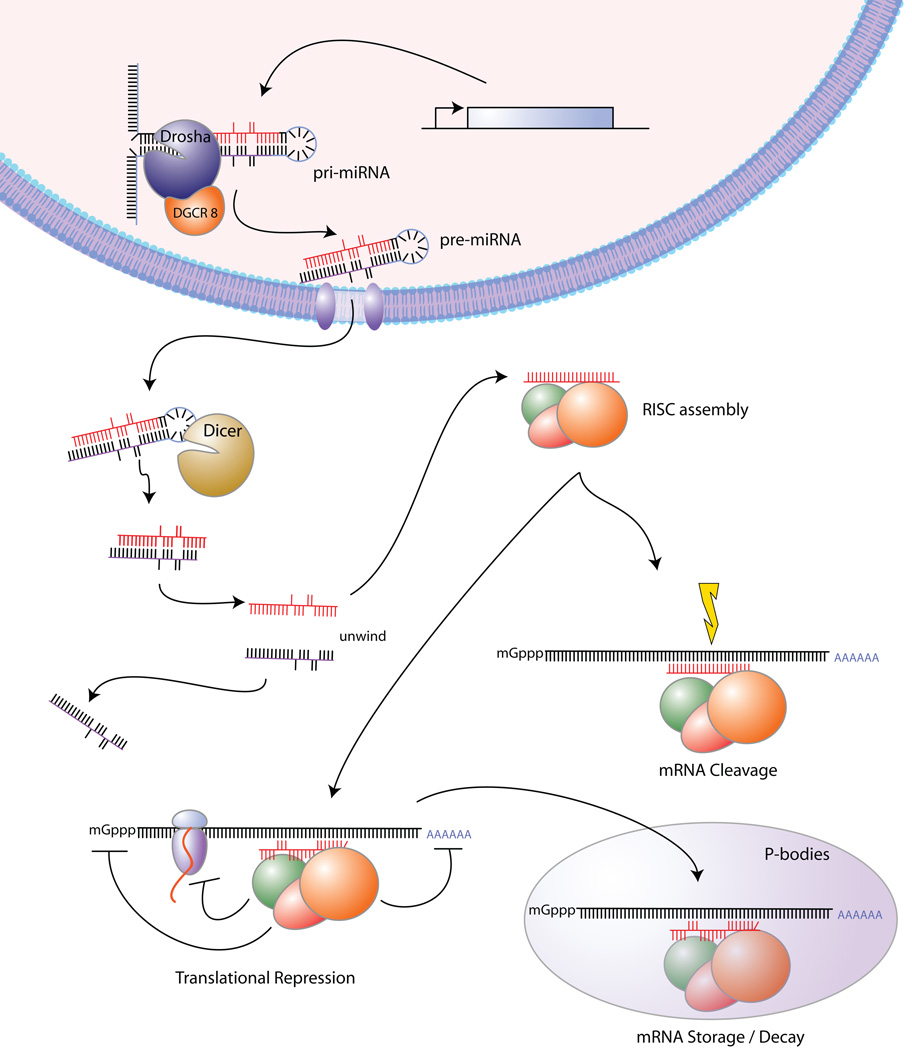
As outlined in Figure 1, miRNAs are transcribed by polymerase II or polymerase III to primary transcripts (pri-miRNAs) in the nucleus. Endonuclease Drosha and its cofactor DGCR8 process pri-miRNA by cutting it at the bottom of its stem loop to ~70-nucleotide precursors (pre-miRNAs). Upon export to the cytoplasm by exportin 5 and Ran-GTP, Dicer generates a ~22 nucleotide miRNA duplex. One strand (mature miRNA) is then preferentially retained in the RNA-induced silencing complex (RISC) and base pairs with specific sequences in their target mRNAs. Depending on the level of complementarity, silencing of the transcript can occur via Argonaut-dependent mRNA cleavage or translational repression or both. The translationally repressed mRNA is either stored in P-bodies or enters the mRNA decay pathway for destruction16.
Figure 1.
miRNA biogenesis and mechanisms of action.
miRNA targeting is primarily through seed-matched sites located within favorable predicted contexts in 3' untranslated regions (UTRs). Both computational and experimental studies show that each miRNA likely targets ~400 mRNAs and that almost half of the mRNAs in mammalian cells are targeted by one or more miRNAs3. As a group, miRNAs may directly regulate expression of over 30% of human and mouse genes and more than 60% of human protein-coding genes have been under selective pressure to maintain pairing to miRNAs3, 17. Furthermore, miRNAs are attractive candidates to be involved in complex diseases such as obesity because the simultaneous coordination of a large number of target genes, potentially accomplished by a single miRNA, may be key to defining specific pathogenic cell states.
Although miRNA expression profiles and functions have been extensively investigated in the hematopoietic and neuronal systems18, 19, little is known about the role of miRNAs in metabolic tissues, particularly adipose tissue10. More recently, several intriguing studies have uncovered critical roles of miRNAs in the development of metabolic tissues such as adipose tissue and pancreas. Many miRNAs are also dysregulated in metabolic tissues from obese animals and humans, and possibly involved in the tissue dysfunction and secondary complications associated with obesity and diabetes. The purpose of this review is to summarize the most recent progress in understanding the roles of miRNAs in metabolic tissues under normal development and obese conditions and to provide a perspective to future research directions in order to utilize miRNAs as diagnostic markers or therapeutic targets.
2. miRNAs in metabolic tissues
2.1 Adipose tissue
Adipose tissue is not only a storage depot of triglycerides, but also an endocrine organ and an important regulator of whole-body energy homeostasis20–22. Adipocyte-specific proteins induced during differentiation, such as adiponectin, resistin, and leptin regulate many aspects of lipid and glucose metabolism in adipose tissue, muscle, and liver, and via actions on the brain that affect food intake23, 24. The development of obesity depends on the coordinated interplay of adipocyte hypertrophy (increased fat cell size), adipocyte hyperplasia (increased fat cell number), and angiogenesis25. Both adipocyte hypertrophy and hyperplasia occur during normal growth phases and the development of obesity. As discussed below, miRNAs are important regulators of fat cell development and insulin sensitivity (Table 1). Many miRNAs are differentially expressed in different fat depots and between normal and obese adipose tissue, likely associated with proper function of adipose tissue.
Table 1.
Mammalian miRNAs regulating adipogenesis and insulin sensitivity.
| miRNA | Function | Model system | Targets | References |
|---|---|---|---|---|
| miR-143 | Pro-adipogenic | Primary human preadipocytes, 3T3-L1 | ERK5/MAPK7(?) | 33, 37 |
| miR-103 | Pro-adipogenic | 3T3-L1 | 33 | |
| miR-200 | Pro-adipogenic | ST2 | 47 | |
| miR-17–92 cluster | Pro-adipogenic | 3T3-L1 | Rb2/p130 | 38 |
| Let-7 | Anti-adipogenic | 3T3-L1 | Hmga2 | 40 |
| miR-27 | Anti-adipogenic | 3T3-L1, OP9 | 39 | |
| miR-29 | Inhibitor of glucose uptake | 3T3-L1 | Insig1(?), Cav2(?) | 52 |
3T3-L1, mouse preadipocyte cell line; ST2 and OP9, mouse bone marrow stromal cells
Validated targets but their participation in adipogeneis or insulin signaling pathway is not clear
2.1.1 miRNA and fat cell development
Adipocytes are derived from multipotent mesenchymal precursor cells that commit to preadipocytes and then either remain quiescent or proceed to become differentiated adipocytes26. The differentiation process is tightly controlled by a combination of multiple transcription factors including peroxisome proliferator-activated receptor gamma (PPARγ) and extracellular hormones such as insulin27–29. The first and best characterized model of adipogenesis in vitro is the 3T3-L1 cell line, a substrain of the Swiss 3T3 mouse cell line30. When treated with a combination of chemicals, post-confluent 3T3-L1 cells undergo clonal expansion, growth arrest and terminal differentiation. Differentiated adipocytes accumulate lipid droplets and express genes associated with lipid metabolism (FABP4), glucose homeostasis (GLUT4), and endocrine functions (adiponectin).
2.1.1.1 miRNA expression
Attempts to catalogue miRNA expression in adipose tissue and during adipogenesis have been carried out using different profiling platforms. Using a miRNA cloning strategy, Gu et al cloned 45 known and 2 novel miRNAs from bovine adipose tissue31. More recently, Liang et al further compared the expression of 345 miRNAs in 40 normal human tissues by quantitative reverse transcription-polymerase chain reaction (RT-PCR)32. Unlike neuronal and muscular tissues, their result did not identify any miRNA expressed exclusively in adipose tissue. One caveat of such tissue profiling studies is that one could easily get fat contamination when isolating other tissues such as breast. Interestingly, miRNA profiling in purified mouse primary adipocytes indentified many adipocyte-enriched miRNAs including miR-103 and miR-143, in comparison to chondrocytes and osteoblasts, which are also derived from mesenchymal progenitor cells33–35.
Using Northern blot analyses, Kajimoto et al profiled ~100 miRNAs in mouse preadipocyte 3T3-L1 cells before and after differentiation and showed that 21 miRNAs were either up- or down-regulated during differentiation36. To provide better sensitivity and coverage, more recent studies have profiled miRNA expression at different stages of 3T3-L1 differentiation using a miRNA microarray approach33, 37, 38,39, 40. In addition, Xie et al compared the expression of 12 selected miRNAs by RT-PCR in enriched mouse primary preadipocytes and adipocytes and suggested that similar changes in miRNA expression occur during in vitro and in vivo adipogenesis33. There studies offer an overview of miRNA regulation during fat cell development and identify many candidate miRNAs for further investigation. Of particular relevance, many miRNAs including let-7, miR-30, 103, 143 and 422b are upregulated during adipogenesis as suggested by multiple independent studies and confirmed by either Northern blots or RT-PCR33, 36, 37, 40. Many of these differentially regulated miRNAs are important regulators of adipogenesis, as discussed below.
2.1.1.2 miRNA function
To infer the global functional role of miRNA in adipose biology, one can disrupt miRNA processing machinery (Figure 1) by conditional knock-out of Dicer or DGCR8 in the adipose tissue, similar to studies performed in skin and lung41–43. So far, such an animal model has not been reported. Nevertheless, in cell culture, knock-down of Drosha by siRNA two days before induction of differentiation abolished 3T3-L1 adipogenesis38.
The first evidence for participation of individual miRNAs in adipogenesis and lipid metabolism came from a genetic screen in Drosophila. Xu et al44 found that deletion of miR-14 results in increased levels of triacylglycerol and diacylglycerol, whereas increases in miR-14 copy number have the opposite effect. Using a similar method, Teleman et al45 demonstrated that homozygous mutations in the miR-278 gene, which is prominently expressed in the fat body of flies, causes a smaller fat body and reduced ratio of total body triglycerides to total protein. This phenotype could be rescued by miR-278 expression. Mutant flies also exhibited hyperglycemia in spite of elevated insulin-like peptide levels due to insulin resistance of the fat body. Thus, miR-278 regulates insulin sensitivity. However, miR-14 and miR-278 have so far been found only in insects and there are no known homologues of these miRNAs in mammals.
More compelling evidence for the role of miRNA in adipogenesis has been obtained in studies of mammalian cells. To screen candidates that have regulatory roles in adipogenesis, Esau et al37 transfected antisense oligonucleotides targeting 86 human miRNAs into primary human preadipocytes. Blocking miR-143 effectively inhibited all five adipocyte differentiation markers by at least 40%, suggesting a pro-adipogenic role of miR-143. Consistent with this, miR-143 expression increased during mouse adipogenesis in vitro and in vivo33. In addition, ectopically expressing miR-143 in 3T3-L1 preadipocytes accelerated fat cell formation33, thus confirming its important role in modulating adipogenesis.
A computational study predicted that the miRNA paralogs miR-103 and miR-107 affect multiple mRNA targets in pathways that involve cellular acetyl-CoA and lipid metabolism46. Interestingly, these miRNAs are derived from introns of the pantothenate kinase gene family members that activate pantothenate for the biosynthesis of coenzyme A. Experimentally, ectopic expression of miR-103 in 3T3-L1 preadipocytes accelerated adipogenesis, as measured both by the upregulation of many adipogenesis markers including PPARγ and FABP4, and by an increase in triglyceride accumulation at an early stage of adipogenesis33. Based on mRNA profiling and computational predictions, potentially important mRNA targets for miR-103 includes several anti-adipogenic factors such as ARNT, FZD1, and RUNX1T1/ETO/MTG8, whose levels are normally downregulated during adipogenesis33.
Many signaling pathways have an effect on adipogenesis28. For example, insulin and BMP2 promotes adipogenesis while Wnt-family proteins and TGFβ repress adipogenesis. Therefore, miRNAs activating or inhibiting any of these signaling pathways are likely to affect adipogenesis. Kennell et al47 identified miR-8 as a negative regulator of Wnt signaling in Drosophila. They further demonstrated that the miR-200 family, homologues of Drosophila miR-8 in mammals, promoted adipogenesis. Retroviral expression of miR-200 cluster in mouse ST2 marrow stromal cells increased lipid accumulation and expression of FABP4 and partially rescued the block of differentiation caused by treatment with recombinant Wnt3a.
Since clonal expansion is one of the key events taking place in early adipogenesis of 3T3-L1 cells in vitro, any miRNAs affecting this process could have a profound effect on adipogenesis. Wang et al38 reported that the miR-17-92 cluster, which promotes cell proliferation in various cancers, is upregulated 2–3 fold during the early clonal expansion stage of 3T3-L1 adipogenesis. Stable overexpression of these miRNAs moderately accelerates adipocyte differentiation in vitro after hormonal stimulation. Additionally, this study provided convincing evidence that Rb2/p130 is a bona fide target of miR-17-92 during adipogenesis. This is not totally unexpected because previous observations suggested a p130:p107 switch during the first 24h after hormonal induction of adipogenesis48.
Another miRNA related to clonal expansion is let-7, which is well known to regulate cell proliferation and differentiation processes in species ranging from C. elegans to human49, 50. Expression of let-7 slightly decreased from day 0 to day 1 and then increased during terminal adipogenesis40. Ectopic expression of let-7 by transfecting pre-let-7 oligonucleotide into 3T3-L1 cells prior to induction of differentiation inhibited clonal expansion as well as terminal differentiation, in part by targeting HMGA240. Interestingly, mice lacking HMGA2 have marked reductions in adipose tissue51. However, the involvement of the miR-17-92 cluster and let-7 during in vivo fat cell development awaits further investigation since clonal expansion is not required for primary preadipocyte differentiation.
Among many downregulated miRNAs during adipogenesis, the miR-27 gene family have been functionally characterized using 3T3-L1 preadipocytes and OP9 mouse bone marrow mesenchymal stem cells39. Overexpression of miR-27 by transfecting miR-27 precursors before adipogenic stimulation specifically inhibited adipocyte formation. Mechanistically, miR-27 prevents the induction of PPARγ and C/EBPα, the two master transcriptional regulators of adipogenesis. Interestingly, although PPARγ contains a putative binding site for miR-27, miR-27 does not repress the level of PPARγ protein in differentiating 3T3-L1 cells if miR-27 was transfected after 2 days after adipogenic stimulation. Therefore, it is plausible that miR-27 inhibits adipogenesis by targeting an unknown gene that prevents the transcriptional induction of PPARγ.
It is interesting to note that overexpression of miRNAs that are upregulated during adipogenesis, namely miR-143, miR-103, miR-17-92, alone without hormonal induction cannot trigger adipogenesis, suggesting that they are not sufficient to initiate differentiation33, 38. Similarly, inhibition of endogenous miR-27 using antisense oligonucleotides is not sufficient to promote adipogenesis39.
2.1.2 miRNA and insulin sensitivity
To identify miRNAs associated with insulin sensitivity, He et al52 examined the miRNA expression profile by miRNA microarray analysis of skeletal muscles from healthy and Goto-Kakizaki rats, a model of type 2 diabetes. miR-29 family members are upregulated in diabetic animals compared to control animals. Northern blot analysis further revealed their upregulation in all three insulin-responsive tissues including adipose tissue, muscle and liver of diabetic rats. Adenovirus-mediated overexpression of miR-29 in 3T3-L1 largely repressed insulin-stimulated glucose uptake, presumably through inhibiting Akt activation. High levels of miR-29 led to insulin resistance mimicking the insulin resistance in cells incubated with high glucose and high insulin. Interestingly, the miR-29 level was upregulated in the presence of high glucose (hyperglycemia) and high insulin (hyperinsulinemia) in 3T3-L1 adipocytes. Two candidate genes Insig1 (insulin-induced gene 1) and Cav2 (caveolin 2) were validated as targets of miR-29 but their participation in insulin signaling pathway is not clear. Further studies are needed to test whether higher level of miR-29 in insulin-responsive tissues result in lower levels of target proteins.
With the observation that individual miRNAs moderately repress many targets, it is tempting to hypothesize that a miRNA family or a miRNA cluster targeting multiple components of the same signaling pathway may be more effective and functionally important than a single miRNA. Xu and Wong performed a computational screen to identify mouse signaling pathways targeted by miRNA clusters53. Most strikingly, one miRNA cluster, mmu-mir-183-96-182 targets Irs1, Rasa1, and Grb2, all of which are located in the insulin signaling pathway. These predictions were further supported experimentally by luciferase report assays. This supports the notion that different members of one miRNA cluster target different components along a signaling pathway and can coordinately control the signal transduction process.
Studies in cancer cells also suggested insulin receptor substrate (IRS)-1 can be repressed at the translation level by miR-145 and miR-12654, 55. Since IRS is an important component of the insulin signaling pathway, it merits further investigation whether these interactions are physiologically relevant and play a role in the pathogenesis of insulin resistance.
2.1.3 miRNAs in different fat depots
Adipose tissue consists of white adipose tissue (WAT) and brown adipose tissue (BAT). While WAT stores excess energy, BAT is very active in energy expenditure and has an anti-obesity function56, 57. This property relies on the expression of a BAT specific protein, UCP-1, which is a proton transporter located in the inner mitochondrial membrane and allows dissipation of the proton electrochemical gradient in the form of heat instead of ATP58.
While BAT persists through life in rodent, it has traditionally been considered insignificant in adult human. In human fetuses and newborns, BAT is found in axillary, cervical, perirenal, and periadrenal regions, but decreases shortly after birth56. However, using morphological studies and positron-emission tomography scanning studies, three independent groups have recently provided conclusive evidence for the existence of active BAT in adults59–61. Importantly, the presence of BAT negatively correlates with both BMI and percentage of body fat, whereas it positively correlates with resting metabolic rate, implicating a role of BAT in preventing obesity in adult human.
White adipose tissue can be further subdivided into subcutaneous fat and visceral fat, which is most closely related to insulin resistance56. Global miRNA expression comparison between subcutaneous and omental fat, a type of visceral fat, from 15 human individuals suggested little differences in these two fat depots overall62; this supports the notion that subcutaneous and visceral fat are indeed two locations of a developmentally homogeneous adipose organ.
Recently, exciting research suggested that white and brown adipocytes originate from different precursors with brown preadipocytes being more closely related to skeletal muscle cells than to white preadipocytes57. Investigation of miRNAs in brown fat has just begun. miR-455, which is low in white pre- and mature adipocyte, was enhanced during brown adipocyte differentiation, similarly to the expression pattern of the brown adipocyte differentiation marker UCP-1 63; thus it may play a role during brown adipocyte differentiation or contribute to the mature brown adipocyte function. Three classical “myogenic” miRNAs, miR-1, miR-133a and miR-206 were absent from white adipocytes but were specifically expressed both in brown pre- and mature adipocytes63. More comprehensive high-throughput expression profiling studies should identify more miRNAs specifically enriched in white fat, brown fat and muscle. Brown fat enriched miRNAs may be related to energy expenditure and/or thermogenesis. In vitro and in vivo manipulation of these brown fat enriched miRNAs, independently or together with transcription factors, may help confer the brown adipocyte properties to white adipocyte or muscle cells, which may be used clinically to treat obesity.
2.1.4 miRNA in obese adipose tissue
Adipose tissue undergoes a dramatic expansion in obesity, which eventually results in adipose tissue dysfunction. As adipose tissue expands, macrophage infiltration in adipose tissue occurs64. Chronic inflammation and hypoxia are two principal features of obese adipose tissue in animals and humans64, 65. Inflammatory cytokines including TNF-α are largely responsible for suppressing many adipocyte-specific genes including PPARγ and adiponectin and reactivating expression of many cell cycle genes, resulting in insulin resistance in obese adipose tissue66.
To compare miRNA expression levels in normal and obese states at a genome-wide scale, Xie et al33 profiled the expression of more than 370 miRNAs in enriched epididymal adipocytes from leptin deficient ob/ob and diet-induced obese (DIO) mice using miRNA microarrays. A total of 71 miRNAs were expressed at significantly different levels in adipocytes from wild type and ob/ob mice of the same gender and age whereas 35 miRNAs were differentially expressed between control and DIO mice. Table 2 lists miRNAs either up- or down-regulated significantly regardless of the obesity model. There was a global positive correlation of miRNA regulation in these two different models of obesity33, suggesting that the changes in the adipocyte miRNA expression profile of ob/ob mice cannot result from leptin deficiency alone, but likely associated with obesity in general.
Table 2.
Principal miRNAs differentially regulated in enriched mouse epididymal adipocytes from both types of obese mice (ob/ob and DIO) compared to wild type mice. Analysis is performed on the miRNA profiling data described by Xie et al 33.
| Up-regulated in obese mice | Down-regulated in obese mice |
|---|---|
| miR-16 miR-24 miR-221 miR-222 miR-223 miR-146b miR-23b miR-27a miR-27b miR-342-3p |
let-7d(*) miR-103 miR-107 miR-145 miR-320 miR-30a* |
Mature miRNAs are referred to by their names in miRBase version 10.1
Most importantly, miRNAs that were induced during adipogenesis were decreased in adipocytes from both types of obese mice and vice versa33. For example, miR-422b, miR-103, miR-30c were induced during adipogenesis but were downregulated in obese adipocytes. Conversely, miR-221 and miR-222 were decreased during adipogenesis but were upregulated in obese adipocytes. These changes are likely associated with the chronic inflammatory environment and elevated TNF-α levels in obese adipose tissue, since they were mimicked by TNF-α treatment of differentiated 3T3-L1 adipocytes33. The remarkable inverse regulatory pattern for many miRNAs during adipogenesis and obesity has important implications for understanding adipose tissue dysfunction in obese mice and humans and the link between chronic inflammation and obesity with insulin resistance.
Independently, Lin et al39 observed the moderate increase of miR-27, a regulator of adipogenesis discussed in the previous section, in epididymal fat tissue of ob/ob mice. They found that miR-27 expression could be regulated by hypoxia, an important extracellular stress associated with obesity. Whether miR-27 is regulated by inflammatory cytokines is still an open question.
In human, significantly lower expression of miR-17-5p, miR-132 and miR-134 was found in omental fat of overweight and obese individuals with newly diagnosed type 2 diabetes compared to those with normal glucose tolerance, whereas the opposite pattern was found for miR-181a62. Expression of miR-17-5p and miR-132 was negatively associated with visceral fat area62. Most strikingly, lower expression of omental miR-132 was correlated with higher macrophage infiltration whereas higher expression of miR-181a was correlated with lower circulating adiponectin62. One possible caveat is that the unfractionated adipose tissues profiled in their study contain many different cell types; thus the difference of certain miRNAs may represent a decrease in the fraction of mature adipocytes in visceral adipose tissue in type 2 diabetes patients, possibly as a result of infiltrating macrophages. Further studies are necessary to dissect causal from correlative relationships and the association needs to be confirmed in larger cohorts.
2.2 Liver
miR-122, a predominant miRNA in the liver, is involved in the regulation of several cholesterol biosynthesis pathway genes67. In vivo inhibition of miRNA-122, in normal and a diet induced obesity mouse model, resulted in a significant decrease in plasma cholesterol levels and improvement in liver steatosis67, 68. In addition, the miR-30 family was recently shown to be required for vertebrate hepatobiliary development, as knockdown of miR-30a in zebrafish larva resulted in defective biliary morphogenesis69. Interestingly, hepatic function was preserved up to 100 days old when Dicer1 was knocked out at birth in the differentiated liver70. This was rather surprising considering the striking effect on cholesterol metabolism when just miR-122 was knocked down. The authors suspected that additional miRNAs contributed to the regulation of metabolic pathways and that global loss of all miRNAs masks the phenotypes uncovered by the unbalanced inhibition of a single component of miRNA-mediated regulation70.
Recently, Li et al71 analyzed the expression of miRNAs in livers of ob/ob mice, streptozotocin (STZ)-induced type 1 diabetic mice and normal C57BL/6 mice by miRNA microarray. Compared to normal C57BL/6 mice, ob/ob mice showed up-regulation of 8 miRNAs (miR-34a, miR-31, miR-103, miR-107, miR-194, miR-335-5p, miR-221, and miR-200a) and down-regulation of 4 miRNAs (miR-29c, miR-451, miR-21 and miR-122) in fatty livers. Up-regulation of miR-34a and down-regulation of miR-122 was found in livers of STZ-induced diabetic mice.
2.3 Pancreas
The role of miRNAs in insulin production (miR-30d), insulin secretion (miR-375, miR-9 and miR-96) and pancreatic islet development (miR-375 and miR-124a) have been reported and extensively reviewed previously72–75. More recently, Poy et al76 reported that mice lacking miR-375 are hyperglycemic, and exhibit increased total pancreatic alpha-cell numbers, decreased beta-cell mass, higher fasting and fed plasma glucagon levels, and increased gluconeogenesis and hepatic glucose output. More interestingly, increased expression of miR-375 is observed in pancreatic islets of leptin-deficient ob/ob mice, which also have increased beta-cell mass76. Genetic deletion of miR-375 from ob/ob mice profoundly diminished the proliferative capacity of the endocrine pancreas and resulted in a severe diabetic state76.
Another study by Lovis et al77 investigated the contribution of miRNAs to fatty acid induced pancreatic beta-cell dysfunction. Prolonged exposure of the beta-cell line MIN6B1 and pancreatic islets to palmitate caused a time- and dose-dependent increase of miR-34a and miR-146. Elevated levels of these miRNAs were also observed in islets of diabetic db/db mice77. Blocking miR-34a or miR-146 activity partially protects palmitate-treated cells from apoptosis77. Therefore, at least part of the detrimental effects of palmitate on beta-cells is caused by alterations in the level of specific miRNAs.
3. Challenges and perspectives
3.1 Functional validation in animal models
As discussed so far, most gain-of-function or loss-of-function studies on miRNA have been carried out in cell lines or primary cell cultures. The efficiency of knocking down miRNAs in cell culture is still not optimized; thus, specific miRNA or miRNA cluster knockout mice, especially conditional knockout mice, are the most powerful loss-of-function approach to assess their role in development and obesity. Fortunately, the Sanger Institute in Cambridge has started an initiative to create a library of knockouts of each of the 500 miRNAs identified in the mouse genome78. The resource will eventually be available to all researchers. Meanwhile, researchers may find some clues about miRNA function from current knockout mice models for protein-coding genes because it is estimated that expression of a miRNA may have been disrupted along with another gene in about 200 knockout cases79. However, it is challenging to knockout all miRNA family members that are regarded to be functionally redundant if these family members are not on the same chromosome. For example, miR-103/107 family members, miR-103-1, miR-103-2 and miR-107, reside on three different chromosomes.
3.2 Combinatorial effect of miRNAs
Beside the fact that each mammalian miRNA regulates a large number of target genes, several different miRNAs can act additively or synergistically at multiple target sites of a single mRNA80. The potential interaction networks connecting miRNAs and mRNAs are enormous and can be further expanded by feedback or feed-forward loops81. The effect of single miRNA may be small and difficult to detect; nonetheless each probably makes an important contribution to the robustness of the development of metabolic tissues and pathogenesis of obesity and associated disorders. Ideally, one should manipulate multiple candidate miRNAs in different combinations rather than changing one miRNA at a time for functional characterization. However, this is labor intensive and technically challenging.
3.3 Therapeutic strategies targeting miRNAs
With experimental evidence beginning to provide functional roles of miRNA in development and metabolism, the need has become apparent to test miRNAs as viable therapeutic targets. Some lessons learned from existing antisense technology and gene therapy approaches can be adapted to manipulate miRNA levels in vivo.
Enhancement of miRNA activity can be accomplished by transfecting synthetic miRNA mimetics or by using plasmids to transcribe miRNAs from endogenous or viral promoters. Repression of miRNA activity can be performed with antisense oligonucleotides with modifications to increase stability and binding specificity. Three major types of modifications includes 2’-O-methyl (2’-O-Me), 2’-O-methoxyethyl (2’-O-MOE) oligonucleotides and locked nucleic acids (LNAs)82. Initial studies exploiting these modified antisense oligonucleotides to repress miRNA expression have shown promising results in animals and non-human primates.
Of relevance to metabolism, Esau et al68 antagonized miR-122, a liver specific miRNA, with 2’-O-MOE oligonucleotides in normal and diet-induced obese mice. When injected intraperitoneally, miR-122 inhibition resulted in reduced plasma cholesterol levels in both normal and diet-induced obese mice. Strikingly, miR-122 inhibition in a diet-induced obesity mouse model significantly improved liver steatosis. Independently, Krutzfeldt et al67 used cholesterol-conjugated 2’-O-Me oligonucleotides (also known as antagomirs) to silence miRNAs. Intravenous administration of antagomirs against miR-122 reduced plasma cholesterol level significantly. The silencing of endogenous miRNAs by antagomirs seems to be specific, efficient, and long-lasting. Most recently, Elmen et al83 used unconjugated LNAs to antagonize miR-122 in non-human primates. Acute administration by intravenous injections in African green monkeys resulted in uptake of LNAs in the cytoplasm of primate hepatocytes and dose-dependent lowering of plasma cholesterol. Silencing of miR-122 was efficient, long-lasting, and reversible without any evidence for LNA-associated toxicities or histopathological changes. Additional examples of the use of antisense oligonucleotides targeting miRNAs have been tabulated in a previous review article82. An alternative approach of silencing miRNAs is the use of miRNA sponges, which contain multiple tandem binding sites for targeted miRNAs, thereby acting as decoys to titrate miRNAs away from their natural targets84. When miRNA sponges are expressed in a plasmid, the effect can be long lasting. Two recent studies have utilized this strategy to inhibit miR-223 and miR-31 in vivo85, 86. One can potentially inhibit multiple miRNAs with a single plasmid by tandem decoy sequences against different miRNAs of interest.
In the future, and with better understanding of the upstream factors controlling miRNA expression, we may be able to therapeutically increase or decrease expression of specific miRNA(s) in disease tissue and thus ameliorate certain disease symptoms.
3.4 Delivery of therapeutic agents
Currently, there is an intense effort to identify agents capable of targeted delivery of nucleic acids to specific tissues and cells87–92. One approach is conjugating a lipophilic moiety or receptor ligand, such as cholesterol in the case of antagomirs, to the oligonucleotide. Alternatively, oligonucleotides can be packed into liposomes, polymers, or nanoparticles that facilitate endocytosis. Recently, combinatorial chemistry has yielded a novel class of lipidoids that may allow for the development of new classes of delivery reagents93.
One challenge is the delivery method to transfer the miRNAs or its inhibitors into the desired tissue. So far, much of the success has been obtained in liver. Some metabolic tissues may be easier accessed and targeted than others in this regard. Circumventing this technical difficulty may involve development of different chemical modifications and conjugates designed to specifically interact with cell-specific membrane proteins.
4. Conclusion
Emerging evidence from both gain- and loss-of-function studies suggests that miRNAs play an important role in metabolic tissue development and function. Expression profiling studies have revealed that some miRNAs are dysregulated in metabolic tissues under the obese state, and possibly involved in the pathogenesis of various metabolic disorders. Further functional characterization of metabolically important miRNAs in vitro and in vivo and better understanding of their mechanism of action may allow for identification of novel therapeutic targets and strategies. The safe, effective and targeted delivery of RNA therapeutics remains an important challenge for clinical development.
5. Expert opinion
The complexity of post-transcriptional regulation by miRNA is still far from understood. Further characterization of metabolically important miRNAs in vivo is likely to shed more light on their functions. Large scale proteomics will help identify targets of a specific miRNA since the effect of miRNAs is often more pronounced at the protein level than at the mRNA level. Improved computational algorithms predicting miRNA targets will aid in this effort as well. The potential interaction networks connecting miRNAs and mRNAs are enormous and complex, thus a system biology approach is necessary to understand the entire RNA regulatory network.
High-throughput miRNA profiling studies have and will continue to reveal miRNA dysregulation in metabolic tissues. However, it is hard to dissect whether change of certain miRNA cause metabolic tissue dysfunction or miRNA is dysregulated as a consequence of metabolic tissue dysfunction. Establishment of the causal relationship will enable better selection of therapeutically potential targets.
Recently, many genome-wide association studies have identified susceptibility loci and gene polymorphisms for obesity and type 2 diabetes94–98. Some of these polymorphisms may create or destroy a putative miRNA target site responsible for the phenotypic variation. As an example, Villuendas et al99 showed that the ACAA-insertion/deletion polymorphism at the 3’UTR of the insulin-like growth factor II receptor (IGF2R) was associated with type 2 diabetes and surrogate markers of insulin resistance. Most strikingly, using luciferase reporter assays, Lv et al100 showed that hsa-miR-657 acts directly at the 3′UTR of the IGF2R and the repression was greater when ACAA was deleted. This finding raise the possibility that the ACAA-insertion/deletion polymorphism may result in the change of IGF2R expression levels at least in part by hsa-miR-657-mediated regulation, contributing to the pathogenesis of type 2 diabetes.
Since miRNAs have been shown to be differentially expressed in healthy and disease states101, it is tempting to use miRNA as biomarkers for diagnosis. Excitingly, Chen et al102 demonstrated that miRNAs were present in serum and plasma of humans and other animals. miRNA levels in serum are stable, reproducible and consistent among individuals of the same species. Using high-throughput sequencing, they identified specific expression patterns of serum miRNAs for lung cancer, colorectal cancer, and diabetes102. In addition, 17 miRNAs were exclusively detected in serum compared to blood cells from diabetic patients102. Although the source of these miRNAs is not yet clear, a serum miRNA fingerprint could complement the current biomarker, C-reactive protein (CRP) for the diagnosis of diabetes. It would be interesting to find out when these serum miRNA expression change respect to disease progression.
In conclusion, miRNAs are promising therapeutic targets for management of obesity and related metabolic disorders. With this great promise, many biotech firms are springing up to develop miRNA-based products that can diagnose, treat, or predict the course of disease103. However, to realize the full potential of miRNA-based therapeutics, more efficient and specific silencing of miRNA and targeted delivery are needed.
Acknowledgements
We thank Tom Dicesare for assistance with graphics. We apologize to colleagues whose work is not discussed here because of length restrictions. H.X is supported by a graduate fellowship from the Singapore-MIT Alliance. Work on this subject in our lab is supported by grant C-382-641-001-091 from SMA and grants DK047618 and DK068348 from NIH.
Abbreviations
- miRNA
microRNA
- RISC
RNA-induced silencing complex
- UTRs
untranslated regions
- PPARγ
peroxisome proliferator-activated receptor gamma
- RT-PCR
reverse transcription-polymerase chain reaction
- IRS
insulin receptor substrate
- WAT
white adipose tissue
- BAT
brown adipose tissue
- DIO
diet-induced obese
- 2’-O-Me
2’-O-methyl
- 2’-O-MOE
2’-O-methoxyethyl
- LNAs
locked nucleic acids
- IGF2R
insulin-like growth factor II receptor
Contributor Information
Huangming Xie, Email: xieh@mit.edu.
Lei Sun, Email: sun@wi.mit.edu.
Harvey F. Lodish, Email: lodish@wi.mit.edu.
Bibliography
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004 Sep 16;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan 23;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008 Sep 4;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008 Sep 4;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 6.Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005 Nov;132(21):4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 8.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006 Dec;12(12):580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008 Feb;8(2):120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 10.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006 Jul;4(1):9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008 Mar;9(3):219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 12.Callis TE, Wang DZ. Taking microRNAs to heart. Trends Mol Med. 2008 Jun;14(6):254–260. doi: 10.1016/j.molmed.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009 Mar;37(4):1035–1048. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008 Jan;18(1):130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005 Jun 9;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009 Jun;21(3):452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009 Jan;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao FB. Posttranscriptional control of neuronal development by microRNA networks. Trends Neurosci. 2008 Jan;31(1):20–26. doi: 10.1016/j.tins.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluiver J, Kroesen BJ, Poppema S, van den Berg A. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006 Nov;20(11):1931–1936. doi: 10.1038/sj.leu.2404387. [DOI] [PubMed] [Google Scholar]
- 20.Rajala MW, Scherer PE. Minireview: The adipocyte--at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology. 2003 Sep;144(9):3765–3773. doi: 10.1210/en.2003-0580. [DOI] [PubMed] [Google Scholar]
- 21.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006 Dec 14;444(7121):847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaffler A, Muller-Ladner U, Scholmerich J, Buchler C. Role of adipose tissue as an inflammatory organ in human diseases. Endocr Rev. 2006 Aug;27(5):449–467. doi: 10.1210/er.2005-0022. [DOI] [PubMed] [Google Scholar]
- 23.Coll AP, Farooqi IS, O'Rahilly S. The hormonal control of food intake. Cell. 2007 Apr 20;129(2):251–262. doi: 10.1016/j.cell.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman JM. The function of leptin in nutrition, weight, and physiology. Nutr Rev. 2002 Oct;60(10 Pt 2):S1–S14. doi: 10.1301/002966402320634878. discussion S68-84, 5-7. [DOI] [PubMed] [Google Scholar]
- 25.Spiegelman BM, Flier JS. Adipogenesis and obesity: rounding out the big picture. Cell. 1996 Nov 1;87(3):377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 26.Ailhaud G, Grimaldi P, Negrel R. Cellular and molecular aspects of adipose tissue development. Annu Rev Nutr. 1992;12:207–233. doi: 10.1146/annurev.nu.12.070192.001231. [DOI] [PubMed] [Google Scholar]
- 27.Feve B. Adipogenesis: cellular and molecular aspects. Best Pract Res Clin Endocrinol Metab. 2005 Dec;19(4):483–499. doi: 10.1016/j.beem.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006 Dec;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 29.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 30.Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell. 1974 Oct;3(2):127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- 31.Gu Z, Eleswarapu S, Jiang H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett. 2007 Mar 6;581(5):981–988. doi: 10.1016/j.febslet.2007.01.081. [DOI] [PubMed] [Google Scholar]
- 32.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009 May;58(5):1050–1057. doi: 10.2337/db08-1299. • Genome-wide miRNA profiling during adipogenesis and in obese adipocytes.
- 34.Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008 Feb 12;105(6):1949–1954. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999 Apr 2;284(5411):143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 36.Kajimoto K, Naraba H, Iwai N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. Rna. 2006 Sep;12(9):1626–1632. doi: 10.1261/rna.7228806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004 Dec 10;279(50):52361–52365. doi: 10.1074/jbc.C400438200. • First miRNA affecting adipogenesis
- 38.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009 Apr;276(8):2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 Regulates 3T3-L1 Adipogenesis. Mol Endocrinol. 2009 Jun;23(6):925–931. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006 Mar;38(3):356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 42.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006 Feb 14;103(7):2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009 Jan 13;106(2):498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003 Apr 29;13(9):790–795. doi: 10.1016/s0960-9822(03)00250-1. • First miRNA affecting fat metabolism
- 45. Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006 Feb 15;20(4):417–422. doi: 10.1101/gad.374406. • miRNA related to insulin resistance in Drosophila
- 46.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007 Jul;91(3):209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richon VM, Lyle RE, McGehee RE., Jr Regulation and expression of retinoblastoma proteins p107 and p130 during 3T3-L1 adipocyte differentiation. J Biol Chem. 1997 Apr 11;272(15):10117–10124. doi: 10.1074/jbc.272.15.10117. [DOI] [PubMed] [Google Scholar]
- 49.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000 Feb 24;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 50.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005 Mar 11;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 51.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995 Aug 31;376(6543):771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 52. He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol. 2007 Nov;21(11):2785–2794. doi: 10.1210/me.2007-0167. • miRNA related to insulin resistance in adipocytes
- 53.Xu J, Wong C. A computational screen for mouse signaling pathways targeted by microRNA clusters. RNA. 2008 Jul;14(7):1276–1283. doi: 10.1261/rna.997708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007 Nov 9;282(45):32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 55.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, et al. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008 Dec 5;377(1):136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 56.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007 Oct 19;131(2):242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 57.Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function--of mice and men. Genes Dev. 2009 Apr 1;23(7):788–797. doi: 10.1101/gad.1779209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004 Jan;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 59.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009 Apr 9;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009 Apr 9;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 61.Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009 Apr 9;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 62. Kloting N, Berthold S, Kovacs P, Schon MR, Fasshauer M, Ruschke K, et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS ONE. 2009;4(3):e4699. doi: 10.1371/journal.pone.0004699. • miRNA profiling in different types of adipose tissue and miRNAs associated with diabetes
- 63.Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J Cell Physiol. 2009 Feb;218(2):444–449. doi: 10.1002/jcp.21621. [DOI] [PubMed] [Google Scholar]
- 64.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest. 2003 Dec;112(12):1785–1788. doi: 10.1172/JCI20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trayhurn P, Wang B, Wood IS. Hypoxia in adipose tissue: a basis for the dysregulation of tissue function in obesity? Br J Nutr. 2008 Aug;100(2):227–235. doi: 10.1017/S0007114508971282. [DOI] [PubMed] [Google Scholar]
- 66.Cawthorn WP, Sethi JK. TNF-alpha and adipocyte biology. FEBS Lett. 2008 Jan 9;582(1):117–131. doi: 10.1016/j.febslet.2007.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005 Dec 1;438(7068):685–689. doi: 10.1038/nature04303. •• First miRNA silencing in vivo
- 68. Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006 Feb;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. • miRNA silencing in vivo
- 69.Hand NJ, Master ZR, Eauclaire SF, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009 Mar;136(3):1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology. 2009 Feb;49(2):618–626. doi: 10.1002/hep.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, et al. Differential expression of MicroRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009 Apr 16; doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hennessy E, O'Driscoll L. Molecular medicine of microRNAs: structure, function and implications for diabetes. Expert Rev Mol Med. 2008;10:e24. doi: 10.1017/S1462399408000781. [DOI] [PubMed] [Google Scholar]
- 73. Tang X, Tang G, Ozcan S. Role of microRNAs in diabetes. Biochim Biophys Acta. 2008 Nov;1779(11):697–701. doi: 10.1016/j.bbagrm.2008.06.010. • Thorough review on miRNAs in diabetes and related complications
- 74.Tang X, Muniappan L, Tang G, Ozcan S. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA. 2009 Feb;15(2):287–293. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008 Mar;389(3):305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 76.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009 Apr 7;106(14):5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang JY, Widmann C, et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008 Oct;57(10):2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dance A. Mouse miRNA library to open. Nature. 2008 Jul 17;454(7202):264. doi: 10.1038/454264c. [DOI] [PubMed] [Google Scholar]
- 79.Osokine I, Hsu R, Loeb GB, McManus MT. Unintentional miRNA ablation is a risk factor in gene knockout studies: a short report. PLoS Genet. 2008 Feb;4(2):e34. doi: 10.1371/journal.pgen.0040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005 May;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 81.Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol. 2007 Jul;3(7):e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Stenvang J, Kauppinen S. MicroRNAs as targets for antisense-based therapeutics. Expert Opin Biol Ther. 2008 Jan;8(1):59–81. doi: 10.1517/14712598.8.1.59. •• Thorough review on miRNAs as therapeutic targets
- 83. Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008 Apr 17;452(7189):896–899. doi: 10.1038/nature06783. • miRNA silencing in primates
- 84.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007 Sep;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gentner B, Schira G, Giustacchini A, Amendola M, Brown BD, Ponzoni M, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009 Jan;6(1):63–66. doi: 10.1038/nmeth.1277. • miRNA inhibition in vivo
- 86. Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009 Jun 12;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. • miRNA inhibition in vivo
- 87.De Paula D, Bentley MV, Mahato RI. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA. 2007 Apr;13(4):431–456. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gray SJ, Samulski RJ. Optimizing gene delivery vectors for the treatment of heart disease. Expert Opin Biol Ther. 2008 Jul;8(7):911–922. doi: 10.1517/14712598.8.7.911. [DOI] [PubMed] [Google Scholar]
- 89.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007 Oct;25(10):1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 90.Cerchia L, Giangrande PH, McNamara JO, de Franciscis V. Cell-specific aptamers for targeted therapies. Methods Mol Biol. 2009;535:59–78. doi: 10.1007/978-1-59745-557-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stewart KM, Horton KL, Kelley SO. Cell-penetrating peptides as delivery vehicles for biology and medicine. Org Biomol Chem. 2008 Jul 7;6(13):2242–2255. doi: 10.1039/b719950c. [DOI] [PubMed] [Google Scholar]
- 92.Marcato PD, Duran N. New aspects of nanopharmaceutical delivery systems. J Nanosci Nanotechnol. 2008 May;8(5):2216–2229. doi: 10.1166/jnn.2008.274. [DOI] [PubMed] [Google Scholar]
- 93.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008 May;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009 Jan;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008 Jun;40(6):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008 May;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007 Jun 1;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 98.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007 Feb 22;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 99.Villuendas G, Botella-Carretero JI, Lopez-Bermejo A, Gubern C, Ricart W, Fernandez-Real JM, et al. The ACAA-insertion/deletion polymorphism at the 3' UTR of the IGF-II receptor gene is associated with type 2 diabetes and surrogate markers of insulin resistance. Eur J Endocrinol. 2006 Aug;155(2):331–336. doi: 10.1530/eje.1.02217. [DOI] [PubMed] [Google Scholar]
- 100.Lv K, Guo Y, Zhang Y, Wang K, Jia Y, Sun S. Allele-specific targeting of hsa-miR-657 to human IGF2R creates a potential mechanism underlying the association of ACAA-insertion/deletion polymorphism with type 2 diabetes. Biochem Biophys Res Commun. 2008 Sep 12;374(1):101–105. doi: 10.1016/j.bbrc.2008.06.102. [DOI] [PubMed] [Google Scholar]
- 101. Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009 Jan;37:D98–D104. doi: 10.1093/nar/gkn714. (Database issue) • miRNA dysregulation in various diseases
- 102. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008 Oct;18(10):997–1006. doi: 10.1038/cr.2008.282. •• miRNAs in diabetic serum from diabetic patients
- 103.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008 Mar 28;319(5871):1782–1784. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]