Abstract
Aneuploidy has a paradoxical effect on cell proliferation. In all normal cells analyzed to date, aneuploidy has been found to decrease the rate of cell proliferation. Yet, aneuploidy is also a hallmark of cancer, a disease of enhanced proliferative capacity, and aneuploid cells are frequently recovered following the experimental evolution of microorganisms. Thus, in certain contexts, aneuploidy may also have growth-advantageous properties. New models of aneuploidy and chromosomal instability have shed light on the diverse effects that karyotypic imbalances have on cellular phenotypes, and suggest novel ways of understanding aneuploidy’s role in development and disease.
Aneuploidy, an incorrect karyotype
Eukaryotic organisms have evolved robust mechanisms to ensure accurate segregation of the genetic material during mitosis. Cell cycle checkpoints delay chromosome segregation until DNA replication has been completed and sister chromatids are properly aligned at the metaphase plate. However, these safeguards occasionally fail, resulting in daughter cells that have gained or lost portions of the genetic material. Since the seminal observations of von Hansemann and Boveri at the beginning of the 20th century, these unequal cell divisions have been associated with developmental defects and cancer [1]. Yet, determining how exactly the aneuploid state impacts cell physiology has remained elusive. In recent years, models of aneuploidy have been developed which demonstrate the existence of common cellular phenotypes that result from karyotypic imbalances in a variety of different organisms (Table 1). Surprisingly, these studies have also revealed certain circumstances in which aneuploidy can actually improve cell proliferation and fitness.
Table 1.
Common phenotypes in aneuploid cells
| Aneuploidy Model | Slow proliferation? | Evidence of proteotoxic stress? | Increased metabolic requirements? | Increased lactate production? | Chromosomal instability? | Recombination defects? | References |
|---|---|---|---|---|---|---|---|
| Disomic budding yeast | Y | Y | Y | - | Y | Y | [32,33,44,95] |
| Disomic fission yeast | Y | ? | ? | - | Y | Y | [34,35,95] |
| Trisomic MEFs | Y | Y | Y | Y | ? | ? | [36,60] |
| CIN fibroblasts | Y | Y | Y | Y | Y | ? | [24–26,60] |
| Down syndrome cells | Y | ? | ? | ? | ? | Y | [23,98–101] |
| Aneuploid cancers | N | Y | Y | Y | Y | Y | [60,66,108,109] |
[?] refers to aneuploid phenotypes which have not yet been tested in the indicated organism or model system.
[-] refers to phenotypes which are unable to be tested in the indicated organism.
A number of excellent reviews have described the cell cycle checkpoints which ensure the fidelity of chromosome segregation [2,3]. Here, we will instead focus on the consequences of aneuploidy, which are only beginning to be understood. For the purposes of this review, we will limit our discussion to aneuploidy, defined as changes in karyotype that are not whole-number multiples of the haploid complement. Aneuploidy differs from polyploidy, in which cells gain a balanced set of all chromosomes. Polyploidy has been reviewed elsewhere, and, unlike aneuploidy, appears to be well-tolerated, particularly in single-cell organisms, plants, and in many metazoan tissues [4,5]. Below, we discuss the costs and benefits of aneuploidy, and suggest mechanisms by which karyotypic imbalances can cause the phenotypes commonly observed in aneuploid cells.
Aneuploidy is detrimental to cell and organismal fitness
Organismal aneuploidy impairs normal development, but some somatic aneuploidy is tolerated in vivo
Whole-organism aneuploidy is the most common cause of miscarriage and mental retardation in humans [6,7]. All human monosomies and 20 out of the 23 possible autosomal trisomies are embryonic lethal. Of the three trisomies which are viable at birth, only one, Trisomy 21/Down syndrome, can survive until adulthood [8]. Similarly, in mice, all autosomal aneuploidies are embryonic lethal, with the exception of Trisomy 19, which dies shortly after birth [9]. Aneuploidy has also been associated with developmental defects and lethality in a variety of other organisms, including maize [10], flies [11], and nematodes [12–14]. Thus, aneuploidy presents a considerable barrier toward successful development.
The consequences of somatic cellular aneuploidy are less well understood. Mammalian neuronal cells in normal adult organisms display low levels of mosaic aneuploidy [15,16], and wide-ranging karyotypic variation, including tetraploidy, octaploidy, and some aneuploidy, is commonly observed in hepatocytes [17,18]. Aneuploid neurons are functional [19], however post-mortem studies in human brains have linked aneuploidy with neurological pathologies like Alzheimer’s disease [20,21]. The functional significance of somatic aneuploidy is unknown, though it may serve as a source of genetic variation in the liver and the brain [16,18]. In vitro work (discussed below) suggests that aneuploid cells in living organisms should exhibit decreased fitness, and indeed reconstitution of the mouse hematopoietic system with trisomic stem cells results in anemia and other blood disorders [22]. However, it may be the case that certain highly specialized cell types, like karyotypically-unstable hepatocytes, have evolved mechanisms to tolerate the detrimental effects of aneuploidy.
Cellular aneuploidy slows the rate of cell proliferation
In addition to its detrimental effects at the organismal level, several lines of evidence demonstrate that aneuploidy decreases the rate of cell proliferation. The first studies on aneuploidy and cell division were performed in fibroblasts from individuals with Down syndrome; these cells were found to divide more slowly than age-matched euploid controls [23]. More recently, several labs have investigated the consequences of aneuploidies which result from genetic ablation of the spindle assembly checkpoint. Cells with a compromised checkpoint display chromosomal instability (CIN), a continuously varying karyotype. While mouse models of CIN are generally tumor prone (see below), individual cells with a CIN phenotype typically exhibit slow proliferation and/or low viability [24–27]. However, we note that some models of CIN have been reported to proliferate at wild-type rates in vitro [28,29]. In many cases, this likely reflects the low levels of aneuploidy in the cell line examined. For instance, Bub1bH/H mouse embryonic fibroblasts (MEFs) do not exhibit a proliferation defect at passage 3, but do exhibit a proliferation defect at passage 7, which presumably results from the increased frequency of aneuploidy after prolonged growth in culture [28]. Alternately, some CIN models interfere with mitotic checkpoint signaling, allowing cells to initiate anaphase after an abbreviated metaphase arrest [29]. In certain instances, the quicker progression through mitosis may conceal the growth-inhibitory effects of aneuploidy.
As these examples illustrate, it can be difficult to distinguish the consequences of aneuploidy from the effects of the CIN-inducing mutations used to generate karyotypic variation. It has therefore proven informative to study aneuploidy in the absence of mutations which compromise chromosome segregation fidelity. Chromosome missegregation events can be generated in otherwise diploid cell lines by transient treatment with spindle poisons; cells which survive this treatment exhibit high levels of aneuploidy without CIN-inducing mutations [30,31]. As with most models of CIN, cells with chemically-induced aberrant karyotypes exhibit poor proliferative capacity. More generally, the effects of single-chromosome aneuploidy have been explored systematically in yeast and mammalian cells. 20 haploid strains of the budding yeast Saccharomyces cerevisiae were generated which carry one or two copies of single yeast chromosomes. All disomic strains were found to proliferate more slowly than an isogenic euploid strain, though the doubling time varied between the disomes [32]. Similarly, all aneuploid strains derived by sporulation of triploid and pentaploid strains displayed impaired proliferation under normal growth conditions in S. cerevisiae [33] and in Schizosaccharomyces pombe [34,35]. In mice, naturally-occurring Robertsonian chromosome fusions have been employed to generate sibling-matched trisomic and euploid MEFs [36]. In all cases, trisomic fibroblasts were found to divide more slowly than euploid controls. Thus, in a variety of aneuploid models, an incorrect karyotype reduces the proliferative capacity of cells.
Why is aneuploidy detrimental to cellular fitness?
Several factors may contribute to the detrimental phenotypes associated with aneuploidy. By definition, aneuploid cells contain different quantities of DNA than euploid cells do. However, it is unlikely that extra DNA alone impairs cell fitness in most cases. Yeast strains carrying large artificial chromosomes containing human or mouse DNA (which presumably encode few or no genes which are expressed in yeast) do not exhibit proliferation defects [32]. An overabundance of specific DNA sequences may confer some toxicity, as yeast cells carrying >10 extra centromeres display a metaphase delay and increased chromosome missegregation [37,38]. However, these effects are not observed when fewer excess centromeres are present, and it is unclear whether the toxicity of specific sequences can explain the sickness frequently observed in cells bearing single extra chromosomes. The most likely explanation for the majority of detrimental phenotypes caused by aneuploidy is the gene dosage hypothesis: gains or losses of whole chromosomes immediately alter the dosage of hundreds of genes in a cell, thereby leading to imbalances in critical proteins (Figure 1a). Several lines of evidence support this hypothesis. First, aneuploid chromosomes appear to be expressed. Tissue from individuals with Down syndrome generally show upregulation of transcripts from chromosome 21 [39,40], and trisomic plants as well as fibroblasts from trisomic mouse embryos display a proportional increase in mRNA levels from the additional chromosomes [36,41]. Aneuploidy correlates with altered transcript levels in yeast [32,42,43], and, importantly, quantitative mass spectrometry demonstrates that aberrant karyotypes cause proportional imbalances in the relative levels of most proteins as well [33,44]. Secondly, the severity of the phenotypes caused by aneuploidy correlates with the number of genes gained or lost. The three human trisomies that survive until birth have the fewest protein-coding genes on them, while the survival of trisomic mouse embryos in utero correlates with the number of genes on the extra chromosome (Figure 1b; [45]). Similarly, in disomic yeast, the delay in cell division correlates with the number of open reading frames on the extra chromosome (Figure 1c), though the presence of particularly toxic genes (i.e. β-tubulin) on small chromosomes can cause disproportionate effects [32,46]. Finally, an increasing body of evidence demonstrates that 2-fold and even 1.5-fold changes in gene copy number can have significant effects on cellular and organismal phenotypes. Down syndrome is associated with a decreased frequency of solid tumor formation [47]; studies in mice suggest that single extra copies of Ets2 and DSCR1 (both located on chromosome 21) may confer protection from tumorigenesis [48,49]. Single extra copies of Sir2 prolong lifespan in yeast [50] and Caenorhabditis elegans [51], while a single extra copy of α-synuclein predisposes individuals to Parkinson’s disease [52]. However, we note that some organisms, including Drosophila melanogaster, appear to maintain mechanisms for dosage compensation which dampen the effects of aneuploidy [53,54]. In these organisms, gene dosage may not be a robust predictor of protein expression levels, and aneuploid phenotypes may result from the overabundance of specific genes which escape attenuation.
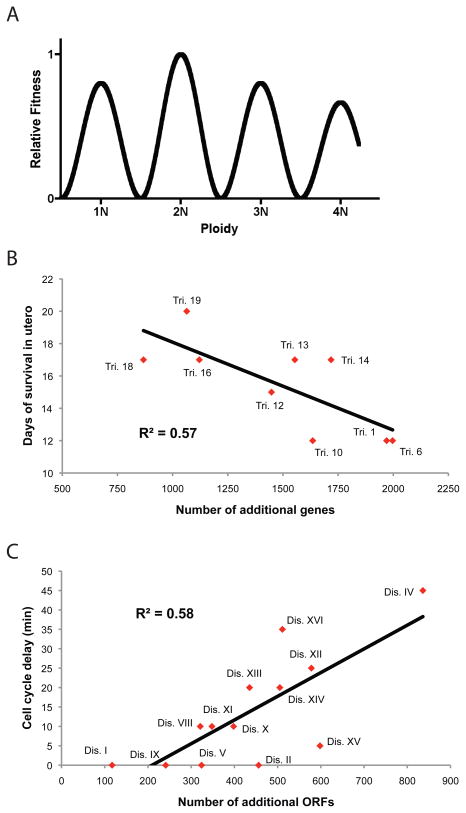
Figure 1. Cellular and organismal fitness vary according to the deviation from euploidy.
A. A model of the effect of ploidy on cell fitness. Cells with balanced sets of all chromosomes are generally healthy, though haploid and polyploid cells are moderately less fit than diploid cells. As cells move away from euploidy, fitness decreases, and greater karyotypic imbalances typically cause more severe phenotypes. Note that transformed cells are able to tolerate a high degree of aneuploidy via mechanisms which are largely unknown. B. The survival in utero of trisomic mouse embryos correlates with the number of genes encoded by the additional chromosome. A linear correlation is plotted against the data (figure adapted from [45]). C. The cell cycle delay in aneuploid yeast strains correlates with the number of open reading frames encoded by the additional chromosome. A linear correlation is plotted against the data (figure adapted from [32]).
How can alterations in gene dosage affect cellular fitness? First, changes in the concentration of a particular protein can directly modulate the efficiency of that protein’s cellular function (Figure 2a). For instance, DSCR1 is a negative regulator of angiogenic signaling, and an additional copy of DSCR1 in a mouse model of Down syndrome blocks tumor formation by inhibiting angiogenesis [48]. Secondly, changes in gene copy number can affect the formation or function of stoichiometry-sensitive complexes (Figure 2b). Proper stoichiometry can be disrupted by both under-expression (i.e., haploinsufficiency; [55]) and over-expression of proteins in complexes. In S. cerevisiae, a single extra copy of the gene encoding the phosphatase Cdc14 delays cell cycle progression, as it is no longer inhibited by its 1:1 stoichiometric binding partner Cfi1/Net1 [56]. Finally, an intriguing recent report suggests that a key determinant of dosage sensitivity is the susceptibility of some proteins to make promiscuous molecular interactions [57]. Genes which are toxic when over-expressed are enriched for those which have many low-affinity binary interaction partners, suggesting that mass-action driven off-target interactions may impair cellular fitness in aneuploid cells (Figure 2c). This type of promiscuous interaction may explain the gain-of-function phenotype in signaling pathways in certain cancers [57]. For instance, when lung cancers with EGFR-activating mutations are treated with an EGFR inhibitor, some cells acquire drug resistance via amplification of the MET oncogene [58,59]. Over-expression of MET leads to activation of kinases downstream of EGFR, which are independent of MET signaling in cells which contain normal levels of MET [58,59]. Thus, promiscuous molecular interactions may significantly alter the cellular interactome when gene copy number is changed.
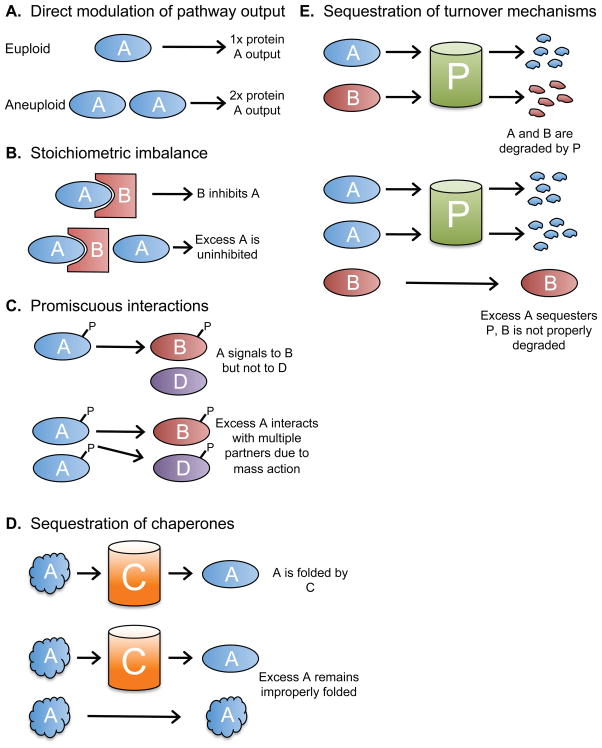
Figure 2. Consequences of changes in gene copy number.
A. An increased dosage of a single gene, such as a rate-limiting enzyme, can increase a cellular pathway’s output or function. B. Altered gene dosage can interfere with the function of stoichiometry-sensitive complexes. C. Protein-protein interactions depend on the concentration of each binding partner. Altered expression of some proteins, such as signal-transduction kinases, may cause promiscuous molecular interactions which alter cellular phenotypes. D. Many proteins require chaperones to fold correctly. If aneuploidy overwhelms cellular chaperones, then misfolded proteins which escape chaperone-dependent folding may form insoluble and potentially cytotoxic aggregates. It is also possible that other essential clients of these chaperones remain unfolded. E. Quality-control mechanisms, including the ubiquitin-proteasome pathway, ensure that misfolded or improperly expressed proteins are rapidly turned over. Regulated protein degradation is also utilized to trigger various cellular programs, including cell cycle progression. The overabundance of certain proteins may interfere with the folding or turnover of other client proteins.
The detrimental phenotypes associated with aneuploidy can also result from the synergistic effects of changing the copy number of several hundred genes at once. Aneuploid strains of yeast are sensitive to conditions that interfere with protein translation, folding, and degradation [32,33,44], and trisomic MEFs and aneuploid cancer cells are killed by the protein-folding inhibitor 17-AAG [60]. These sensitivities are largely independent of the identity of the additional chromosome, suggesting that aneuploidy generally challenges a cell’s ability to maintain protein homeostasis. While the consequences of over-expressing any one protein may be unique, cells utilize a limited number of quality-control mechanisms for protein folding and turnover. For instance, most proteins which contain a WD40 domain require the eukaryotic chaperonin TRiC/CCT to correctly fold [61]. In the absence of sufficient chaperone capacity to accommodate over-expressed proteins, other chaperone clients may remain unfolded, leading to loss-of-function phenotypes and the formation of potentially cytotoxic aggregates (Figure 2d). In yeast, misfolded proteins exact a fitness cost, even when the misfolded proteins represent less than 0.1% of the total cellular proteome [62]. Thus, aneuploidy may impair cell proliferation via an accumulation of improperly folded or aggregated proteins.
Misfolded proteins, as well as properly-folded proteins which are present in excess, can also impinge on cellular mechanisms for protein turnover. It has been demonstrated that cells ensure the integrity of some stoichiometric complexes by rapidly degrading over-expressed free subunits, such as histones [63] and ribosomal proteins [64,65]. Accordingly, not all proteins in aneuploid cells are proportionally over-expressed, and most dosage-compensated proteins are members of protein complexes [44]. Thus, aneuploidy-induced stoichiometric imbalances may severely stress the proteasome (Figure 2e). In order to understand how cells cope with the anti-proliferative effects of aneuploidy, disomic yeast strains were grown continuously for 14 days, then genetic alterations which improved their proliferative capacity were identified by whole-genome sequencing [44]. Interestingly, several different disomic strains developed mutations in the proteasome pathway, and two strains independently acquired loss-of-function mutations in the ubiquitin-specific protease UBP6. Deletion of UBP6 was found to improve the growth rates of four disomic strains, and quantitative mass spectrometry demonstrated that loss of UBP6 led to an attenuation in the levels of proteins overproduced due to aneuploidy [44]. These results suggest that proteotoxic stress is a key source of aneuploidy’s anti-proliferative effects.
Lastly, aneuploidy has been found to significantly alter metabolism and increase cellular energy needs. Aneuploid yeast and MEFs are less efficient at converting nutrients into biomass than euploid cells are [32,36]. This could result from cells wasting energy by translating and then turning over excess proteins from the additional chromosomes. Consequently, aneuploid fibroblasts and cancer cells display broad sensitivities to drugs that interfere with cellular energy homeostasis [60]. It has also been noted that aneuploid cells produce significantly more lactate during proliferation than euploid cells do [26,36], a phenotype which they share with cancer cells [66]. The underlying cause of these metabolic alterations is not known, but the resultant energy stress is a likely limit on the proliferation of aneuploid cells. Additionally, the metabolic similarities between aneuploid primary cells and cancer cells suggest that cancer cells have not fully escaped the stresses associated with aneuploidy.
Aneuploidy is frequently associated with enhanced cell proliferation
Despite the seemingly detrimental effects of the massive gene dosage imbalances which result from aneuploidy, changes in karyotype appear to confer proliferative advantages in certain circumstances. Most notably, aneuploidy is a hallmark of cancer, a disease of increased cell proliferation [67]. Greater than 90% of solid tumors and 75% of hematopoietic cancers have gained or lost entire chromosomes [68]. Several lines of evidence suggest that aneuploidy has a causal role in tumorigenesis, and is not simply a byproduct of transformation. First, aneuploidy appears prior to or coincident with malignant transformation in a variety of contexts, including in human patients [69–71] and mouse models of cancer [72,73]. Secondly, tumor genotyping has identified clonal mutations or genetic alterations in various cancers which compromise the fidelity of chromosome segregation [74–76]. Thirdly, inherited mutations in the spindle checkpoint component BubR1 result in mosaic aneuploidy and predispose individuals to cancer, suggesting a link between faulty spindle checkpoint function and tumorigenesis [77]. Lastly, transgenic mice with elevated rates of chromosome missegregation are generally tumor prone [26,76,27,78]. These studies suggest that changes in karyotype can contribute to cellular transformation, a process that enhances the proliferative capacity of a cell.
Certain aneuploidies also confer a proliferative advantage during growth or experimental evolution in microorganisms. S. cerevisiae cells grown in nutrient-limiting conditions or deprived of a cytokinetic motor develop aneuploidies [42,79,80], as do pathogenic strains of Candida albicans grown in the presence of the anti-fungal drug fluconazole [81,82]. In the yeast deletion collection, ~8% of all strains have spontaneously gained extra chromosomes [43]. Lastly, in a comprehensive analysis of 38 aneuploid strains derived from triploid or pentaploid meiosis, some aneuploid strains were found to exhibit improved fitness under extreme culture conditions, including growth at low temperature and treatment with 4-NQO [33]. Thus, it is clear that alterations in karyotype can improve cellular fitness under certain circumstances.
How is aneuploidy beneficial?
As with the detrimental consequences of aneuploidy, most of the growth-advantageous properties of aneuploidy likely result from changes in gene copy number. In particular, many benefits of aneuploidy can apparently be explained by the change in copy number of one or a few genes. In studies of nutrient-limited yeast, cells acquire aneuploidies that confer extra copies of the transporters that take up the scarce nutrient [79,80]. Fluconazole resistance in C. albicans can be caused by the gain of an isochromosome containing TAC1, a transcriptional regulator of drug-efflux pumps, and ERG11, the target of fluconazole [82]. Decreasing the copy number of TAC1 and ERG11 abolishes the protective effects of aneuploidy. Among the aneuploid strains which arose during construction of the yeast deletion collection, many of the strains were found to harbor extra copies of a chromosome containing a paralogue of the gene which was deleted, suggesting a dosage-dependent rescue of cell proliferation [43]. Moreover, in the evolution of cytokinesis-defective yeast, improved growth due to aneuploidy can be phenocopied in euploid strains by increasing the dosage of a transcription factor and a signaling kinase that are present on a frequently-gained chromosome [42].
Changes in copy number of a few genes may also explain the tumorigenic properties of aneuploidy. Karyotypic alterations can lead to the gain in copy number of growth-promoting oncogenes and loss of tumor-suppressor loci. For instance, cervical cancers frequently gain extra copies of chromosome 3, which contains the human telomerase gene TERC [70,83], while leukemias frequently gain chromosome 8, which contains the MYC transcription factor [84,85]. It is believed that these genes are largely responsible for the recurrent gain of these chromosomes because in many cancers these loci are focally amplified [86–89]. Chromosome loss can also promote cellular transformation by decreasing the dosage of tumor suppressors [78,90]. Interestingly, loss of heterozygosity at tumor suppressors is frequently “copy-neutral,” i.e. a duplication event occurs to maintain euploid dosage levels of all genes on the affected chromosome [78,91,92]. This may arise due to selective pressure to protect cancer cells from the deleterious effects of haploinsufficiency, or from imbalances in other genes required for proliferation.
Recent evidence has also suggested that indirect consequences of aneuploidy could contribute to the improved growth phenotype associated with karyotypic imbalances. It has been hypothesized that the protein imbalances caused by aneuploidy could interfere with a cell’s basal mechanisms for ensuring genomic integrity, and this aneuploidy-induced genomic instability could speed the development of potentially growth-promoting genetic alterations [45,93,94]. A causative link between aneuploidy and genomic instability has recently been demonstrated in yeast: aneuploid strains of S. cerevisiae and S. pombe were found to display various forms of genomic instability, including hyper-recombination, elevated levels of chromosome missegregation, increased mutation, and an inability to repair genotoxic damage [95]. Aneuploidy-induced genomic instability may extend to mammalian cells as well: the chromosomal instability of chemically-transformed Chinese hamster embryo cells was found to vary according to their initial karyotype, with highly aneuploid cells exhibiting the greatest degree of instability [96]. Aneuploid cells lacking p53 were also found to be karyotypically unstable [97]. Finally, trisomic cells derived from individuals with Down syndrome are sensitive to ionizing radiation and other genotoxic agents, suggesting that DNA repair is compromised in these cells [98–101]. Genomic instability has been shown to confer a proliferative advantage during evolutionary competition and may cause or contribute to cellular transformation [102–105]. It is therefore possible that aneuploidy-induced genomic instability could play a causal role in the improved growth observed in cancer cells. Whether aneuploidy can actually speed the evolutionary process, and whether aneuploidy-induced genomic instability contributes to the instability that is characteristic of cancer cells [106,107], remain to be tested.
Concluding Remarks
Aneuploidy represents a gross challenge to cellular homeostasis: by simultaneously changing the copy number of several hundred genes at once, aneuploidy can vastly alter normal cellular functions. The consequences of this dysregulation are apparent in the defects caused by aneuploidy during cell proliferation and development. Yet, it is for this same reason that certain genetic and environmental stresses can be overcome via aneuploidy. When one chromosome contains a gene that has a dosage-dependent effect on cell growth (such as an oncogene or a transporter of a scarce nutrient), chromosome missegregation represents a relatively easy way by which the copy number of that gene can be increased. This aneuploidy exerts a high cost on the cell, particularly by increasing the burden on energy and protein homeostasis. However, cells can evolve mechanisms that shield them from the detrimental consequences of aneuploidy, and this adaptation may be accelerated by aneuploidy-induced genomic instability. Nonetheless, the pathways which are commonly stressed in aneuploid cells represent attractive targets for the development of chemotherapeutics that could potentially be useful in treating a broad spectrum of aneuploid cancers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Works Cited
- 1.Hardy PA, Zacharias H. Reappraisal of the Hansemann-Boveri hypothesis on the origin of tumors. Cell Biology International. 2005;29:983–992. doi: 10.1016/j.cellbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Schvartzman JM, et al. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 4.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 5.Lee HO, et al. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassold T, et al. Human aneuploidy: Incidence, origin, and etiology. Environ Mol Mutagen. 1996;28:167–175. doi: 10.1002/(SICI)1098-2280(1996)28:3<167::AID-EM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 7.Brown S. Miscarriage and Its Associations. Semin Reprod Med. 2008;26:391–400. doi: 10.1055/s-0028-1087105. [DOI] [PubMed] [Google Scholar]
- 8.Pai GS, et al. Handbook of Chromosomal Syndromes. 1. Wiley-Liss; 2002. [Google Scholar]
- 9.Epstein CJ. Mouse monosomies and trisomies as experimental systems for studying mammalian aneuploidy. Trends in Genetics. 1985;1:129–134. [Google Scholar]
- 10.McClintock B. A Cytological and Genetical Study of Triploid Maize. Genetics. 1929;14:180–222. doi: 10.1093/genetics/14.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsley DL, et al. SEGMENTAL ANEUPLOIDY AND THE GENETIC GROSS STRUCTURE OF THE DROSOPHILA GENOME. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkin J. X chromosome dosage and gene expression in Caenorhabditis elegans: Two unusual dumpy genes. Mol Gen Genet. 1983;192:452–458. [Google Scholar]
- 13.Hodgkin J, et al. Nondisjunction Mutants of the Nematode Caenorhabditis Elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sigurdson DC, et al. An X-autosome fusion chromosome of Caenorhabditis elegans. Mol Gen Genet. 1986;202:212–218. doi: 10.1007/BF00331639. [DOI] [PubMed] [Google Scholar]
- 15.Rehen SK, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proceedings of the National Academy of Sciences. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yurov YB, et al. Aneuploidy and Confined Chromosomal Mosaicism in the Developing Human Brain. PLoS ONE. 2007;2:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S. Hepatic polyploidy and liver growth control. Semin Cancer Biol. 2000;10:161–171. doi: 10.1006/scbi.2000.0317. [DOI] [PubMed] [Google Scholar]
- 18.Duncan AW, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingsbury MA, et al. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci U S A. 2005;102:6143–6147. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iourov IY, et al. Aneuploidy in the normal, Alzheimer’s disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiol Dis. 2009;34:212–220. doi: 10.1016/j.nbd.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Iourov IY, et al. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Human Molecular Genetics. 2009;18:2656–2669. doi: 10.1093/hmg/ddp207. [DOI] [PubMed] [Google Scholar]
- 22.Herbst EW, Winking H. Adoptive transfer of the hematopoietic system of trisomic mice with limited life span: stem cells from six different trisomies are capable of survival. Dev Genet. 1991;12:415–422. doi: 10.1002/dvg.1020120606. [DOI] [PubMed] [Google Scholar]
- 23.Segal DJ, McCoy EE. Studies on Down’s syndrome in tissue culture. I. Growth rates protein contents of fibroblast cultures. J Cell Physiol. 1974;83:85–90. doi: 10.1002/jcp.1040830112. [DOI] [PubMed] [Google Scholar]
- 24.Kops GJPL, et al. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babu JR, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proceedings of the National Academy of Sciences. 2010;107:14188–14193. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver BAA, et al. Aneuploidy Acts Both Oncogenically and as a Tumor Suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 29.van Ree JH, et al. Overexpression of the E2 ubiquitin–conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. The Journal of Cell Biology. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres EM, et al. Effects of Aneuploidy on Cellular Physiology and Cell Division in Haploid Yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 33.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa O, Yanagida M. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Current Genetics. 1985;9:463–470. [Google Scholar]
- 35.Niwa O, et al. Growth arrest and chromosome instability in aneuploid yeast. Yeast. 2006;23:937–950. doi: 10.1002/yea.1411. [DOI] [PubMed] [Google Scholar]
- 36.Williams BR, et al. Aneuploidy Affects Proliferation and Spontaneous Immortalization in Mammalian Cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Futcher B, Carbon J. Toxic effects of excess cloned centromeres. Mol Cell Biol. 1986;6:2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Runge KW, Zakian VA. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol Cell Biol. 1989;9:1488–1497. doi: 10.1128/mcb.9.4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao R, et al. Global up-regulation of chromosome 21 gene expression in the developing down syndrome brain. Genomics. 2003;81:457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 40.FitzPatrick DR, et al. Transcriptome analysis of human autosomal trisomy. Human Molecular Genetics. 2002;11:3249–3256. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 41.Huettel B, et al. Effects of Aneuploidy on Genome Structure, Expression, and Interphase Organization in Arabidopsis thaliana. PLoS Genet. 2008:4. doi: 10.1371/journal.pgen.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rancati G, et al. Aneuploidy Underlies Rapid Adaptive Evolution of Yeast Cells Deprived of a Conserved Cytokinesis Motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 44.Torres EM, et al. Identification of Aneuploidy-Tolerating Mutations. Cell. 2010 doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torres EM, et al. Aneuploidy: Cells Losing Their Balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katz W, et al. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasle H, et al. Risks of leukaemia and solid tumours in individuals with Down’s syndrome. The Lancet. 2000;355:165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 48.Baek KH, et al. Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature. 2009;459:1126–1130. doi: 10.1038/nature08062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sussan TE, et al. Trisomy represses ApcMin-mediated tumours in mouse models of Down’s syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 50.Kaeberlein M, et al. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & Development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 52.Ibáñez P, et al. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 53.Devlin RH, et al. Autosomal dosage compensation Drosophila melanogaster strains trisomic for the left arm of chromosome 2. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:1200–1204. doi: 10.1073/pnas.79.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McAnally AA, Yampolsky LY. Widespread transcriptional autosomal dosage compensation in Drosophila correlates with gene expression level. Genome Biol Evol. 2010;2:44–52. doi: 10.1093/gbe/evp054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papp B, et al. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 56.Kaizu K, et al. Fragilities Caused by Dosage Imbalance in Regulation of the Budding Yeast Cell Cycle. PLoS Genet. 2010;6:e1000919. doi: 10.1371/journal.pgen.1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vavouri T, et al. Intrinsic Protein Disorder and Interaction Promiscuity Are Widely Associated with Dosage Sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 58.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engelman JA, et al. MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 60.Tang Y-C, et al. Identification of Aneuploidy-Selective Antiproliferation Compounds. Cell. 2011 doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siegers K, et al. TRiC/CCT cooperates with different upstream chaperones in the folding of distinct protein classes. EMBO J. 2003;22:5230–5240. doi: 10.1093/emboj/cdg483. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Geiler-Samerotte KA, et al. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proceedings of the National Academy of Sciences. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gunjan A, Verreault A. A Rad53 Kinase-Dependent Surveillance Mechanism that Regulates Histone Protein Levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 64.Abovich N, et al. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3429–3435. doi: 10.1128/mcb.5.12.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warner JR, et al. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985;5:1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vander Heiden MG, et al. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 68.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Current Opinion in Cell Biology. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 69.Balaban GB, et al. Karyotypic evolution in human malignant melanoma. Cancer Genetics and Cytogenetics. 1986;19:113–122. doi: 10.1016/0165-4608(86)90378-x. [DOI] [PubMed] [Google Scholar]
- 70.Heselmeyer-Haddad K, et al. Genomic Amplification of the Human Telomerase Gene (TERC) in Pap Smears Predicts the Development of Cervical Cancer. Am J Pathol. 2005;166:1229–1238. doi: 10.1016/S0002-9440(10)62341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rubin CE, et al. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology. 1992;103:1611–1620. doi: 10.1016/0016-5085(92)91185-7. [DOI] [PubMed] [Google Scholar]
- 72.Conti CJ, et al. Aneuploidy, an early event in mouse skin tumor development. Carcinogenesis. 1986;7:1845–1848. doi: 10.1093/carcin/7.11.1845. [DOI] [PubMed] [Google Scholar]
- 73.Danielsen HE, et al. Specific gain of chromosome 19 in preneoplastic mouse liver cells after diethylnitrosamine treatment. Carcinogenesis. 1991;12:1777–1780. doi: 10.1093/carcin/12.10.1777. [DOI] [PubMed] [Google Scholar]
- 74.Barber TD, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proceedings of the National Academy of Sciences. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cahill DP, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 76.Sotillo R, et al. Mad2 Overexpression Promotes Aneuploidy and Tumorigenesis in Mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 78.Baker DJ, et al. Whole Chromosome Instability Caused by Bub1 Insufficiency Drives Tumorigenesis through Tumor Suppressor Gene Loss of Heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dunham MJ, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Selmecki A, et al. Aneuploidy and Isochromosome Formation in Drug-Resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Selmecki A, et al. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68:624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 83.Cao Y, et al. Increased copy number of the TERT and TERC telomerase subunit genes in cancer cells. Cancer Sci. 2008;99:1092–1099. doi: 10.1111/j.1349-7006.2008.00815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kibbelaar RE, et al. Detection of trisomy 8 in hematological disorders by in situ hybridization. Cytogenet Cell Genet. 1991;56:132–136. doi: 10.1159/000133069. [DOI] [PubMed] [Google Scholar]
- 85.Oudat R, et al. Detection of Trisomy 8 in Philadelphia Chromosome-Positive CML Patients Using Conventional Cytogenetic and Interphase Fluorescence in situ Hybridization Techniques and its Relation to c-myc Involvement. Ann Clin Lab Sci. 2001;31:68–74. [PubMed] [Google Scholar]
- 86.Soder AI, et al. Amplification, increased dosage and in situ expression of the telomerase RNA gene in human cancer. Oncogene. 1997;14:1013–1021. doi: 10.1038/sj.onc.1201066. [DOI] [PubMed] [Google Scholar]
- 87.Yokoi S, et al. TERC Identified as a Probable Target within the 3q26 Amplicon That Is Detected Frequently in Non-Small Cell Lung Cancers. Clinical Cancer Research. 2003;9:4705–4713. [PubMed] [Google Scholar]
- 88.Jennings BA, Mills KI. c-myc locus amplification and the acquisition of trisomy 8 in the evolution of chronic myeloid leukaemia. Leuk Res. 1998;22:899–903. doi: 10.1016/s0145-2126(98)00097-6. [DOI] [PubMed] [Google Scholar]
- 89.La Farina M, et al. Two distinct amplification events of the c-myc locus in a colorectal tumour. J Cell Physiol. 2008;217:34–39. doi: 10.1002/jcp.21469. [DOI] [PubMed] [Google Scholar]
- 90.Baker DJ, van Deursen JM. Chromosome missegregation causes colon cancer by APC loss of heterozygosity. Cell Cycle. 2010;9:1711–1716. doi: 10.4161/cc.9.9.11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beroukhim R, et al. Inferring Loss-of-Heterozygosity from Unpaired Tumors Using High-Density Oligonucleotide SNP Arrays. PLoS Comput Biol. 2006;2:e41. doi: 10.1371/journal.pcbi.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gondek LP, et al. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111:1534–1542. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holliday R. Chromosome error propagation and cancer. Trends Genet. 1989;5:42–45. doi: 10.1016/0168-9525(89)90020-6. [DOI] [PubMed] [Google Scholar]
- 94.Matzke M. Does the intrinsic instability of aneuploid genomes have a causal role in cancer? Trends in Genetics. 2003;19:253–256. doi: 10.1016/s0168-9525(03)00057-x. [DOI] [PubMed] [Google Scholar]
- 95.Sheltzer J, et al. Aneuploidy drives genomic instability in yeast. Science. doi: 10.1126/science.1206412. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Duesberg P, et al. Genetic instability of cancer cells is proportional to their degree of aneuploidy. PNAS. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. Journal of Cell Biology. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lambert B, et al. DNA repair and frequency of X-ray and u. v.-light induced chromosome aberrations in leukocytes from patients with Down’s syndrome. Ann Human Genet. 1976;39:293–303. doi: 10.1111/j.1469-1809.1976.tb00133.x. [DOI] [PubMed] [Google Scholar]
- 99.Iijima K, et al. Bleomycin-induced chromosomal aberrations and sister chromatid exchanges in Down lymphocyte cultures. Hum Genet. 1984;66:57–61. doi: 10.1007/BF00275187. [DOI] [PubMed] [Google Scholar]
- 100.Otsuka F, et al. (May) Hypersensitivity to ionizing radiation in cultured cells from down syndrome patients. Journal of the Neurological Sciences. 69:103–112. doi: 10.1016/0022-510x(85)90011-5. [DOI] [PubMed] [Google Scholar]
- 101.Shafik HM, et al. Chromosomal radiosensitivity of Down syndrome lymphocytes at different stages of the cell cycle. Hum Genet. 1988;78:71–75. doi: 10.1007/BF00291238. [DOI] [PubMed] [Google Scholar]
- 102.Cahill DP, et al. Genetic instability and darwinian selection in tumours. Trends in Cell Biology. 1999;9:M57–M60. [PubMed] [Google Scholar]
- 103.Thompson DA, et al. Ploidy Controls the Success of Mutators and Nature of Mutations during Budding Yeast Evolution. Current Biology. 2006;16:1581–1590. doi: 10.1016/j.cub.2006.06.070. [DOI] [PubMed] [Google Scholar]
- 104.Sniegowski PD, et al. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 105.Giraud A, et al. Costs and Benefits of High Mutation Rates: Adaptive Evolution of Bacteria in the Mouse Gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 106.Lengauer C, et al. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 107.Bielas JH, et al. Human cancers express a mutator phenotype. Proceedings of the National Academy of Sciences. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lengauer C, et al. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 109.Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31:955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]