Abstract
Traumatic injury to the central nervous system results in increased expression and deposition of chondroitin sulfate proteoglycans (CSPGs) that are inhibitory to axonal regeneration. Transforming growth factor– β (TGF-β) has been implicated as a major mediator of these changes, but the mechanisms through which TGF-β regulates CSPG expression are not known. Using lentiviral expressed Smad-specific shRNA we show that TGF-β induction of CSPG expression in astrocytes is Smad dependent. However, we find a differential dependence of the synthetic machinery on Smad2 and/or Smad3. TGF-β induction of neurocan and xylosyl transferase 1 required both Smad2 and Smad3, whereas induction of phosphacan and chondroitin synthase 1 required Smad2 but not Smad3. Smad3 knockdown selectively reduced induction of chondroitin-4-sulfotransferase 1 and the amount of 4-sulfated CSPGs secreted by astrocytes. Additionally, Smad3 knockdown in astrocytes was more efficacious in promoting neurite outgrowth of neurons cultured on the TGF-β treated astrocytes. Our data implicate TGF–β-Smad3 mediated induction of 4-sulfation as a critical determinant of the permissiveness of astrocyte secreted CSPGs for axonal growth.
Keywords: Smad3, Smad2, transforming growth factor-β, glial scar, astrocytes, cell culture
Introduction
Most axons in the adult central nervous system (CNS) fail to regenerate following traumatic injury (Fitch & Silver 2008). Regenerative failure is due both to a poor growth capacity of the neurons themselves as well as to the formation of a glial scar, which further inhibits regrowth. The glial scar is comprised of a dense meshwork of hypertrophied astrocytic processes, other glial cells and their deposited extracellular matrix (ECM) molecules including chondroitin sulfate proteoglycans (CSPGs) (Galtrey & Fawcett 2007, Sherman & Back 2008). CSPGs consist of a core protein with covalently attached sulfated glycosaminoglycan (GAG) side chains. The expression of many CSPG core proteins, as well as the synthesis and sulfation of the attached GAG chains, is substantially increased after injury (Asher et al. 2000, Asher et al. 2002, Dobbertin et al. 2003, Tang et al. 2003). Moreover, animals treated with the enzyme chondroitinase ABC (cABC), that degrades GAG chains, showed enhanced neuronal regrowth and recovery of function after brain and spinal cord injury (Bradbury et al. 2002, Moon et al. 2001, Lee et al. 2010), suggesting that growth inhibition is predominantly due to the sulfated GAG chains. Understanding the molecular mechanisms regulating CSPG deposition after injury may enable therapies to reduce their synthesis and hence provide a more permissive environment for axon regeneration.
TGF-β is a central mediator initiating formation of the glial scar and deposition of CSPGs (Logan & Berry 1999, Logan et al. 1999, Rimaniol et al. 1995, Lagord et al. 2002). Expression of TGF-β1, and its receptors, TβRI and TβRII, is markedly upregulated after injury (Mctigue 2000, Rimaniol et al. 1995). At the earliest stages of the injury response, TGF-β1 is released from platelets and secreted from cells of the monocyte/macrophage lineage (Logan et al. 1992, Nichols et al. 1991 ); at later time points TGF–β is expressed by microglia, astrocytes and neurons around the injury site (Wang et al. 2007, Makwana et al. 2007, Mctigue 2000). Inhibition of TGF-β1 or TGF-β2 with neutralizing antisera significantly reduced matrix deposition and fibrogenic scarring around a wound (Logan et al. 1994, Logan et al. 1999) suggesting that interference with TGF-β signaling could be a powerful approach to reduce scar formation and hence facilitate neuronal regeneration. TGF-β induces many of the genes encoding CSPG core proteins or enzymes regulating GAG chain synthesis (Asher et al. 2000, Hamel et al. 2005, Smith & Strunz 2005, Gris et al. 2007, Wang et al. 2008).
TGF-β signals through transmembrane serine/threonine protein kinase receptors (Kang et al. 2009). Ligand-receptor activation leads to phosphorylation of many downstream targets including the Smad transcription factors (reviewed by Ross & Hill 2008, Moustakas & Heldin 2009). The receptor activated Smads (R-Smads), Smad2 and Smad3, are phosphorylated directly by the activated TGF-β receptor, TβRI, complex with the co-Smad Smad4 and translocate to the nucleus to activate transcription (Dijke & Hill 2004). TGF-β can activate many other pathways including the Erk, JNK, p38 and PI3 Kinase pathways, and NF-kB and Rho GTPase signaling in a Smad independent manner (Moustakas & Heldin 2005, Zhang 2009).
We have previously shown that Smad3 null mice display more rapid wound closure and reduced scar formation after cortical stab injury (Wang et al. 2007). However, as Smad2 null mice die in utero, the role of Smad2 in mediating TGF-β signals within CNS is unknown (Weinstein et al. 1998, Waldrip et al. 1998, Nomura & Li 1998). We therefore undertook to determine the relative importance of Smad2 and Smad3 signaling in the TGF-β mediated induction of CSPG core proteins and synthetic enzymes in astrocytes in vitro. We show that Smad proteins are critically important for induction of CSPG production in response to TGF-β, but that there is a differential effect of Smad2 and Smad3. While reduction of either Smad reduced the actions of TGF-β to decrease astrocyte matrix permissiveness, Smad3 reduction had a higher efficacy. Interestingly, we found only one gene, encoding the enzyme chondroitin-4-sulfotransferase-1 (C4ST-1), which increases 4-sulfation of GAG chains, that was dependent on Smad3 and not Smad2. Thus, our data suggest that TGF-β signaling through Smad3 may lead to a less permissive environment for neuronal growth by increasing the amount of 4-sulfated GAG chains.
Materials and Methods
Materials
Cell culture reagents and fetal bovine serum were obtained from Invitrogen Life Technologies (Carlsbad, CA) and culture plates from Costar (Corning, NY). Recombinant human TGF-β was purchased from Peprotech (Rocky Hill, NJ). Primers were synthesized on a PE Applied Biosystems 394 synthesizer by the USUHS in-house oligonucleotide facility. The following primary antisera were used: Smad2 (3103) and Smad3 (9523, Cell Signaling Technology; Danvers, MA); CS-56 (C8035, Sigma-Aldrich; St Louis, MO); actin (SC47778, Santa Cruz Biotechnology, Inc, Santa Cruz, CA); phosphacan (MAB5210, Millipore; Billerica, MA); brevican (610894, BD Biosciences, San Jose, CA); 2H6 (370710-1 Northstar Bioproducts, East Falmouth, MA) Guinea pig anti-mouse neurocan was a kind gift from Dr. Dieter Zimmermann (Zurich, Switzerland). Chondroitinase ABC was obtained from Associates of Cape Cod (Northstar Bioproducts, East Falmouth, MA).
Cell culture
All animal care and procedures were approved by the Institutional Animal Care and Use Committee at Uniformed Services University and performed in accordance with their guidelines. Mixed cortical glial cultures were prepared from neonatal (1–3 day old) C57BL/6 mouse brain according to a published protocol (Armstrong 1998). Briefly, the cerebral hemispheres were dissected out, their meninges removed, and cortices collected in MEM-HEPES. They were minced with sterile scalpel blades and incubated with papain (20 units/ml), DNAse (20 μg/ml), EDTA (0.15 mg/ml) and L-cysteine (0.16 mg/ml) for 30 minutes at 37°C. The enzyme solution was aspirated after quick centrifugation and the tissue was further incubated in the same solution for another 30 minutes before addition of serum-containing medium stopped digestion. The disrupted tissue was triturated, the cells centrifuged and the resulting pellet was suspended in Dulbecco’s modified Eagle medium containing heat inactivated fetal bovine serum (FBS, 10%), and gentamycin (25 mg/ml). Cells were seeded in 75 cm2 flasks and incubated with 5% CO2 in air. When confluent (10 – 14 days later), astrocytes were purified by vigorous shaking overnight (190 rpm, 37°C) after a one hour pre shake to remove microglia. The adherent cells were plated onto 6-well plates (750,000 cells/well) in serum-containing medium and two to three days later they were infected with lentiviral stocks. Astrocyte purity was assessed at over 95% purity by counting the percentage of cells that expressed glial fibrillary acidic protein. Approximately 20 h after infection, the medium was changed. Three days after infection, cells were washed once and incubated overnight in serum free medium. On the fourth day, they were treated with TGF-β (10 ng/ml) for a further three days.
C8D1A cells were grown and maintained in Dulbecco’s modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum, glutamax (1%) at 37°C with 5% CO2.
Lentivirus preparation and infection of astrocytes
Lentiviral shRNA sequences specifically targeting murine Smad2 or Smad3 in pLKO.1 vector were obtained from Open Biosystems (Huntsville, AL); the control vector (pLKO.1) was from Sigma-Aldrich (St Louis, MO). Recombinant lentivirus was generated from HEK-293T cells transfected with ViraPower™ Packaging Mix (Invitrogen) and the target plasmid using Lipofectamine 2000 (Invitrogen) according to manufacturer’s recommendations. Virus-containing medium was collected and filtered through 0.45 μm filters before concentrating 12 fold by ultracentrifugation at 50 000 g (2h, 4°C). The viral pellet was resuspended in Opti-MEM, aliquoted and stored at −80°C. Each viral preparation was tested both on C8D1A (mouse astrocyte cell line) cells and primary astrocytes for specific knockdown of target gene expression by western blotting.
Preparation of conditioned media and cell lysates
Astrocyte conditioned medium was placed into a tube containing complete protease inhibitors (Cocktail Set V, Calbiochem, La Jolla, CA). The conditioned medium was then centrifuged at 2000 g for 10 min to remove cell debris and concentrated to one-tenth of its original volume using an Amicon Ultra-15 NMWL100K centrifugal filter device (Millipore). The protein content was measured using BCA protein assay reagent (Pierce). Chondroitinase ABC digestion was performed on one-half of the concentrated sample using 0.01 U/ml conditioned medium for 3hr at 37°C. The digestion was stopped by boiling the sample in gel loading buffer.
For the analysis of Smad expression, cells (astrocytes or C8D1A) were washed twice in ice cold PBS before lysing in RIPA buffer containing protease inhibitors for 30 minutes at 4°C. Lysates were cleared of cellular debris by centrifugation at 16000g for 20 minutes at 4°C . All protein samples were stored at −80°C until needed.
Western Blotting
Equal amounts of protein from conditioned medium or cell lysate were loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide gels. Gels were run and transferred to nitrocellulose membranes using published protocols (Asher et al. 2000, Tang et al. 2003). Nitrocellulose membranes were probed with various primary antisera before probing with the appropriate HRP-linked secondary antisera and developed with Supersignal ECL reagent (Pierce). Chemiluminescence signals were detected using a Fuji LAS-3000 image acquisition system equipped with a cooled CCD camera. Immunoreactivity for each protein was quantified using Fuji Image analysis software (Multi Gauge). Blots of cell lysate were reprobed with β-actin as a loading control.
Quantitative RT-PCR
Total RNA was extracted from astrocytes using the RNeasy mini kit (Qiagen). Genomic DNA was removed by RNase free DNAse I treatment according to the manufacturer’s protocol. RNA was subsequently reverse transcribed using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) and quantitative PCR (QPCR) was performed on MyiQ Real Time PCR Detection System (Bio-Rad, Hercules, CA) using SYBR Green master mix (Applied Biosystems). Primers specific to mouse sequences were designed using Primer Express software (Applied Biosystems) to incorporate intron/exon boundaries. The primers employed for QPCR are listed in Table 1. Amplicon specificity was confirmed by melting curve analysis. Serial dilution of mouse brain cDNA was used to obtain standard curves for various primers. Expression of all genes was normalized to the levels of expression of the internal reference gene cyclophilin or GAPDH. To ensure no contamination or amplification of DNA we included controls without template, or with no reverse transcriptase within each QPCR run. Duplicate QPCR replicates were run on triplicate biological samples. Quantification of relative changes in mRNA levels between control and treated samples was calculated using the delta delta threshold cycle (ΔΔCt) method. ΔCt was calculated by subtracting the Ct values for the target gene from the Ct values for the internal reference gene expression. ΔΔCt was calculated by subtracting the ΔCt for TGF- β samples from the ΔCt for untreated cells in the same experiment. Fold change was then calculated as 2−ΔΔCt. Data from three to five independent experiments was used for analysis of relative gene expression.
Table 1.
Primers used for real time QPCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Neurocan | GAGAGAGATTGCAGGCGCCGAGCTG | CTCGGTGCTGGGAGAATCCTTCATC |
| Phosphacan | TTGGCTCCTACTATCAACATCCTCC | AGCTCATCCCTCTCAGCAGCTGAAG |
| Brevican | CTCGATTTCGTTAGGGGACA | ATTTCCACCTGGTTTTGCTG |
| XTI | TCAAGCCTCAGGACTTCCATCG | CTTTCGGGCAAAGAAGGTGGGC |
| XTII | GTCTGGATTGACCCCACCTA | AGGCTCAGTGGAGGCTTGTA |
| CSGalNacT1 | TCTGTTCAGTCAGTATAACCCCG | CCAAAATCCCGTTTCCTTCTTTATG |
| ChyS-1 | GAGAGAACTTCTGCATGGGC | CAATGTGTGGTGCCATTCTCC |
| C4ST1 | GAAGAGGCTCATGATGGTCC | GAGAGAGTAGACCGTCTGCC |
| C6ST1 | GGATTCCACCTTTTCCCATCTG | TGCCCTGCTGGTTGAAGAAC |
Axonal outgrowth assay
Astrocyte cultures were grown to confluence, infected with lentiviral particles, and subsequently treated with TGF-β. After three days, the media was carefully replaced with Neurobasal medium containing B27 supplement, 25 mM KCl, 2 mM Glutamine and penicillin/streptomycin (1%). Cerebellar granule neurons (CGN) were prepared from 5–7 day old C57/BL6 mice as described previously (Wang et al. 2008) and plated onto the confluent monolayer of astrocytes at a density of 3X104 cells/well. Approximately 20 h later, cells were fixed with 4% paraformaldehyde; neurons were stained with anti-βIII-tubulin (T8860, Sigma; 1:200) and astrocytes with anti-GFAP (Z0334, DAKO; 1:500). Cover slips were washed in PBS and incubated in appropriate secondary antibodies conjugated either to Alexa-488 or Alexa-568 (1:200; Invitrogen). Fluorescence images (at least 20 images per condition) were acquired on an Olympus BX61 microscope with attached CCD camera. Relative axonal length was measured by stereological assessment as previously described (Ronn et al. 2000; Wang et al. 2008) using ImageJ (NIH). Briefly, a counting grid with six lines was superimposed onto each image of CGN-astrocyte coculture. The ratio of the number of neurite intersection points with a line, to the number of cell bodies within the grid provided the relative axonal length for each condition.
Statistical Analysis
All data are expressed as mean ± s.e.m. Data were analyzed using InStat software (Graphpad) either by unpaired students t test, to compare two samples, or by one way ANOVA with bonferroni post test when comparing three or more samples. A P value less than 0.05 was considered statistically significant. Analysis used and statistical significance for each data set are provided in the figure legends.
Results
Lentiviral ShRNA infection efficiently and specifically knocked down either Smad2 or Smad3
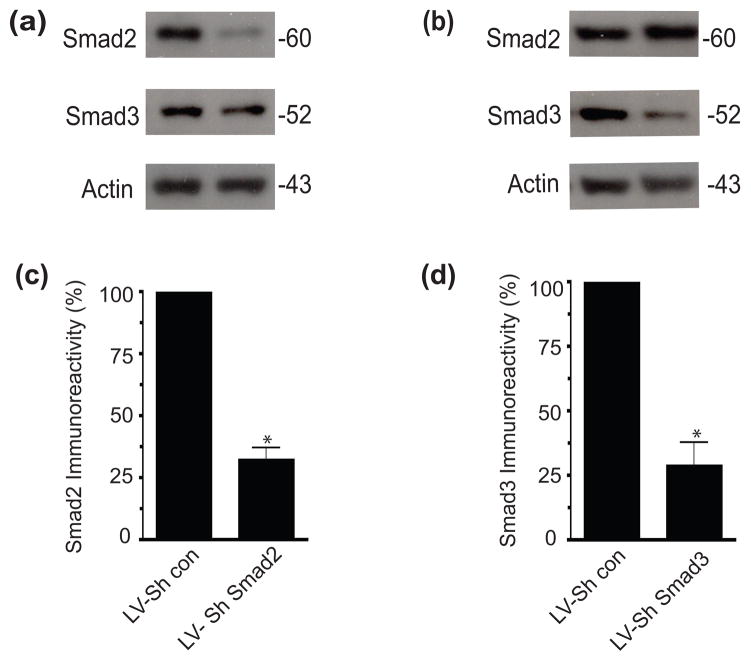
To understand more precisely the different requirements for Smad2 and Smad3 in TGF-β mediated induction of CSPGs, we utilized lentiviruses expressing short hairpin RNA (LV-ShRNA) that targeted specific Smad proteins (TRC, Open Biosystems). Five different lentiviruses for either Smad2 or Smad3 were tested for efficacy and specificity in the murine astrocyte cell line, C8D1A. The lentivirus with the strongest knockdown of Smad2 (LV-ShSmad2) reduced its expression to 32% of control, while the strongest knockdown of Smad3 (LV-ShSmad3) reduced Smad3 expression to 28% of control cells (Figure 1). Similar results were obtained when these constructs were transduced in astrocyte cultures (data not shown). The knockdown was Smad specific as LV-ShSmad2 did not alter expression of Smad3, nor did LV-ShSmad3 alter Smad2 levels (Figure 1). Lentivirus containing the control vector (pLKO.1) did not alter expression of either Smad.
Figure 1.
Lentivirus expressing ShRNA for Smad2 or Smad3 specifically knocks down Smad expression in C8D1A cells. Four days after lentiviral infection of C8D1A cells, cells were lysed and Smad expression analyzed. Representative western blots showing (a) specific Smad2 knockdown and (b) specific Smad3 knockdown after lentiviral infection with Smad-specific shRNA. Quantification (c &d) of Smad immunoreactivity normalized to actin (mean ±SEM, n=3). (*p<0.05: t-test.).
Knockdown of Smad2 or Smad3 differentially regulate CSPG core proteins
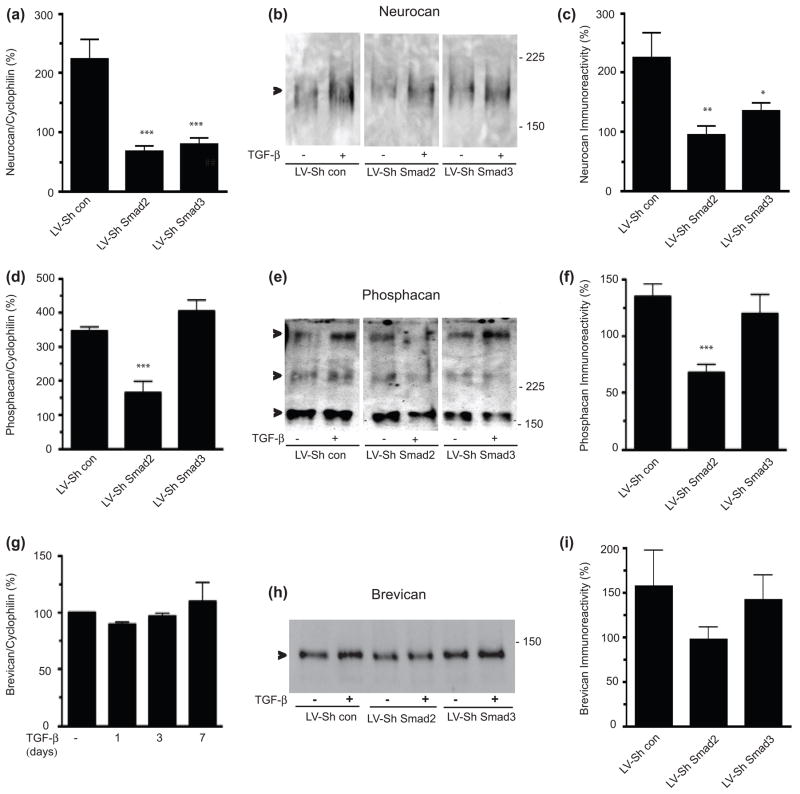
We next sought to determine whether Smad proteins were necessary for TGF- β mediated induction of CSPG core protein expression in primary astrocyte cultures. Transduction of cultures with LV-ShRNA for either Smad2 or Smad3 blocked TGF-β mediated induction of neurocan mRNA and protein (Figure 2a–c). However, transduction of cells with LV-ShSmad2, but not LV-ShSmad3, led to a decrease of TGF-β mediated upregulation of phosphacan mRNA and protein (Figure 2d–f). TGF-β did not induce brevican mRNA in our hands (Figure 2g), but brevican protein was induced 157 ± 41 % by TGF-β. Infection with either lentiviral ShSmad vector did not significantly reduce the TGF–β induction of brevican, although there was a trend towards reduction with LV-ShSmad2 infection (Figure 2h,i). Thus, TGF–β mediated induction of core CSPG gene expression is Smad dependent, although with differential dependence on Smad2 and/or Smad3.
Figure 2.
Differential effects of Smad2 or Smad3 on the TFG-β-mediated increase in CSPG core proteins. Quantification of neurocan (a), phosphacan (d) and brevican (g) mRNA expression by QPCR in astrocytes treated with TGF-β (10 ng/ml) for 3 days in the presence or absence of LV-ShSmad2 or LVShSmad3. mRNA expression was normalized to cyclophillin and presented as percent expression in TGF-β treated/untreated samples (mean ± s.e.m., n = 3–5 independent experiments), and compared to the LV-control. Representative Western blot showing neurocan (b), phosphacan (e) and brevican (h) expression in chABC-treated conditioned media harvested from astrocytes treated with TGF-β for 3 days in the presence of LV-ShSmad2 or LVShSmad3. Immunoreactivity was quantified using LAS3000 imaging system (Fuji Film). (c) Quantification of neurocan (c), phosphacan (f) and brevican (i) expression by western blot (mean ± s.e.m., n=3–7). Data are presented as percentage of expression in TGF-β treated vs untreated samples and compared to the LV-control. (*p<0.05 **p< 0.01, ***p<0.001; ANOVA with bonferroni post test).
Effects of LV-ShRNA for Smad2 or Smad3 on enzymes involved in GAG synthesis and modification
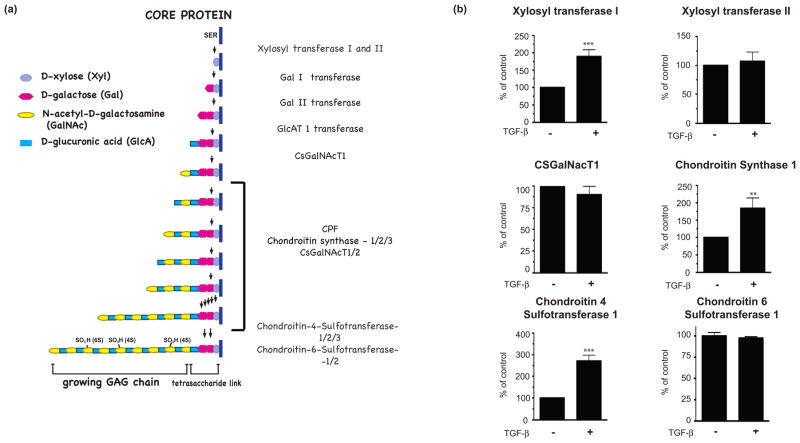
The CSPG GAG chains contribute to the inhibition of axonal growth (Powell et al. 1997, Wang et al. 2008). Therefore, understanding the mechanisms through which GAG chains are synthesized may enable interventions to reduce their synthesis after injury. Biosynthesis of the GAG chain is performed by many different enzymes (Figure 3a). Xylosl transferase (XT) mediates the rate-limiting step of GAG chain synthesis: adding the initial xylose residue to a serine residue of the core protein (Gotting et al. 2000). The tetrasaccharide linker, shared by all sulfated proteoglycans, is completed by the addition of two galactose molecules and one glucuronic acid (Prydz & Dalen 2000). The bulk of the GAG chain is composed of repeating disaccharides of N-acetyl galactosamine and glucuronic acid, which may be added by any one of six separate enzymes (Prydz & Dalen 2000, Zhang 2010). Finally, the GAG chain is sulfated at different positions within the disaccharide units by additional enzymes (Kusche-Gullberg & Kjellen 2003). We analyzed several important enzymes involved in GAG chain synthesis, either because they had previously been shown to be induced by TGF-β (C4ST-1, xylosyl transferase-1 (XT-1)) or because they play a critical role in GAG chain synthesis (xylosyl transferase 2, CsGalNacT-1, chondroitin synthase 1 (ChyS-1), chondroitin 6 sulfotransferase -1) (Wang et al. 2008, Gris et al. 2007, Yin et al. 2009). We found that TGF-β treatment of primary astrocytes for 3 days induced mRNA expression of XT-1, Chys-1 and C4ST-1 but not XT-2, C6ST-1 or CsGalNacT-1 (Figure 3b).
Figure 3.
(a) Schematic showing the synthesis of the glycosaminoglycan chains of CSPGs. (b) The effect of TGF-β treatment on mRNA expression of XT-1, XT-2, CSGalNacT-1, ChSy-1, C4ST-1 and C6ST-1. RNA was prepared from astrocytes after TGF-β (10 ng/ml) treatment for three days and quantitative RT-PCR was performed, mRNA expression was normalized to cyclophillin or GAPDH and presented as percent expression in TGF-β treated vs untreated samples (mean ± s.e.m., n = 3–5 independent experiments). (**p< 0.01; ***p< 0.001; t-test).
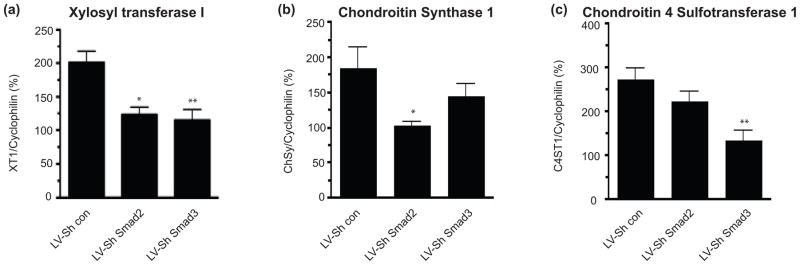
The TGF-β induced increase in XT-I mRNA was blocked by either LV-ShSmad2 or LV-ShSmad3 (Figure 4a). However, the TGF-β mediated increase in ChyS-1, an enzyme that mediates the polymerization of the GAG chain (Kitagawa et al. 2003, Kitagawa et al. 2001), was reduced by LVShSmad2 but not by LV-ShSmad3 (Figure 4b). C4ST-1 adds a sulfate group to the C4 position of N-acetyl-D-galactosamine residuesin the GAG chain (Habuchi 2000). 4-sulfated GAGs play an important role in axonal growth regulation and are upregulated after injury to CNS (Wang et al. 2008). Transduction of astrocytes with LV-ShSmad3, but not LV-ShSmad2, significantly blocked TGF-β mediated up-regulation C4ST-1 (Figure 4c). Our data therefore show that, similar to CSPG core proteins, TGF-β induction of CSPG GAG chain synthetic enzymes is also Smad dependent but there is a differential requirement for Smad2 and/or Smad3. Thus, although TGF-β causes a coordinate upregulation in both CSPG core proteins and synthetic enzymes, leading overall to a large increase in deposition of CSPGs after injury, there does not appear to be a single common mechanism of upregulation of these different proteins.
Figure 4.
Effects of Smad specific knockdown on TGF-β induced increase in XT-1, ChSy-1 and C4ST-1 expression. Quantification of XT-1 (a), ChSy-1 (b) and C4ST-1 (c) mRNA expression by QPCR in astrocytes treated with TGF-β (10 ng/ml) for 3 days in the presence or absence of LV-ShSmad2 or LVShSmad3. mRNA expression was normalized to cyclophillin and presented as percent expression in TGF-β treated vs untreated samples (mean ± s.e.m., n = 3–5 independent experiments). (*p<0.05 **p< 0.01; ANOVA with bonferroni post test).
Knock down of Smad3 blocks TGF-β mediated inhibition of neurite outgrowth
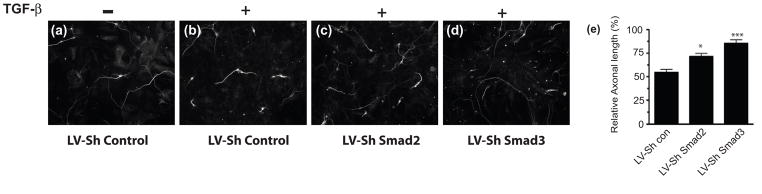
Astrocytes treated with TGF-β produce a less permissive matrix for neurite growth than untreated astrocytes (Wang et al. 2008). This difference can be assayed by determining the length of neurites on monolayers of astrocytes that had been pre-treated with TGF-β. In order to determine the functional significance of Smad2 and Smad3 knockdown, we examined the ability of neurons to extend axons on an astrocyte monolayer after different treatments. Astrocytes were first infected with either LV-control ShRNA, LV-ShSmad2 or LV-ShSmad3 and then treated with TGF-β for three days. We confirmed the efficiency of knock down of Smad2 and Smad3 (data not shown). Cerebellar granule neurons were then plated on the astrocytes. Cultures were fixed and stained 20 hours later. As previously shown (Wang et al. 2008), TGF-β treatment of astrocyte cultures resulted in a dramatic decrease in the length of axons compared to untreated cultures (Figure 5a,b). Transduction with either LV-ShSmad2 or LV-ShSmad3 reduced the inhibition produced by TGF-β, with LV-ShSmad3 infected astrocytes being the most permissive for axonal growth (Figure 5). Indeed, with Smad3 knocked down in astrocytes, TGF–β treatment did not reduce the ability of astrocytes to support neurite outgrowth. These results show that TGF-β mediated induction of inhibitory ECM utilizes the Smad signaling pathway. This induction may be blocked by knockdown of Smad2 or Smad3, but that knockdown of Smad3 has a greater effect on the inhibitory nature of the CSPG containing ECM.
Figure 5.
Smad-specific knockdown relieves the TGF-β mediated inhibition of neurite growth. Astrocytes were infected with (a;b) LV-Sh control (c) LV-ShSmad2; or (d) LV-ShSmad3 for three days before treatment with (b, c, d) TGF-β (10ng/ml) for a further three days. The media was changed and cerebellar granule neurons were plated onto the astrocyte monolayers. 20 hours later, cells were fixed, neurons visualized by immunohistochemistry and their relative axonal length was measured. Scale bar; 50 μM. (e) Graph of the effects of Smad knockdown on axonal growth (mean ± s.e m., of three independent experiments). Relative axonal length in each condition is compared to that in the untreated LV-Sh Control. Knockdown of specific Smad proteins significantly relieves the TGF-β mediated inhibition of axonal growth (*p< 0.05; ***p<0.001; ANOVA with bonferroni post test).
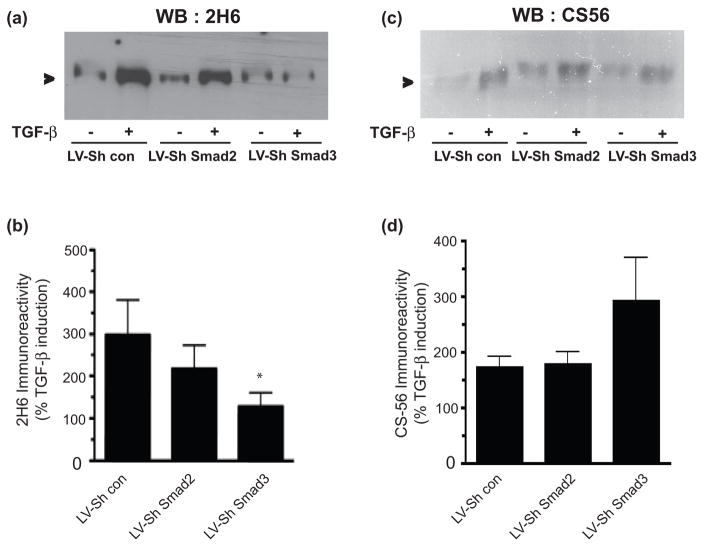
As our functional assay pointed to the importance of Smad3 in mediating the inhibitory nature of the astrocyte secreted ECM, we reexamined our results to determine which TGF-β induced genes/proteins were dependent on Smad3 but not on Smad2. Surprisingly, we found only one gene that had this Smad3- specific dependence: C4ST-1 (Figure 4c). To determine whether reduction in expression of C4ST-1 led to a reduction in the amount of 4-sulfated CSPGs, we utilized a 4-sulfation specific antiserum, 2H6 (Yamamoto et al. 1995) to examine conditioned media harvested from TGF-β treated astrocyte cultures infected with the different lentiviral constructs. In astrocytes with Smad3 silenced, there was minimal TGF-β mediated induction of the epitope recognized by 2H6, in comparison with astrocytes infected either with the control virus or the LV-ShSmad2 virus (Figure 6a,b). Thus, knockdown of Smad3 prevents both TGF-β mediated induction of C4ST-1 and 4-sulfated proteoglycans (Figures 4 and 6). In contrast, knock down of either Smad2 or Smad3 did not change TGF-β induced increases in immunoreactivity for the CSPG specific antiserum CS56 (Figure 6c,d). The exact epitope recognized by CS56 is not known, but it is commonly used as a pan-CSPG GAG chain antiserum, and is thought to recognize a disaccharide repeat that contains both 4S and 6S motifs (Ito et al. 2005). Thus, knockdown of Smad3 leads to a reduction in the amount of 4-sulfated GAG chains on proteoglycans produced by astrocytes, and this correlates with a more permissive astrocyte ECM for neuronal growth (Figure 5), despite no reduction in overall CSPG GAG chains as determined by CS56 immunoreactivity (Figure 6c,d). We therefore conclude that TGF-β mediated signaling through Smad3 has a critical role in determination of CSPG bioactivity, through its regulation of C4ST-1 and the amount of 4-sulfated proteoglycans.
Figure 6.
Smad3 knockdown inhibits the amount of TGF-β induced 4-sulfation in astrocytes. Representative western blot showing 2H6 (a), CS56 (b) expression in conditioned media harvested from astrocytes treated with TGF-β (10ng/ml) for 3 days in the presence of LV-ShSmad2 or LV-ShSmad3. Quantification of westerns for (b) 2H6 and (d) CS56 immunoreactivity (mean ± s.e.m., n=3–4). Data are presented as percentage of expression in TGF-β treated vs untreated samples and compared to the LV-Sh Control. (*p<0.05; ANOVA with bonferroni post test).
Discussion
TGF-β coordinately regulates transcription and secretion of many extracellular components of the glial scar after traumatic injury in the CNS. Many of the actions of TGF-β are mediated through the canonical TGF-β-Smad2/3 signaling pathway (reviewed by Ross & Hill 2008; Moustakas & Heldin 2009). By selectively targeting the individual Smads, we have shown that CSPG deposition in astrocytes in response to TGF-β is regulated through this pathway. Surprisingly, the genes involved in core protein synthesis or glycosylation are differentially regulated by Smad2 and/or Smad3 to modify the production and composition of the astrocyte extracellular matrix and thus its biological activity.
Deposition of CSPGs after injury contributes significantly to the inhibition of axonal regeneration by the glial scar, and blockade of TGF-β in vivo can reduce glial scar formation and CSPG deposition (Logan et al. 1994, Logan et al. 1999). In vitro, TGF-β models these actions by producing a reactive phenotype in astrocytes, including increasing the production of an inhibitory extracellular matrix comprised largely of CSPGs (Asher et al. 2000, Hamel et al. 2005, Gris et al. 2007, Wang et al. 2008). Our data confirm the induction of C4ST-1 (Figure 4c) and neurocan (Figure 2a–c) in astrocytes by TGF-β. In addition, we now show that XT-1 and ChyS-1 are also increased in response to TGF-β. Neurocan is a core protein that carries inhibitory GAG chains. XT-1 catalyzes the first step in GAG chain synthesis, while ChyS-1 regulates the extension of the disaccharide chain. The consistent result of these changes would be an increase in production of GAG chains. It has previously been shown that targeting XT-1 with a DNA enzyme can increase neurite outgrowth (Hurtado et al. 2008), and reduction in the synthesis of chondroitin polymerizing factor, an enzyme with a similar function as ChyS-1, also led to a more permissive substrate for neurite growth (Laabs et al. 2007). On the other hand, previous work did not show a significant induction of phosphacan in response to TGF-β (Dobbertin et al. 2003). This difference may be due to the source of astrocytes or the antibodies used in the previous study.
While neurite outgrowth improved after knocknown of either Smad2 or Smad3, Smad3 knockdown had a greater effect than did knockdown of Smad2. This is likely due to the selective actions of the respective Smad downstream of TGF-β in altering the production of particular CSPG components. Thus, the increase in neurocan core protein production was equally affected by either knockdown, while Smad2 knockdown selectively blunted the increase in phosphacan. Likewise, XT-1 induction was reduced equally. In contrast, C4ST-1 was the only gene that was dependent on Smad3 and not Smad2 for TGF-β mediated inhibition, while phosphacan and ChyS-1 were more potently targeted by Smad2. Phosphacan can be growth promotional to neurons culture, primarily via the DSD-1 carbohydrate epitope on the attached GAG chains (Faissner et al, 1994). A reduction in phosphacan expression with Smad2 knockdown may serve to augment, rather than antagonize, the growth inhibitory effects of TGF-β, which might explain the difference between knockdown of the two Smads.
The selective actions of Smad2 and Smad3 in regulating phenotypic changes in response to TGF-β is not restricted to astrocytes. Thus, in a keratinocyte cell line the TGF-β induced increase in transglutaminase-2 expression was reduced by knockdown of Smad3 but not Smad2 (Kretschmer et al. 2003). In mouse embryonic fibroblasts derived from Smad2 or Smad3 null mice, TGF-β induced changes were differentially dependent on Smad2 and/or Smad3: TGF-β autoinduction and induction of Smad7 and c-fos were Smad3 dependent, whereas TGF-β-induced expression of MMP-2 was dependent on Smad2 and not Smad3 (Piek et al. 2001). Interestingly, the absence of Smad3, but not Smad2, led to an inability of TGF-β to induce transcription through the standard TGF-β reporters 3TP-lux, or (smad binding element)4-lux, whereas the absence of Smad2 resulted in a lack of TGF-β induced expression of the activin responsive element-luciferase reporter (Piek et al. 2001). These differential affinities of Smad2 and Smad3 for specific DNA binding sites, leads to their non-redundant functions, and has the advantage of selectively altering gene expression in situations of TGF-β activation or inhibition.
Knockdown of Smads reduced the level of 4-sulfated GAG chains as detected by the 2H6 antibody in Western blotting. However, we did not find a similar reduction in the GAG chains as detected by the CS-56 antibody. The exact epitope recognized by CS-56 is a mixture of sulfated disaccharide residues (as discussed by Ito et al., (2005)), while the 2H6 epitope is found on the 4-sulfated Cs-A unit (GlcUA-GalNAc(4S)) (Yamamoto et al. 1995). The large reduction in C4ST-1 and 4-sulfation of GAG chains by Smad3 knockdown is thus consistent with our proposal for a major role of a specific pattern of GAG chain sulfation, especially 4-sulfation, rather than overall levels of GAGs, in mediating the inhibitory actions of CSPGs. The lack of correlation between the amount of epitope recognized by CS-56 and the growth ability of neurites on the same CSPGs indicate the limitations of utilizing the CS-56 antisera alone, for CSPG GAG chain identification. The inability of Smad2 or Smad3 knockdown to alter the amount of CS-56 epitope detected after TGF–β induction is surprising as a number of CSPGs core proteins and synthetic enzymes are reduced in these astrocytes. It is possible that the total amount of GAG sidechains on CSPGs are not significantly reduced due to the presence of other CSPGs that may be unaffected by TGF–β treatment and that contain high amounts of GAG chain, for example aggrecan.
In summary, we have shown that Smad proteins are necessary for TGF-β-mediated induction of CSPG core proteins and modifying enzyme genes in astrocytes. Smad3-mediated induction of 4-sulfation of CSPG GAG chains seems particularly important to the ability of CSPGs to inhibit neurite growth on their astrocyte matrix. Ultimately, the deposition of CSPGs after injury relies on multiple signals to several cell types. Identifying the critical epitope or mediator of the inhibitory function of CSPGs will allow better targeting of mechanisms to prevent its synthesis, or its interactions with neurons. Delineating the mechanism through which TGF-β induces this mediator will also allow us to intervene more specifically to disrupt this induction, and prevent deposition of specific molecules after injury.
Acknowledgments
This work was supported by a grant from the Maryland Spinal Cord Injury Research Board (AJS) and the Intramural Research Program of the National Heart, Lung and Blood Institute (P.Y., Y.K. and H.M.G). We thank Dr. Dours Zimmermann (Zurich, Switzerland) for the generous gift of neurocan antibody. We are grateful to past and present members of the Symes and Geller laboratories for their helpful comments and suggestions. The opinions and assertions contained herein are the private opinions of the authors and are not to be construed as reflecting the views of the Uniformed Services University of the Health Sciences or the US Department of Defense. The authors report no conflicts of interest.
References
- Armstrong RC. Isolation and characterization of immature oligodendrocyte lineage cells. Methods. 1998;16:282–292. doi: 10.1006/meth.1998.0685. [DOI] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Fidler PS, et al. Neurocan is upregulated in injured brain and in cytokine-treated astrocytes. J Neurosci. 2000;20:2427–2438. doi: 10.1523/JNEUROSCI.20-07-02427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher RA, Morgenstern DA, Shearer MC, Adcock KH, Pesheva P, Fawcett JW. Versican is upregulated in CNS injury and is a product of oligodendrocyte lineage cells. J Neurosci. 2002;22:2225–2236. doi: 10.1523/JNEUROSCI.22-06-02225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- Dijke Pt, Hill CS. New insights into TGF-[beta]-Smad signalling. Trends in Biochemical Sciences. 2004;29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Dobbertin A, Rhodes KE, Garwood J, Properzi F, Heck N, Rogers JH, Fawcett JW, Faissner A. Regulation of RPTPbeta/phosphacan expression and glycosaminoglycan epitopes in injured brain and cytokine-treated glia. Mol Cell Neurosci. 2003;24:951–971. doi: 10.1016/s1044-7431(03)00257-4. [DOI] [PubMed] [Google Scholar]
- Faissner A, Clement A, Lochter A, Streit A, Mandl C, Schachner M. Isolation of a neural chondroitin sulfate proteoglycan with neurite outgrowth promoting properties. J Cell Biol. 1994;126:783–799. doi: 10.1083/jcb.126.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch MT, Silver J. CNS injury, glial scars, and inflammation: Inhibitory extracellular matrices and regeneration failure. Exp Neurol. 2008;209:294–301. doi: 10.1016/j.expneurol.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtrey CM, Fawcett JW. The role of chondroitin sulfate proteoglycans in regeneration and plasticity in the central nervous system. Brain Res Rev. 2007;54:1–18. doi: 10.1016/j.brainresrev.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Gotting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K. Molecular cloning and expression of human UDP-d-Xylose:proteoglycan core protein beta-d-xylosyltransferase and its first isoform XT-II. J Mol Biol. 2000;304:517–528. doi: 10.1006/jmbi.2000.4261. [DOI] [PubMed] [Google Scholar]
- Gris P, Tighe A, Levin D, Sharma R, Brown A. Transcriptional regulation of scar gene expression in primary astrocytes. Glia. 2007;55:1145–1155. doi: 10.1002/glia.20537. [DOI] [PubMed] [Google Scholar]
- Habuchi O. Diversity and functions of glycosaminoglycan sulfotransferases. Biochim Biophys Acta. 2000;1474:115–127. doi: 10.1016/s0304-4165(00)00016-7. [DOI] [PubMed] [Google Scholar]
- Hamel MG, Mayer J, Gottschall PE. Altered production and proteolytic processing of brevican by transforming growth factor beta in cultured astrocytes. J Neurochem. 2005;93:1533–1541. doi: 10.1111/j.1471-4159.2005.03144.x. [DOI] [PubMed] [Google Scholar]
- Hurtado A, Podinin H, Oudega M, Grimpe B. Deoxyribozyme-mediated knockdown of xylosyltransferase-1 mRNA promotes axon growth in the adult rat spinal cord. Brain. 2008;131:2596–2605. doi: 10.1093/brain/awn206. [DOI] [PubMed] [Google Scholar]
- Ito Y, Hikino M, Yajima Y, Mikami T, Sirko S, von Holst A, Faissner A, Fukui S, Sugahara K. Structural characterization of the epitopes of the monoclonal antibodies 473HD, CS-56, and MO-225 specific for chondroitin sulfate D-type using the oligosaccharide library. Glycobiology. 2005;15:593–603. doi: 10.1093/glycob/cwi036. [DOI] [PubMed] [Google Scholar]
- Kang JS, Liu C, Derynck R. New regulatory mechanisms of TGF-[beta] receptor function. Trends in Cell Biology. 2009;19:385–394. doi: 10.1016/j.tcb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Izumikawa T, Uyama T, Sugahara K. Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. J Biol Chem. 2003;278:23666–23671. doi: 10.1074/jbc.M302493200. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Uyama T, Sugahara K. Molecular cloning and expression of a human chondroitin synthase. J Biol Chem. 2001;276:38721–38726. doi: 10.1074/jbc.M106871200. [DOI] [PubMed] [Google Scholar]
- Kretschmer A, Moepert K, Dames S, Sternberger M, Kaufmann J, Klippel A. Differential regulation of TGF–β signaling through Smad2, Smad3 and Smad4. Oncogene. 2003;22:6748–6763. doi: 10.1038/sj.onc.1206791. [DOI] [PubMed] [Google Scholar]
- Kusche-Gullberg M, Kjellen L. Sulfotransferases in glycosaminoglycan biosynthesis. Curr Opin Struct Biol. 2003;13:605–611. doi: 10.1016/j.sbi.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Laabs TL, Wang H, Katagiri Y, McCann T, Fawcett JW, Geller HM. Inhibiting glycosaminoglycan chain polymerization decreases the inhibitory activity of astrocyte-derived chondroitin sulfate proteoglycans. J Neurosci. 2007;27:14494–14501. doi: 10.1523/JNEUROSCI.2807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagord C, Berry M, Logan A. Expression of TGFbeta2 but not TGFbeta1 correlates with the deposition of scar tissue in the lesioned spinal cord. Mol Cell Neurosci. 2002;20:69–92. doi: 10.1006/mcne.2002.1121. [DOI] [PubMed] [Google Scholar]
- Lee H, McKeon RJ, Bellamkonda RV. Sustained delivery of thermostabilized chABC enhances axonal sprouting and functional recovery after spinal cord injury. Proc Natl Acad Sci U S A. 2010;107:3340–3345. doi: 10.1073/pnas.0905437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan A, Berry M. Transforming growth factor beta and CNS scarring. In: Berry M, Logan A, editors. CNS injuries: Cellular responses and pharmacological strategies. CRC Press; London: 1999. pp. 151–168. [Google Scholar]
- Logan A, Berry M, Gonzalez AM, Frautschy SA, Sporn MB, Baird A. Effects of transforming growth factor beta 1 on scar production in the injured central nervous system of the rat. Eur J Neurosci. 1994;6:355–363. doi: 10.1111/j.1460-9568.1994.tb00278.x. [DOI] [PubMed] [Google Scholar]
- Logan A, Frautschy SA, Gonzalez AM, Sporn MB, Baird A. Enhanced expression of transforming growth factor beta 1 in the rat brain after a localized cerebral injury. Brain Res. 1992;587:216–225. doi: 10.1016/0006-8993(92)91000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan A, Green J, Hunter A, Jackson R, Berry M. Inhibition of glial scarring in the injured rat brain by a recombinant human monoclonal antibody to transforming growth factor–β2. Eur J Neurosci. 1999;11:2367–2374. doi: 10.1046/j.1460-9568.1999.00654.x. [DOI] [PubMed] [Google Scholar]
- Makwana M, Jones LL, Cuthill D, et al. Endogenous transforming growth factor beta 1 suppresses inflammation and promotes survival in adult CNS. J Neurosci. 2007;27:11201–11213. doi: 10.1523/JNEUROSCI.2255-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mctigue D. Localization of Transforming Growth Factor-β1 and Receptor mRNA after Experimental Spinal Cord Injury. Experimental Neurology. 2000;163:220–230. doi: 10.1006/exnr.2000.7372. [DOI] [PubMed] [Google Scholar]
- Moon LD, Asher RA, Rhodes KE, Fawcett JW. Regeneration of CNS axons back to their target following treatment of adult rat brain with chondroitinase ABC. Nat Neurosci. 2001;4:465–466. doi: 10.1038/87415. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Non-Smad TGF-{beta} signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- Nichols NR, Laping NJ, Day JR, Finch CE. Increases in transforming growth factor–β mRNA in hippocampus during response to entorhinal cortex lesions in intact and adrenalectomized rats. J Neurosci Res. 1991;28:134–139. doi: 10.1002/jnr.490280114. [DOI] [PubMed] [Google Scholar]
- Nomura M, Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- Piek E, Ju WJ, Heyer J, et al. Functional characterization of transforming growth factor beta signaling in Smad2- and Smad3-deficient fibroblasts. J Biol Chem. 2001;276:19945–19953. doi: 10.1074/jbc.M102382200. [DOI] [PubMed] [Google Scholar]
- Powell EM, Fawcett JW, Geller HM. Proteoglycans Provide Neurite Guidance at an Astrocyte Boundary. Mol Cell Neurosci. 1997;10:27–42. doi: 10.1006/mcne.1997.0629. [DOI] [PubMed] [Google Scholar]
- Prydz K, Dalen KT. Synthesis and sorting of proteoglycans. J Cell Sci. 2000;113(Pt 2):193–205. doi: 10.1242/jcs.113.2.193. [DOI] [PubMed] [Google Scholar]
- Rimaniol AC, Lekieffre D, Serrano A, Masson A, Benavides J, Zavala F. Biphasic transforming growth factor–β production flanking the pro-inflammatory cytokine response in cerebral trauma. Neuroreport. 1995;7:133–136. [PubMed] [Google Scholar]
- Ronn LC, Ralets I, Hartz BP, Bech M, Berezin A, Berezin V, Moller A, Bock E. A simple procedure for quantification of neurite outgrowth based on stereological principles. J Neurosci Methods. 2000;100:25–32. doi: 10.1016/s0165-0270(00)00228-4. [DOI] [PubMed] [Google Scholar]
- Ross S, Hill CS. How the Smads regulate transcription. The International Journal of Biochemistry & Cell Biology. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Sherman LS, Back SA. A 'GAG' reflex prevents repair of the damaged CNS. Trends Neurosci. 2008;31:44–52. doi: 10.1016/j.tins.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Smith GM, Strunz C. Growth factor and cytokine regulation of chondroitin sulfate proteoglycans by astrocytes. Glia. 2005;52:209–218. doi: 10.1002/glia.20236. [DOI] [PubMed] [Google Scholar]
- Tang X, Davies JE, Davies SJ. Changes in distribution, cell associations, and protein expression levels of NG2, neurocan, phosphacan, brevican, versican V2, and tenascin-C during acute to chronic maturation of spinal cord scar tissue. J Neurosci Res. 2003;71:427–444. doi: 10.1002/jnr.10523. [DOI] [PubMed] [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- Wang H, Katagiri Y, McCann TE, et al. Chondroitin-4-sulfation negatively regulates axonal guidance and growth. J Cell Sci. 2008;121:3083–3091. doi: 10.1242/jcs.032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Moges H, Bharucha Y, Symes A. Smad3 null mice display more rapid wound closure and reduced scar formation after a stab wound to the cerebral cortex. Exp Neurol. 2007;203:168–184. doi: 10.1016/j.expneurol.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Weinstein M, Yang X, Li C, Xu X, Gotay J, Deng CX. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor Smad2. Proc Natl Acad Sci U S A. 1998;95:9378–9383. doi: 10.1073/pnas.95.16.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Atoji Y, Oohira A, Suzuki Y. Immunohistochemical localization of chondroitin sulfate in the forestomach of the sheep. Eur J Histochem. 1995;39:265–272. [PubMed] [Google Scholar]
- Yin J, Sakamoto K, Zhang H, et al. Transforming growth factor–β1 upregulates keratan sulfate and chondroitin sulfate biosynthesis in microglias after brain injury. Brain Res. 2009;1263:10–22. doi: 10.1016/j.brainres.2009.01.042. [DOI] [PubMed] [Google Scholar]
- Zhang L. Glycosaminoglycan (GAG) biosynthesis and GAG-binding proteins. Prog Mol Biol Transl Sci. 2010;93:1–17. doi: 10.1016/S1877-1173(10)93001-9. [DOI] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF–β signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]