Abstract
PAX5 encodes a master regulator of B-cell development. It fuses to other genes associated with acute lymphoblastoid leukemia (ALL). These fusion products are potent dominant-negative (DN) inhibitors of wild-type PAX5 resulting in a blockade of B-cell differentiation. Here, we show that multimerization of PAX5 DNA-binding domain (DBD) is necessary and sufficient to cause extremely stable chromatin binding and DN-activity. ALL-associated PAX5-C20S results from fusion of the N-terminal region of PAX5 including its paired DBD, to the C-terminus of C20orf112, a protein of unknown function. We report that PAX5-C20S is a tetramer which interacts extraordinarily stably with chromatin as determined by fluorescence recovery after photobleaching (FRAP) in living cells. Tetramerization, stable chromatin-binding and DN-activity all require a putative five-turn amphipathic α-helix at the C-terminus of C20orf112, and does not require potential co-repressor binding peptides elsewhere in the sequence. In vitro, the monomeric PAX5 DBD and PAX5-C20S binds a PAX5-binding site with equal affinity when it is at the center of an oligonucleotide too short to bind to more than one PAX5 DBD. But PAX5-C20S binds the same sequence with tenfold higher affinity than the monomeric PAX5 DBD when it is in a long DNA molecule. We suggest that the increased affinity results from interactions of one or more of the additional DBDs with neighboring non-specific sites in a long DNA molecule, and that this can account for the increased stability of PAX5-C20S chromatin binding compared to wt PAX5, resulting in DN-activity by competition for binding to PAX5-target sites. Consistent with this model, the ALL-associated PAX5 fused to ETV6 or the multimerization domain of ETV6 SAM results in stable chromatin binding and DN-activity. In addition, PAX5 DBD fused to artificial dimerization, trimerization, and tetramerization domains result in parallel increases in the stability of chromatin binding and DN-activity. Our studies suggest that oncogenic fusion proteins that retain the DBD of the transcription factor and the multimerization sequence of the partner protein can act in a DN fashion by multimerizing and binding avidly to gene targets preventing the normal transcription factor from binding and inducing expression of its target genes. Inhibition of this multimeriztion may provide a novel therapeutic approach for cancers with this or similar fusion proteins.
Keywords: PAX5 fusions, DNA binding, multimerization stable
Introduction
Gene fusions caused by chromosomal translocations contribute to the pathogenesis of many human neoplasms (Edwards, 2010). Most rearrangements are one of two types: 1) “promoter swapping,” which juxtaposes proto-oncogenes to abnormal transcriptional regulatory elements, leading to aberrant timing and/or expression level of a normal protein, or 2) fusions of two genes leading to expression of a chimeric fusion protein with abnormal function (Gasparini et al., 2007). Most oncogenic chimeric genes in the second class involve tyrosine kinases or transcription factors (TFs) (Mitelman et al., 2007). For oncogenic TF fusions, the resulting chimeric protein usually retains the TF DNA-binding domain (DBD) and results in either enhanced or reduced expression of target genes of the wild-type (wt) TF, promoting proliferation or blocking differentiation (O’Neil and Look, 2007). In many cases, oncogenic TF-fusions exert dominant-negative (DN)-activity over the wt TF expressed at a similar level from an unrearranged allele in the same cell (Pabst and Mueller, 2007). In many of these examples, oncogenic TF-fusions may act as DNs because the fusion proteins contain repression domains that recruit co-repressors to target genes, inhibiting their expression despite co-binding of the wt protein to repeated binding sites in the same control regions (Grignani et al., 1998; Lin et al., 1998). However, in some cases, oncogenic DN-TFs are generated from fusions of DBDs to structural proteins (e.g. elastin (Bousquet et al., 2007) or a myosin (Liu et al., 1993)) that have oligomerization domains, but no known repression domains. How oligomerization contributes to aberrant transcriptional activity is incompletely understood (So and Cleary, 2004).
PAX5 encodes a TF essential for B-cell differentiation that activates a number of B-cell specific genes and represses several genes specific for other hematopoietic lineages (Cobaleda et al., 2007). We and others have cloned a number of PAX5-fusion genes from acute lymphoblastic leukemia (ALL) cells (Table S1), including the two most commonly studied here, PAX5-C20orf112 (An et al., 2008; Kawamata et al., 2008) and PAX5-ETV6 (Cazzaniga et al., 2001). In each case, the generated fusion proteins retain the N-terminal PAX5 DBD which is a paired domain that binds DNA as a monomer (Xu et al., 1995), and lack the C-terminal PAX5 activation domain (Dorfler and Busslinger, 1996). These fusion products are able to block the expression of PAX5 target genes and are associated with a block in B cell differentiation (Bousquet, M et al. 2006; Cobeleda C, 2007; Fazio et al, 2008; Kawamata et al, 2008). Here, we show that multimerization of the PAX5-DBD by the fusion partner is necessary and sufficient for DN-activity. Multimerization greatly increases the stability of the protein-chromatin interaction in living cells, probably through cooperative binding of the multimerized DBD to neighboring, non-specific low affinity DNA sites. The resulting high affinity binding of the fusion protein competes with wt PAX5 binding to target genes. Inhibiting multimerization of the DBD interferes with stable chromatin binding and the resulting DN activity. Consequently, this would likely be a useful therapeutic strategy for either ALL with PAX5-fusions or other cancers with similar fusion proteins.
Results
Structural determinants of DN-activity
Two types of fusion genes associated with ALL involve PAX5 and C20orf112, PAX5-C20S resulting in an in-frame fusion of PAX5 exon 5 to C20orf112 exon 8 (Fig. 1A), and PAX5-C20L generating an in-frame fusion of PAX5 exon 8 to C20orf112 exon 3 (An et al., 2008; Kawamata et al., 2008). We choose to focus on PAX5-C20S because it represents the smallest region of C20 still associated with development of ALL. Analysis of the C20orf112 sequence in PAX5-C20S (Fig. 1B) revealed three regions that may contribute to DN-activity: a putative binding motif for the CtBP co-repressor (Chinnadurai, 2002), a Ser-Thr-Pro rich region, and the last 35 amino acids (aa) predicted to adopt an α-helical secondary structure (http://bioinf.cs.ucl.ac.uk/psipred/). The CtBP motif and the putative α-helical domain are highly conserved in C20orf112 orthologs from insects to humans (Fig. S1). To determine which regions of C20S are responsible for PAX5-C20S DN-activity over wt PAX5, we performed transient transfection assays with a luciferase reporter with three PAX5-binding sites derived from the murine CD19 promoter (Dorfler and Busslinger, 1996). Experiments were performed with expression vectors for wt PAX5, PAX5-C20S, and derivatives of PAX5-C20S with yellow fluorescent protein (YFP) fused to their N-termini (Fig. 2A) in preparation for in vivo fluorescence studies described below. The YFP-N-terminal fusion had little effect on either activation by PAX5-wt or the DN-activity of PAX5-C20S (compare Figs. 2A and S2). DN-activity was observed even though YFP-PAX5-C20S was expressed at lower levels than YFP-PAX5-wt (Fig. S3). Deletion of the CtBP binding motif and the Ser-Thr-Pro rich regions of YFP-PAX5-C20S, but retention of the putative α-helical region (YFP-PAX5-C20-MD, Fig. 1B), did not diminish DN-activity (Fig. 2A). In contrast, deletion of the putative α-helical region, but retention of the CtBP binding motif and the Ser-Thr-Pro region (YFP-PAX5-C20ΔMD, Fig. 1B) nearly eliminated DN-activity (Fig. 2A), indicating that the proposed α-helical region of C20S is required for suppression of wt PAX5 activity. The slight reduction in luciferase observed with YFP-PAX5-C20ΔMD was likely due to competition for binding to the reporter by the deleted protein that lacks an activation domain.
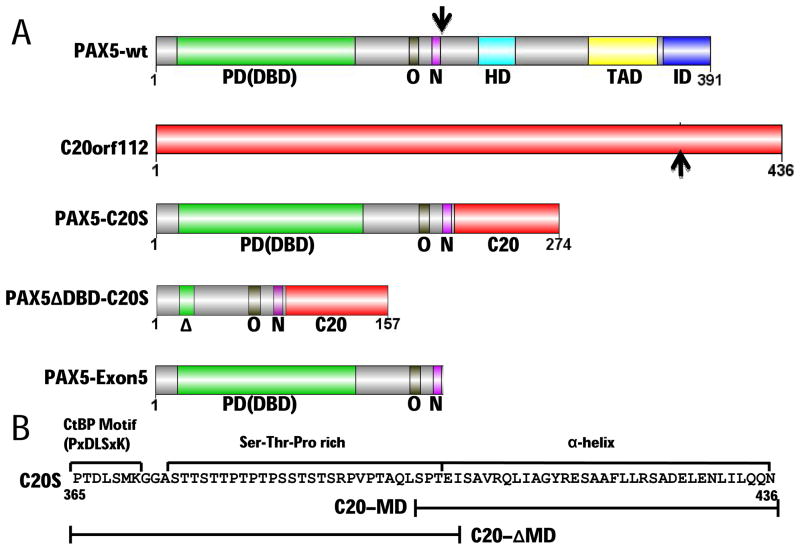
Figure 1. Structural determinants of PAX5-C20orf112 oncogenic fusion protein.
(A) Domains of PAX5 (Cobaleda et al., 2007), C20orf112, PAX5-C20S, PAX5ΔDBD-C20S with the paired DBD deleted, and PAX5-Exon5 (encoded by PAX5 exons 1–5, the portion of PAX5 in PAX5-C20S). The PAX5 DNA-binding domain [PD (DBD)], an octapeptide motif bound by Groucho-family co-repressors (O), and a nuclear localization sequence (N) are included in PAX5-C20S while its partial homeodomain (HD), activation domain (TAD), and inhibitory domain (ID) are not. Black arrows indicate points of fusion. (B) Analysis of C20orf112 sequence in PAX5-C20S reveals three regions that may contribute to DN activity: a CtBP binding motif (P×DLS×K), a serine-threonine-proline (Ser-Thr-Pro)-rich region, and a putative α-helical region. Constructs that contain bracketed parts of C20S are YFP-PAX5-C20-MD that fuses only the α-helical region, and YFP-PAX5-C20ΔMD that fuses only the P×DLS×K motif and the Ser-Thr-Pro rich region.
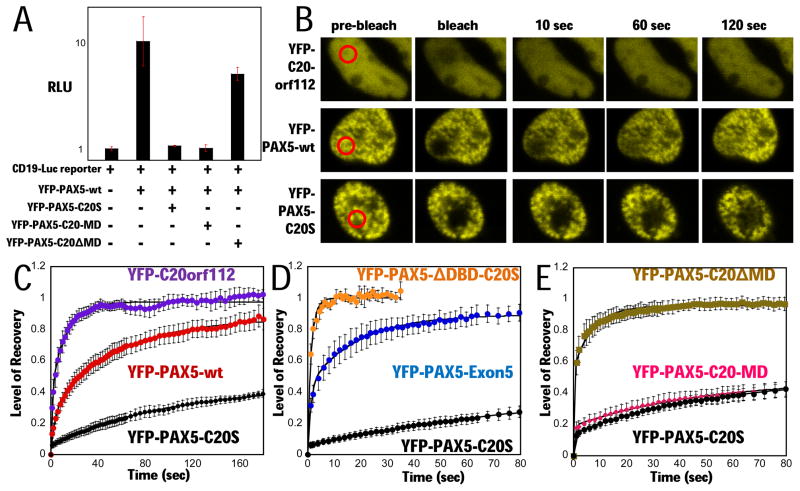
Figure 2. PAX5-C20S suppresses wt PAX5 activity and causes extremely slow nuclear mobility in living cells.
(A) A YFP-PAX5-wt vector and the luciferase reporter were either transfected or co-transfected with vectors for YFP-PAX5-C20S, YFP-PAX5-C20-MD, or YFP-PAX5-C20ΔMD and luciferase assayed (RLU). (B) Fluorescence micrographs from FRAP assays in 293T cells. Red circled areas were photobleached regions, and fluorescence signal in each area was sequentially measured. FRAP assays of (C) YFP-PAX5-C20S (black circles, N=4, t1/2 ~ 200 sec), YFP-C20orf112 (purple circles, N=4, t1/2 ~ 3 s) and YFP-PAX5-wt (red circles, N=7, t1/2 ~ 24 s) in 293T cells, (D) YFP-PAX5-ΔDBD-C20S (orange circles, N=3, t1/2 ~ 1 s), YFP-PAX5-C20S (black circles, N=4, t1/2 ~ 200 s), and YFP-PAX5-Exon5 (blue circles, N=3, t1/2 ~ 4 s) in 293T cells, (E) YFP-PAX5-C20ΔMD (gold squares, N=6, t1/2 ~ 1 s), YFP-PAX5-C20-MD (pink diamonds, N= 7, t1/2 ~200 s), and YFP-PAX5-C20S (black circles, N=5, t1/2 ~200 s) in H1299 cells. Error bars here and in subsequent FRAP assays represent standard error of the mean.
Results from chromatin immunoprecipitation (ChIP) assays using antibody specific for the C-terminus of wt PAX5 indicated that PAX5-C20S expression significantly reduced binding of wt PAX5 to the BLK PAX5-target gene in Nalm6 B-cell leukemia cells, even though the two proteins were expressed at similar levels (Kawamata et al., 2008). We speculated that the C20orf112 C-terminal region might induce oligo- or multimerization of the PAX5 DBD, increasing PAX5-C20S DNA binding affinity by cooperative binding by two or more DBDs in the multimer to neighboring sites in chromosomal DNA. High affinity binding by the fusion might then compete for binding by wt PAX5 at equal concentrations. To test this hypothesis, we performed Fluorescence Recovery After Photobleaching (FRAP) experiments in living cells to measure the stability of chromatin-binding by wt PAX5 and PAX5-C20S in vivo. Fluorescence signals from most TF-fluorescent protein fusions recover quickly after photobleaching a region of the nucleus (t1/2 < 10 sec) (Phair et al., 2004). However, wt PAX5 displayed unusually stable binding with a t1/2 ~ 24 sec (Figs. 2B, 2C), consistent with a recent report that several members of the PAX family TFs in general exhibit slow recovery times (Elvenes et al., 2010). Remarkably, the leukemogenic PAX5-C20S fusion exhibited extremely slow recovery in the FRAP assay (estimated t1/2 ~200 sec), while YFP-labeled C20orf112 displayed a t1/2 ~ 3 sec (Figs. 2B and 2C).
To examine which regions of the fusion protein are responsible for its unusually slow nuclear mobility, we performed FRAP assays with several deletions of PAX5-C20S fused to YFP. Deletion of the PAX5-DBD (Fig. 1A) showed that it is required for stable binding (Fig. 2D), suggesting that low PAX5-C20S nuclear mobility requires binding to DNA in chromatin. The DBD was also required for DN-activity in the reporter assay (Fig. S4A). All of these YFP-labeled constructs localized to the nucleus (Fig. S4B). FRAP assays with YFP-PAX5-C20ΔMD and YFP-PAX5-C20MD (Fig. 2E) showed that the putative α-helical region at the C-terminus is required and sufficient for the decreased mobility of PAX5-C20S; decreased mobility does not require either the CtBP binding motif or the Ser-Thr-Pro rich region. FRAP was also performed with YFP fused to the region of PAX5 in PAX5-C20S encoded by exons 1–5 (“PAX5-Exon 5” (Fig. 1A) which gave a t½ ≈ 4 sec (Fig. 2D). Consequently, the region of PAX5 C-terminal to the portion in PAX5-C20S is required for the slow nuclear mobility of wt PAX5. However, this cannot contribute to the stability of PAX5-C20S chromatin-binding since it is absent from the fusion protein. Also, a potential octapeptide Groucho repression motif in exon 5 of PAX5 (“O” in Fig. 1A), does not contribute significantly to the DN-activity of PAX5-C20S, since mutation Y179E in PAX5 disrupts interactions with the Groucho proteins (Berhard et al., 2000) and had no effect on the stable chromatin binding of PAX5-C20S and only a minor effect on the DN-activity of either PAX5-C20S or the PAX5-p53tetra fusion (Figs. S5A, B). Thus, the putative C-terminal α-helical region of PAX5-C20S greatly increases the stability of chromatin-binding and is required for DN-activity.
We also examined the product of the PAX5-ETV6 fusion gene, the most frequently reported PAX5-fusion in ALL (Table S1). The portion of ETV6 fused to the N-terminus of PAX5 contains a SAM domain whose crystal structure reveals a head-to-tail polymer (Kim et al., 2001) that we speculated might result in stable chromatin binding as we observed for PAX5-C20S. Indeed, both YFP fused to PAX5-ETV6 and to the N-terminal region of PAX5 fused to the SAM domain alone (YFP-PAX5-SAM) exhibited stable chromatin binding comparable to that of YFP-PAX5-C20S (estimated t1/2 ≈200 sec) (Fig. S5C). Reporter gene assays showed that the SAM domain alone was sufficient to generate a potent DN-activity equivalent to that of PAX5-ETV6 (Fig. S5D).
PAX5-C20S has a C-terminal homo-multimerization domain
A helical-wheel projection (Fig. 3A) of the C20S C-terminal sequence revealed 4–3 hydrophobic heptad repeats, consistent with an amphipathic helix. To determine if the predicted amphipathic α-helix is involved in PAX5-C20S oligomerization, we expressed separate fusions of YFP and CFP to C20orf112 residues 391–436 (“C20HL”) in transfected 293T cells and performed fluorescence resonance energy transfer (FRET) assays by flow cytometry, a sensitive method for detecting the interaction of two proteins in vivo in thousands of individual cells (Siegel et al., 2000). YFP-C20HL and CFP-C20HL showed strong FRET signals, similar to the positive control of a CFP-YFP tandem fusion (Fig. 3B). FRET was also observed using YFP-PAX5-C20S and CFP-PAX5-C20S (Fig. 3B), suggesting that PAX5-C20S fusion proteins associate through an α-helical region at their C-termini.
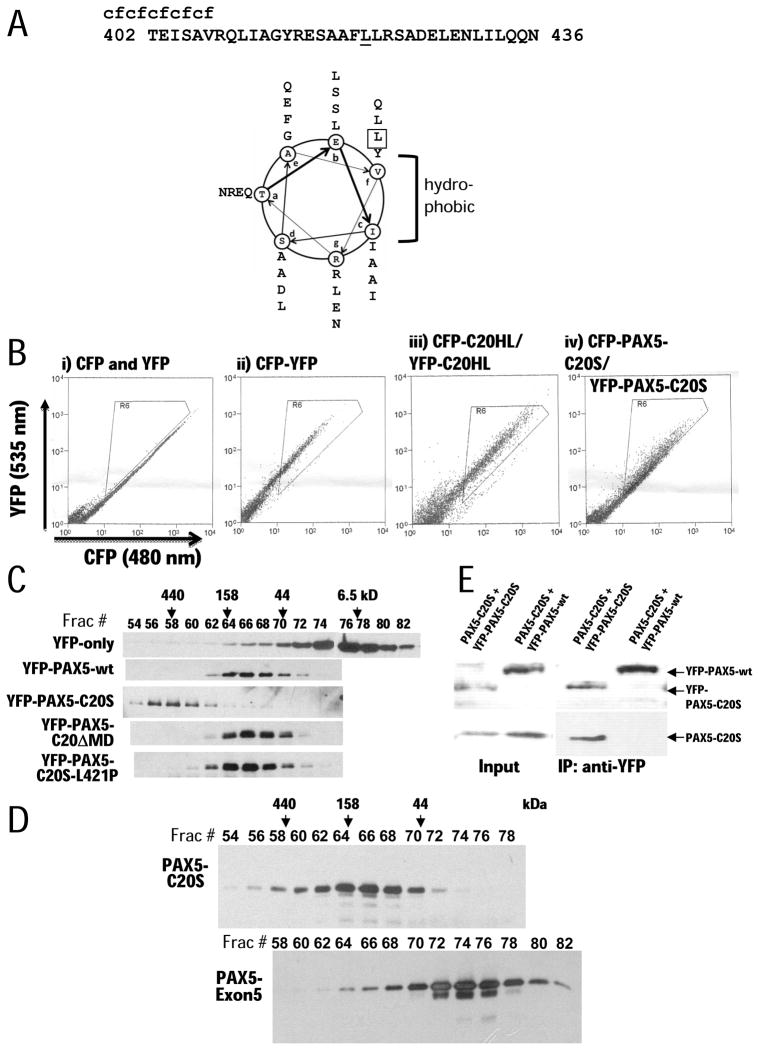
Figure 3. α-helical region of C20S mediates multimerization.
(A) Helical wheel projection of the predicted α-helix (aa 402–436). Heptad positions are labeled by letters ‘a’ through ‘g’ in the wheels. Hydrophobic residues occur at positions ‘c’ and ‘f’. The leucine at position 421 mutated to proline is boxed in projection and underlined in the sequence
(B) Results of FRET assays. CFP or YFP fusion proteins were transiently expressed in 293T cells, and FRET signals were detected by flow cytometry. Cells were excited with a 436 nm laser, and emission signals were measured at 480 nm (x axis) and 535 nm (y axis). When FRET occurs, yellow signals (535 nm, y axis) increase. Signals in the triangular areas indicate cells with FRET signals. In a negative control (i), independent CFP and YFP proteins were co-expressed and produced no significant FRET signal (rare signals in the triangular area). A YFP-CFP fusion protein was expressed and used as a positive control (ii), showing that most cells exhibited strong FRET signals. (iii) Expression of YFP-C20HL and CFP-C20HL produced FRET signals comparable to the YFP-CFP fusion positive control. (iv) Expression of YFP-PAX5-C20S and CFP-PAX5-C20S produced a FRET signal in most cells of lower magnitude because of the greater distance between the YFP and CFP.
(C) Gel filtration analysis of YFP, YFP-PAX5, YFP-PAX5-C20S, YFP-PAX5-C20-ΔMD, and YFP-PAX5-C20S-L421P. Proteins were expressed in H1299 cells, extracted from nuclei, and fractionated by gel filtration. Column fractions were subjected to immunoblotting with anti-YFP antibody. Fraction numbers are indicated above panel with protein standards eluted at specific volumes as indicated by arrows.
(D) Gel filtration of PAX5-C20S-His6 and PAX5-Exon5-His6 expressed in and purified from E. coli.
(E) Co-immunoprecipitation assays. (left panel: Input) PAX5-C20S was co-expressed with either YFP-PAX5-C20S or YFP-PAX5-wt in 293T cells. Expression of each protein was confirmed by immunoblotting cell extracts with YFP-specific antibody (top panel) or antibody specific for the C20 C-terminus (bottom panel). (Right panel) extracts were immunoprecipitated using YFP-antibody and immunoblotted as for the input panels.
To confirm oligomerization of PAX5-C20S, YFP-fusion proteins in nuclear extract (Dignam et al., 1983) from the same cells used for the FRAP studies were subjected to gel filtration chromatography (Fig. 3C). YFP and YFP-PAX5-wt eluted as predicted for monomeric proteins. However, YFP-PAX5-C20S eluted much earlier than YFP-PAX5-wt, consistent with formation of a multimer. Purified PAX5-C20S-His6 (31.2 kD) expressed in E. coli eluted just after the peak of Aldolase (158 kD), consistent with a tetramer (Fig. 3D). In contrast, when PAX5-Exon5-His6, containing only the portion of PAX5 included in PAX5-C20S (25.4 kD), was similarly expressed in and purified from E. coli, it eluted as expected for a monomer (Fig. 3D). Interestingly, in gel filtration the YFP-PAX5-SAM fusion eluted in multiple fractions including the void volume (>2 MDa) indicating that it can form polymeric species from dimer to high-order polymers (data not shown), consistent with the crystal structure of the SAM domain.
We also co-expressed PAX5-C20S with either YFP-PAX5-C20S or YFP-PAX5-wt in 293T cells and performed co-immunoprecipitation assays with anti-YFP antibody. As expected from the results indicating that PAX5-C20S forms a tetramer, PAX5-C20S co-immunoprecipitated with YFP-PAX5-C20S. However, importantly, PAX5-C20S failed to co-immunoprecipitate with YFP-PAX5-wt (Fig. 3D), indicating that while PAX5-C20S forms homo-multimers, it does not interact with monomeric wt PAX5 expressed in the same cell. Thus, the DN-activity of PAX5-C20S does not result from the formation of mixed multimers between wt PAX5 and the oncogenic, multimeric fusion protein.
Multimerization of PAX5 and DN-activity
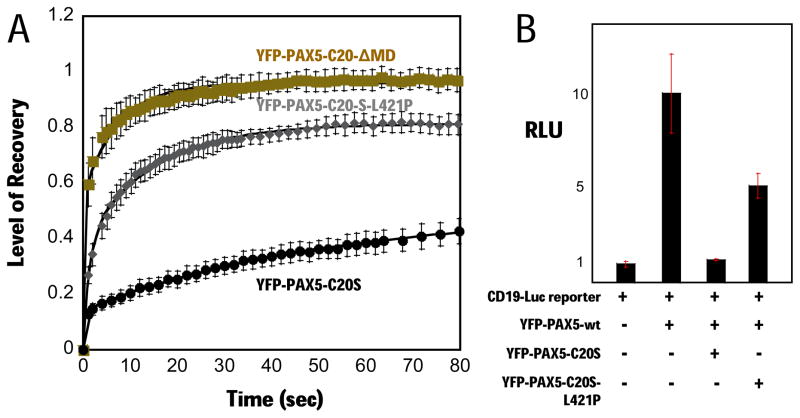
Mutation of leucine 421 (Fig. 3A) to proline near the middle of the proposed C-terminal amphipathic helix of C20orf112 is predicted to greatly reduce the propensity of this sequence to form interacting alpha helices [using Marcoil (Delorenzi and Speed, 2002), Paircoil2 (McDonnell et al., 2006), and COILS/PCOILS (Gruber et al., 2005) programs]. Gel filtration showed that this mutation converts YFP-PAX5-C20S to a monomer (Fig. 3C). Consistent with the model that stable chromatin binding and DN-activity of PAX5-C20S results from multivalent interactions between several DBDs in a PAX5-C20S multimer and several sites in DNA, the L421P mutation caused both a substantial decrease in FRAP t1/2 (Fig. 4A) and a prominent loss of DN-activity (Fig. 4B).
Figure 4. Disruption of C20S multimerization abrogates suppression of PAX5 activity and stable chromatin binding.
(A) FRAP assay comparing YFP-PAX5-C20-ΔMD (gold squares, N=5, t1/2 ~ 2 s), YFP-PAX5-C20S-L421P (gray diamonds, N=5, t1/2 ~ 5 s), and YFP-PAX5-C20S (black circles, N=4, t1/2 ~ 200 s) at 24 h post-transfection in H1299 cells.
(B) YFP-PAX5-wt and the luciferase reporter were co-transfected alone, or with YFP-PAX5-C20S, YFP-PAX5-C20-MD, YFP-PAX5-C20-ΔMD, and YFP-PAX5-C20S-L421P, and luciferase activity assayed.
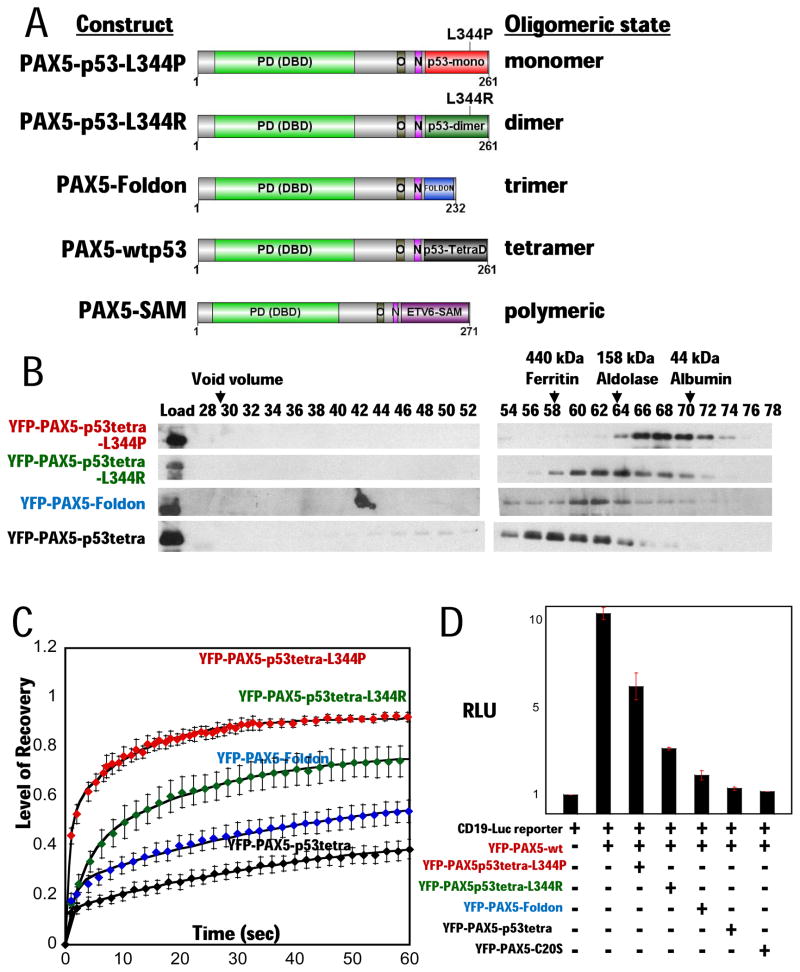
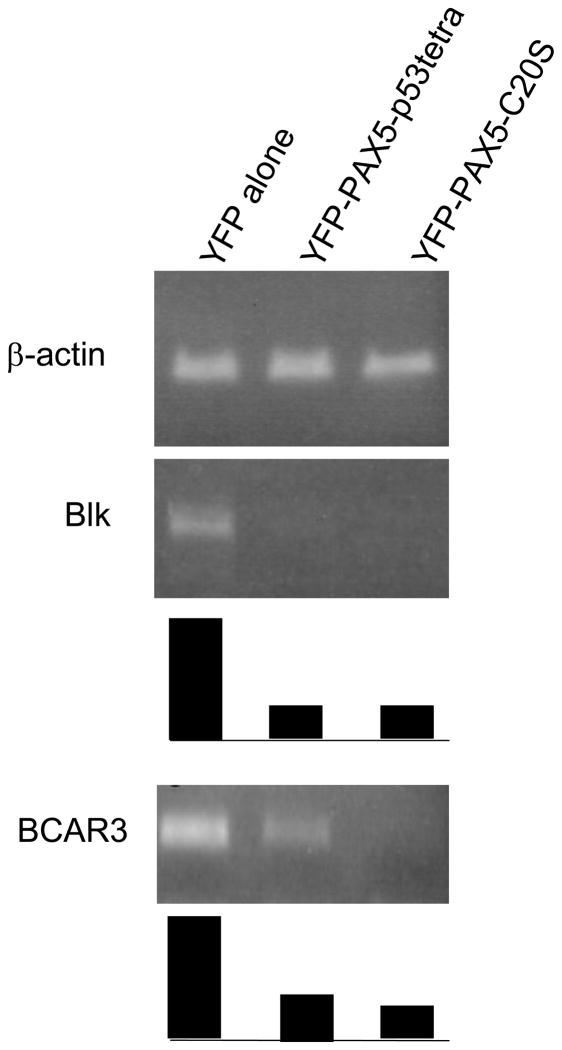
To test the hypothesis further, we fused the N-terminal region of PAX5 to previously characterized protein oligomerization domains known to form either a dimer, trimer, or tetramer (Fig. 5A). The p53 tetramerization domain (p53tetra, aa 305–360) has been studied extensively and shown to behave as either a monomer or dimer with single point mutants L344P and L344R, respectively (Davison et al., 2001). In the exhaustively studied p53 protein, the tetramerization domain is not known to function directly in transcriptional repression by interacting with co-repressors. The foldon domain is a trimerization module from the C-terminal 27 aa of phage T4 fibritin (Letarov et al., 1999). The fusion proteins expressed in p53-minus H1299 cells eluted from a gel filtration column according to their predicted oligomeric states (Fig. 5B). The peaks of YFP-PAX5-C20S and YFP-PAX5-p53tetra eluted in the same fractions (Figs. 3C and 5B), consistent with the conclusion that the C20orf112 C-terminal region is a tetramerization domain based on gel filtration of PAX5-C20S-His6 expressed in E. coli (Fig. 3D). Importantly, FRAP assays of these artificial multimers showed increasing stability of protein-chromatin interactions in vivo in parallel with the increasing number of PAX5-DBDs in the multimer (Fig. 5C). The t1/2 increased by ~2-3-fold as the oligomeric state of the fusion increased successively from monomer to dimer, etc. Significantly, luciferase reporter assays showed that DN-activity increased in parallel with the number of monomers in the multimer and the resulting increase in stability of chromatin binding (Figs. 5D). Fusion of the p53 tetramerization domain generated DN-activity comparable to that of PAX5-C20S. Further, to confirm if this artificial fusion PAX5 which tetramerizes via the p53 tetramerization domain suppressed PAX5 downstream target genes, we transfected either YFP-PAX5-p53tetra, YFP-PAX5-C20s, or YFP constructs into Daudi cells (Burkit B-cell lymphoma cell line). YFP-positive cells were sorted and expression levels of PAX5 downstream target genes (Schebesta A et al. 2007) were examined by semi-quantitative RT-PCR. Expression levels of two major PAX5 downstream target genes (Blk and BCAR3) (Schebesta A et al. 2007) were suppressed by YFP-PAX5-p53tetra as strongly as by YFP-PAX5-C20s (Fig. 5E).
Figure 5. Multimerization of PAX5 fusions mediates suppression of wt PAX5 activity and stable chromatin binding.
(A) Structure of PAX5-fusions: PAX5-p53tetra-L344P (monomer), PAX5-p53tetra-L344R (dimer), PAX5-Foldon (trimer), and PAX5-p53tetra (tetramer) domain. The fusions include the same N-terminal region of PAX5 in PAX5-C20S fused to the oligomerization domain of the fusion partner, p53 (aa 305–360) and foldon (aa 458–484). PAX5-SAM fusion contains the same region of PAX5 fused to the ETV6 SAM multimerization domain.
(B) Gel filtration analysis of YFP-PAX5-fusions. Proteins were expressed in H1299 cells, extracted, fractionated by gel filtration, and detected by immunoblotting with anti-YFP antibody.
(C) FRAP assays with YFP-PAX5 fusions in H1299 cells. FRAP recovery curves of YFP-PAX5-L344P (red diamonds, N= 3, t1/2 ~ 3 s), YFP-PAX5-L344R (green diamonds, N=3, t1/2 ~ 14 s), YFP-PAX5-Foldon (blue diamonds, N=5, t1/2 ~ 50 s), and YFP-PAX5-wtp53tetra (black diamonds, N=5, t1/2 ~ 200 s) show that increasing multimerization of the PAX5 DNA binding domain results in increasing stability of chromatin association.
(D) YFP-PAX5wt and reporter gene were co-transfected either alone or with YFP-PAX5-L344P, YFP-PAX5-L344R, YFP-PAX5-Foldon, YFP-PAX5-wtp53tetra, or YFP-PAX5-C20S in p53-minus H1299 cells. Increasing multimerization of the PAX5 DNA binding domain caused increasing suppression of YFP-PAX5wt activity.
(E) Semi-quantitative RT-PCR. Expression levels of two major PAX5 downstream target genes, Blk and BCAR3, as well as beta-actin (internal control) were examined by semi-quantitative RT-PCR. Products were amplified from cDNA of Burkit B-cell line, Daudi transfected with either YFP, YFP-PAX5-p53tetra, or YFP-PAX5-C20S (PCR cycle for Beta-actin: 25; PCR cycles for Blk: 30; PCR cycles for BCAR3: 33). Ten microliter of each PCR product was run in 2% agarose gel. Relative semi-quantitative levels of each PCR product to beta-actin PCR product are graphed uner each panel of PCR bands; levels of PCR products from Daudi cells transfected with YFP alone were regarded as 1.
PAX5-C20S binds DNA in vitro with ten-fold higher affinity than a monomeric PAX5 DBD
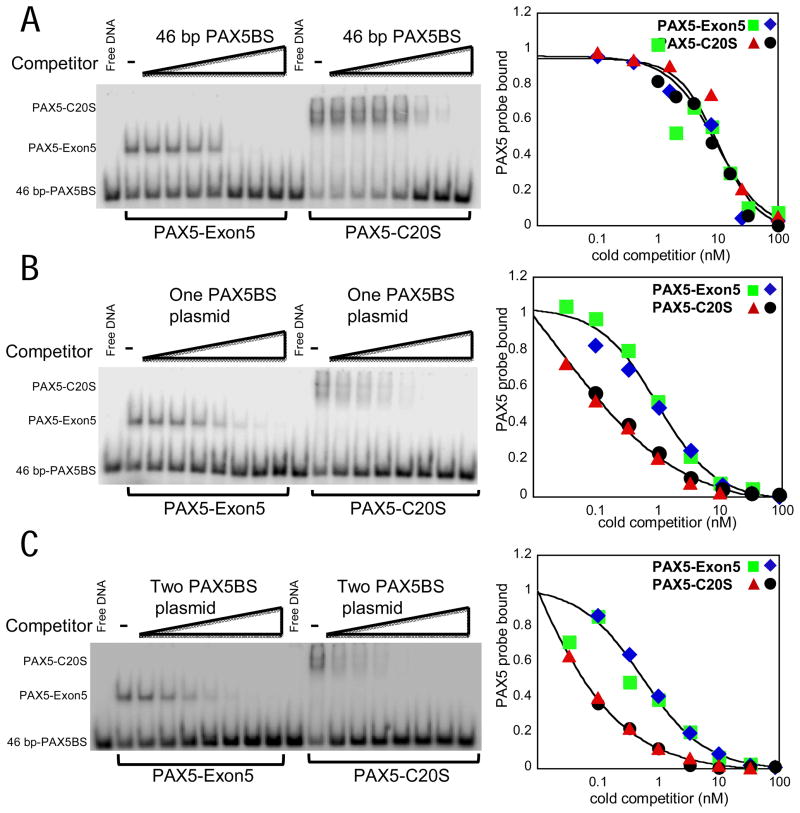
The unusual stability of chromatin binding by PAX5-C20S and PAX5-ETV6 might be accounted for by multivalency, i.e. the binding of multiple DBDs in the same stable multimer to nearby specific and non-specific sites in chromosomal DNA. Since KDs of TF DBDs for non-specific sites are often in the 10−4 to −6 M range, the affinity of such a multimer would increase as the number of DNA-binding domains in the multimer increases. Such an effect would be missed in the earlier reported in vitro assays of DNA-binding that did not show a difference in affinity between wt PAX5 and PAX5-C20S (Kawamata et al., 2008) because the short oligonucleotide probe used could be bound by only a single DNA-binding domain. To test this possibility, we performed in vitro DNA binding assays (EMSAs) using recombinant PAX5 proteins purified from E. coli and long unlabeled DNA molecules in competition experiments with a short 32P-labeled oligonucleotide probe containing a single PAX5 binding site. We were not able to express full-length wt PAX5 at a high level in E. coli, and consequently compared PAX5-C20S to the portion of PAX5 that is fused to C20orf112 in PAX5-C20S (Fig. 1A, PAX5-Exon5). As expected for a tetramer, PAX5-C20S generated a slower mobility EMSA band than the monomeric PAX5 paired DNA-binding domain in PAX5-Exon5 (Fig. 6). The PAX5-C20S DNA-protein complexes resolved into two bands, probably due to binding of one or two oligonucleotide probes to separate DBDs in the tetramer. Failure to observe three or four bands may indicate that steric restraints restrict binding to more than two oligonucleotides probes.
Figure 6. PAX5-C20S tetramer has greater affinity in vitro for a PAX5 site in a long DNA molecule than monomeric PAX5-Exon5.
(A) EMSA using a 46 bp 32P-labeled high affinity PAX5 binding site (PAX5BS) derived from the CD19 promoter (Czerny et al., 1993). Increasing amounts of unlabeled 46-bp PAX5BS (0.1, 0.4, 1.6, 6.4, 26, 100, 400 nM) were added before addition of protein to the binding reaction. The fraction of labeled 46-bp PAX5BS probe bound by protein (PAX5-Exon5 or PAX5-C20S) was calculated and plotted to the right relative to the amount bound without competitor. Results from two independent assays are indicated by the green squares and blue diamond symbols for PAX5-Exon5 and the red triangles and black circles for PAX5-C20. (B, C) Performed as in (A) except the unlabeled competitor DNA (0.1, 0.3, 1.0, 3, 10, 30, 100 nM) was a 5.15 kb plasmid with one or two copies (separated by 2 kb) of the 46 bp PAX5BS cloned into it. The data were fit by the variable slope sigmoid equation (see Experimental Procedures, Supplemental Materials).
To assay differences in binding affinity to short and long DNA molecules, we added increasing amounts of cold competitor DNAs to a labeled 46 bp probe before the addition of protein. The 46 bp labeled probe is bound by a single PAX5 paired DBD that protects 28 bp at the center of the probe from DNase I digestion (Czerny et al., 1993; Xu et al., 1995). When the unlabeled probe was added as competitor, it competed equally the binding of the labeled probe to the PAX5-Exon 5 monomer and the PAX5-C20S tetramer (Fig. 6A). However, significantly, when a 5.15 kb plasmid with a single PAX5 binding site was used as competitor, competition to 50% probe binding was observed at one-tenth the competitor concentration for PAX5-C20S compared to PAX5-Exon5 (Fig. 6B), indicating that PAX5-C20S has ten-fold higher affinity for a PAX5 site in a long DNA molecule, like cellular DNA, than a monomeric PAX5-DBD. Since the plasmid contains a single specific PAX5-binding site, the increased affinity probably results from interactions of one or more of the additional DBDs in the tetramer with non-specific sites in the long DNA. The plasmid competed probe binding to the monomeric PAX5-Exon5 at a lower concentration than the cold 46 bp probe (50% probe binding at 1 versus 9 nM) because in the binding reactions with the plasmid, competition also occurs from the high number of low affinity non-specific sites in the plasmid—every bp of the 5.15 kb plasmid constitutes the first bp of another non-specific site.
A plasmid with two PAX5-binding sites separated by 2 kb competed for binding to the monomeric PAX5-Exon5 at about one-half the concentration of the plasmid with a single PAX5 binding site (Fig. 6C) as expected, since the concentration of specific sites is two times higher for the same concentration of plasmid. The increased affinity of PAX5-C20S for the plasmid with two PAX5 sites remained 10-fold higher than for monomeric PAX5-Exon5. Similar results were observed for the luciferase reporter plasmid with three high affinity PAX5 binding sites separated by 49 bp between the centers of the sites (data not shown). These results support the conclusion that the increased affinity of PAX5-C20S results from interactions of one or more of the three extra DBDs in the PAX5-C20S tetramer with a neighboring non-specific site or sites. Otherwise, one would have expected a much greater increase in affinity for plasmids with two and three specific sites compared to the plasmid with a single specific site.
Inhibition of PAX5-C20S DN-activity by expression of the C20S tetramerization domain
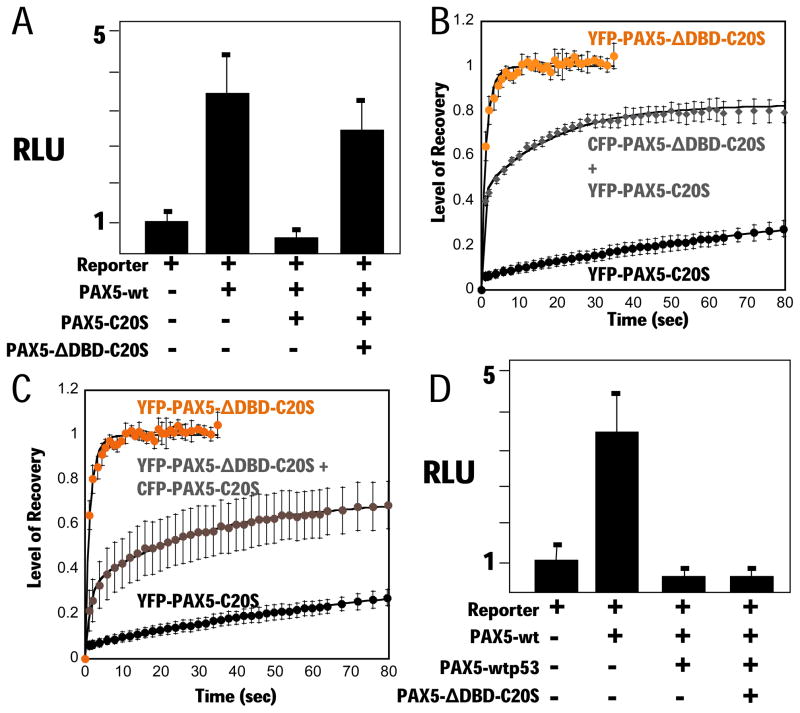
Our data support the model that tetramerization of the PAX5 DBD via the C-terminal region of C20S causes high affinity binding of the oncogenic TF-fusion to its cognate DNA binding sites, competing for binding by wt PAX5. This model suggests that co-expression of the C20orf112 tetramerization domain not fused to a PAX5-DBD would disrupt tetramerization of the DBD via hetero-oligomerization, and consequently interfere with PAX5-C20S DN-activity. To test this, we co-expressed PAX5-ΔDBD-C20S with PAX5-C20S, using a four-fold excess of the PAX5-ΔDBD-C20S vector. DN activity was nearly eliminated as measured by reporter assay (Fig. 7A). A similar result has been reported for disruption of both the tetramerization and transforming activity of RUNX1-ETO by expression of an ETO multimerization domain (Wichmann et al., 2007). FRAP assays with CFP-PAX5-ΔDBD-C20S in the same cells as YFP-PAX5-C20S confirmed that the stability of YFP-PAX5-C20S association with chromatin was decreased considerably, although it is unclear why recovery of fluorescence plateaued at ~80% (Fig. 7B). Similarly, the rapid nuclear mobility of PAX5-ΔDBD-C20S (t1/2 ~ 1 s) was decreased substantially by co-expression of an equal amount of CFP-PAX5-C20S (t1/2 ~ 25 s, Fig. 7C). The specificity of this effect was demonstrated in experiments with YFP-PAX5-p53tetra whose DN-activity was not affected by PAX5-ΔDBD-C20S (Fig. 7D). These data confirm that multimerization of the PAX5 DBD causes stable chromatin binding, and demonstrate that disruption of the multimerization interface is sufficient to reverse PAX5-C20S DN-activity.
Figure 7. The C20S tetramerization domain suppresses the dominant negative activity of PAX5-C20S.
(A) Vectors for PAX5-wt, PAX5-C20S, and PAX5-C20SΔDBD and the luciferase reporter were transfected into 293T cells as indicated and luciferase assayed after 24 h. FRAP assays for (B) YFP-PAX5-ΔDBD-C20S (orange circles, N=3, t1/2 ~ 1 s), YFP-PAX5-C20S co-expressed with four-fold more CFP-PAX5-ΔDBD-C20S (YFP FRAP gray diamonds, N=7, t1/2 ~ 4 s), and YFP-PAX5-C20S (black circles, N=4, t1/2 ~ 200 s), and for (C) YFP-PAX5-ΔDBD-C20S (orange circles, N=3, t1/2 ~ 1 s), YFP-PAX5-ΔDBD-C20S and an equal amount of CFP-PAX5-C20S (gray circles, N=8, t1/2 ~ 25 s), and YFP-PAX5-C20S (black circles, N=4, t1/2 ~ 200 s). (D) PAX5-wtp53tetra was expressed with wt PAX5 and PAX5 reporter gene either with or without PAX5-C20SΔDBD and luciferase assayed.
Discussion
The requirement for induced oligomerization of oncogenic TF fusions for DN-activity was originally identified in studies of RARα and RUNX1 fusions (Lin and Evans, 2000; Minucci et al., 2000). Some of the oligomerization domains fused to the RARα DBD contribute to DN activity by enhancing recruitment of co-repressors, such as SMRT, binding partner RXR, and by relaxing the DNA binding specificity compared to wt RARα/RXR (Kamashev et al., 2004; Lin and Evans, 2000; Martens et al., 2010; Minucci et al., 2000; Zhou et al., 2006). Here, we present evidence that for DN PAX5-fusions, oligomerization of the DNA binding domain (DBD) alone is sufficient to suppress activation by wt PAX5, and consequently, to contribute to leukemogenesis, without invoking recruitment of co-repressors. We propose that PAX5-DBD oligomerization to a tetramer (PAX5-C20S, PAX5-p53tetra) or higher order multimer (PAX5-ETV6 SAM domain) results in an ~10-fold increase in affinity for PAX5 binding sites through interactions of the additional DBDs in the oligomer with neighboring non-specific sites in DNA, competing with wt PAX5 to target genes.
Gel filtration chromatography of recombinant PAX5-C20S purified from E. coli indicated that the fusion protein forms a stable tetramer. Further evidence that PAX5-C20S is a tetramer comes from the observation that YFP-PAX5-C20S co-eluted from the gel filtration column with a YFP-fusion of the same N-terminal region of PAX5 to the well-characterized p53 tetramerization domain. This led to a great increase in the stability of chromatin-binding by the fusion proteins compared to wt PAX5 as determined by FRAP assays in living cells. A similar finding was noted when the DNB region of PAX5 was fused to the multermization region of p53.
In vitro DNA-binding assays revealed that PAX5-C20S has ~10-fold higher affinity for a PAX5-binding site in plasmid DNA compared to the monomeric PAX5 DBD. Consequently, the increased stability of chromatin binding by PAX5-C20S may be accounted for largely by the increase in DNA-binding affinity of the tetramer compared to the monomer. This increased DNA-binding affinity probably involves cooperative binding of more than one DBD in the PAX5-C20S tetramer to DNA, since no difference in affinity was observed for an oligonucleotide containing the same high affinity PAX5-binding site, but which was too short to interact with more than one DBD. Further, the increase in affinity is probably due to interactions between DNA-binding domains in the multimer and neighboring non-specific sites in a long DNA molecule since it was observed with a plasmid containing a single high affinity consensus PAX5-binding site.
In hypothesis confirming studies, we noted no change in the stability of chromatin-binding assayed by FRAP by mutation of potential co-repressor binding sites in the PAX5-C20S sequence. But, a single leucine to proline mutation (L421P) in the predicted amphipathic α-helical tetramerization domain inhibited YFP-PAX5-C20S stable multimerization and greatly decreased the stability of binding to chromatin. Strikingly, when the PAX5 N-terminal region was fused to p53 domains or fibritin that generated either a dimer, a trimer, or a tetramer, the stability of chromatin binding and the DN-activity of the fusion protein increased in parallel. Consequently, the increased stability of chromatin-binding by PAX5-C20S compared to wt PAX5 seems best explained by the increased DNA-binding affinity of the tetramer compared to the monomer, without invoking binding by a co-repressor. Our experiments with another PAX5 fusion (PAX5-ETV6) suggest that it too behaves in as a DN in a similar fashion.
Earlier, we observed by chromatin immunoprecipitation that PAX5-C20S expressed at a similar concentration to endogenous wt PAX5 in cultured B-cells, significantly reduced binding of wt PAX5 to its cognate binding site in the BLK gene (Kawamata et al., 2008). The results discussed above strongly suggest that this decrease is due to competition for binding to the same site by PAX5-C20S, which binds to a PAX5-site in chromosomal DNA with ~10-fold higher affinity than the monomeric PAX5 DBD in wt PAX5.
Earlier FRAP studies of other fusion proteins associated with hematopoietic malignancies have been reported including RARα fusions to either PML, PLZF, NPM, NuMA, or STAT5b in acute promyelocytic leukemia (APL) (Dong et al., 2004; Huang et al., 2008), RUNX1 fusions to ETO and MTG16 in acute myeloid leukemia (AML) (Qiu et al., 2006), and, recently, a PAX5-PML fusion (Qiu et al., 2010). In most cases, only modest decreases in nuclear mobility compared to the wt TF were observed. Only in the cases of PML-RARα and PAX5-PML was the decrease in nuclear mobility as dramatic as observed here for PAX5-C20S and PAX5-ETV6. Moreover, as for PAX5-C20S, the unusually slow nuclear mobility of these fusions to PML is dependent on an amphipathic helical domain contributed by the PML fusion partner (Huang et al., 2008; Qiu et al., 2010) that oligomerizes into a tetramer (Antolini et al., 2003). In other cases of ALL, PAX5 is fused to other partner genes that provide multimerization domains, including FOXP1 and the extracellular protein Elastin (Strefford et al., 2009). Many of the other PAX5-fusion partners associated with ALL (ZNF521, JAK2, BRD1, DACH1, DACH2, GOLGA6, and KIF3B) contain putative, as yet uncharacterized oligomerization domains. Our findings are probably applicable to many other oncogenic fusions containing the DBD of a transcription factor and a multimerization domain of the other partner. Since disruption of PAX5-DBD multimerization nearly eliminated the DN-activity of PAX5-C20S, pharmacologic interference with multimerization of the PAX5-fusion proteins, or indeed any similar oncogenic fusion might be a novel therapeutic approach.
Experimental Procedures
(Detailed procedures in Supplementary Information)
Cell lines and transfection
293T and H1299 (p53−/−) cells were transfected with Effectene (Qiagen) or Lipofectamine 2000 (Invitrogen) according to the manufacturers’ protocols. Burkitt B-cell line, Daudi, was maintained in RPMI1640 media with 10% fetal calf serum. Six microgram of YFP, YFP-PAX5-p53tetra, or YFP-PAX5-C20S constructs were transfected into 3 × 106 Daudi cells using Amaxa Nucleofector (Solution L and Condition A20) according to the manufacturer’s recommendation (Lonza, Basel, Switzerland).
Vectors
Wt PAX5 cDNA was cloned into the pMSCV vector (Clontech). All YFP or CFP containing constructs were cloned into pcDNA3.1 (Invitrogen).
Luciferase reporter assays
Three PAX5 recognition sites derived from the murine CD19 promoter were placed in front of firefly luciferase in pGL2-basic (Promega) (Dorfler and Busslinger, 1996) with pRenilla-luciferase-SV40 (Promega) used as a transfection efficiency control. Luciferase activities were measured using Promega Dual Luciferase reporter assay kit.
FRAP
In these experiments, a region of fluorescent proteins within the nucleus is irreversibly bleached by a short, intense laser pulse, and recovery of fluorescence in the bleached region is monitored over time. After photobleaching, bleached molecules dissociate from their binding sites and are replaced by non-bleached fluorescent proteins that diffuse from outside the bleached region. Proteins that are highly mobile generate fast recovery curves while proteins tightly bound to anchored sites in the nucleus, such as chromosomes, recover slowly (Sprague and McNally, 2005). Live 293T or H1299 cells were imaged 24 h post-transfection (800 ng vector per dish) with a Leica TCS SP2 AOBS single photon confocal microscope with the stage and 63×1.4-numerical-aperture oil immersion objective placed in an enclosed chamber pre-equilibrated to 37°C with continuous flow of 5% CO2 over the microscope stage.
FRET
293T were transfected using Effectene. Living cells were suspended in PBS and assays performed on FACStar as described (Siegel et al., 2000) with 436 nm laser excitation and emission detected with 480 nm (x axis) and 535 nm (y axis) filters.
EMSAs
Electrophoretic mobility shift assays were performed with a 46-bp probe containing one PAX5 binding site as described (Czerny et al., 1993) using recombinant proteins purified by Ni+2 affinity and gel filtration chromatography.
Cell sorting and semi-quantitative RT-PCR
YFP positive transfected Daudi cells were sorted by FACStar Cell Sorter (Becton, Dickinson and company, Franklin Lakes, NJ). Total RNA was extracted and complementary DNA was genereated using SuperScript® III One-Step RT-PCR system (Invitrogen, Carlsbad, CA). Expression levels of beta actin in each samples were examined by 25 cycles of PCR reaction; expression levels of two PAX5 downstream target genes, Blk and BCAR3 genes, were also examined by 30 and 32 cycles of PCR amplification, respectively. Ten microliter of each of the PCR products were run in 2% gel. PCR products were photographed and density of each PCR band was measured by NIH image software (http://rsb.info.nih.gov/nih-image/index.html).
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01CA026038-32, and U54CA143930, and A* STAR Investigator Grant to H.P.K and R37CA25235 to A.J.B., the Tower Cancer Foundation grant and NIH grant GM008243 to N.K., and ACS postdoctoral fellowship PF0804801GMC to M.A.P. We thank Kolja A. Wawrowsky in the Confocal Microscopy Core Unit and Patricia Lin in the FACS Core Unit for their technical assistance. FRAP experiments were performed at the California Nano Systems Institute Advanced Light Microscopy/Spectroscopy (CNSI-ALMS) at UCLA.
References
- An Q, Wright SL, Konn ZJ, Matheson E, Minto L, Moorman AV, et al. Variable breakpoints target PAX5 in patients with dicentric chromosomes: a model for the basis of unbalanced translocations in cancer. Proc Natl Acad Sci U S A. 2008;105:17050–4. doi: 10.1073/pnas.0803494105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolini F, Lo Bello M, Sette M. Purified promyelocytic leukemia coiled-coil aggregates as a tetramer displaying low alpha-helical content. Protein Expr Purif. 2003;29:94–102. doi: 10.1016/s1046-5928(03)00004-4. [DOI] [PubMed] [Google Scholar]
- Bousquet M, Broccardo C, Quelen C, Meggetto F, Kuhlein E, Delsol G, et al. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood. 2007;109:3417–23. doi: 10.1182/blood-2006-05-025221. [DOI] [PubMed] [Google Scholar]
- Cazzaniga G, Daniotti M, Tosi S, Giudici G, Aloisi A, Pogliani E, et al. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001;61:4666–70. [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–61. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–25. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- Dong S, Stenoien DL, Qiu J, Mancini MA, Tweardy DJ. Reduced intranuclear mobility of APL fusion proteins accompanies their mislocalization and results in sequestration and decreased mobility of retinoid X receptor alpha. Mol Cell Biol. 2004;24:4465–75. doi: 10.1128/MCB.24.10.4465-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfler P, Busslinger M. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. Embo J. 1996;15:1971–82. [PMC free article] [PubMed] [Google Scholar]
- Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. Embo J. 2000;19:2292–303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PA. Fusion genes and chromosome translocations in the common epithelial cancers. J Pathol. 2010;220:244–54. doi: 10.1002/path.2632. [DOI] [PubMed] [Google Scholar]
- Elvenes J, Sjottem E, Holm T, Bjorkoy G, Johansen T. Pax6 localizes to chromatin-rich territories and displays a slow nuclear mobility altered by disease mutations. Cell Mol Life Sci. 67:4079–94. doi: 10.1007/s00018-010-0429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazio G, Palmi C, Rolink A, Biondi A, Cazzaniga G. PAX5/TEL Acts as a Transcriptional Repressor Causing Down-modulation of CD19, enhances Migration to CXCL12, and Confers Survival Advantage in pre-Bl Cells. Cancer Research. 2008;68:181–189. doi: 10.1158/0008-5472.CAN-07-2778. [DOI] [PubMed] [Google Scholar]
- Frohling S, Dohner H. Chromosomal abnormalities in cancer. N Engl J Med. 2008;359:722–34. doi: 10.1056/NEJMra0803109. [DOI] [PubMed] [Google Scholar]
- Gasparini P, Sozzi G, Pierotti MA. The role of chromosomal alterations in human cancer development. J Cell Biochem. 2007;102:320–31. doi: 10.1002/jcb.21481. [DOI] [PubMed] [Google Scholar]
- Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–8. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- Gruber M, Soding J, Lupas AN. REPPER--repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 2005;33:W239–43. doi: 10.1093/nar/gki405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Qiu J, Chen G, Dong S. Coiled-coil domain of PML is essential for the aberrant dynamics of PML-RARalpha, resulting in sequestration and decreased mobility of SMRT. Biochem Biophys Res Commun. 2008;365:258–65. doi: 10.1016/j.bbrc.2007.10.184. [DOI] [PubMed] [Google Scholar]
- Kamashev D, Vitoux D, De The H. PML-RARA-RXR oligomers mediate retinoid and rexinoid/cAMP cross-talk in acute promyelocytic leukemia cell differentiation. J Exp Med. 2004;199:1163–74. doi: 10.1084/jem.20032226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata N, Ogawa S, Zimmermann M, Niebuhr B, Stocking C, Sanada M, et al. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc Natl Acad Sci U S A. 2008;105:11921–6. doi: 10.1073/pnas.0711039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, et al. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. Embo J. 2001;20:4173–82. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Evans RM. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol Cell. 2000;5:821–30. doi: 10.1016/s1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–4. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–4. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- Martens JH, Brinkman AB, Simmer F, Francoijs KJ, Nebbioso A, Ferrara F, et al. PML-RARalpha/RXR Alters the Epigenetic Landscape in Acute Promyelocytic Leukemia. Cancer Cell. 2010;17:173–85. doi: 10.1016/j.ccr.2009.12.042. [DOI] [PubMed] [Google Scholar]
- McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–8. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell. 2000;5:811–20. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- O’Neil J, Look AT. Mechanisms of transcription factor deregulation in lymphoid cell transformation. Oncogene. 2007;26:6838–49. doi: 10.1038/sj.onc.1210766. [DOI] [PubMed] [Google Scholar]
- Pabst T, Mueller BU. Transcriptional dysregulation during myeloid transformation in AML. Oncogene. 2007;26:6829–37. doi: 10.1038/sj.onc.1210765. [DOI] [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, et al. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Wong J, Tweardy DJ, Dong S. Decreased intranuclear mobility of acute myeloid leukemia 1-containing fusion proteins is accompanied by reduced mobility and compartmentalization of core binding factor beta. Oncogene. 2006;25:3982–93. doi: 10.1038/sj.onc.1209431. [DOI] [PubMed] [Google Scholar]
- Qiu JJ, Chu H, Lu X, Jiang X, Dong S. The reduced and altered activities of PAX5 are linked to the protein-protein interaction motif (coiled-coil domain) of the PAX5-PML fusion protein in t(9;15)-associated acute lymphocytic leukemia. Oncogene. 2010 doi: 10.1038/onc.2010.473. [DOI] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B-cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Siegel RM, Chan FK, Zacharias DA, Swofford R, Holmes KL, Tsien RY, et al. Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Sci STKE. 2000;2000:pl1. doi: 10.1126/stke.2000.38.pl1. [DOI] [PubMed] [Google Scholar]
- So CW, Cleary ML. Dimerization: a versatile switch for oncogenesis. Blood. 2004;104:919–22. doi: 10.1182/blood-2004-03-0992. [DOI] [PubMed] [Google Scholar]
- Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Strefford JC, An Q, Harrison CJ. Modeling the molecular consequences of unbalanced translocations in cancer: lessons from acute lymphoblastic leukemia. Cell Cycle. 2009;8:2175–84. doi: 10.4161/cc.8.14.9103. [DOI] [PubMed] [Google Scholar]
- Wichmann C, Chen L, Heinrich M, Baus D, Pfitzner E, Zornig M, et al. Targeting the oligomerization domain of ETO interferes with RUNX1/ETO oncogenic activity in t(8;21)-positive leukemic cells. Cancer Res. 2007;67:2280–9. doi: 10.1158/0008-5472.CAN-06-3360. [DOI] [PubMed] [Google Scholar]
- Xu W, Rould MA, Jun S, Desplan C, Pabo CO. Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–50. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- Zhou J, Peres L, Honore N, Nasr R, Zhu J, de The H. Dimerization-induced corepressor binding and relaxed DNA-binding specificity are critical for PML/RARA-induced immortalization. Proc Natl Acad Sci U S A. 2006;103:9238–43. doi: 10.1073/pnas.0603324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.