Abstract
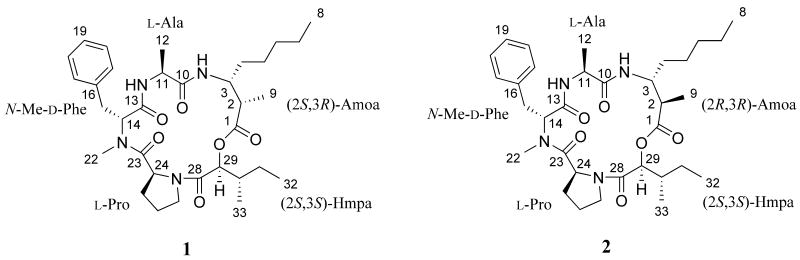
NMR-guided fractionation of a non-polar extract of a Florida Keys collection of Lyngbya sp. resulted in the isolation of two novel epimeric cyclic depsipeptides, porpoisamides A (1)and B (2). The planar structures of these compounds were determined using NMR spectroscopic techniques. The absolute configurations of amino and hydroxy acid subunits were assigned by enantioselective HPLC analysis. These compounds showed weak cytotoxicity towards HCT-116 colorectal carcinoma and U2OS osteosarcoma cells. The porpoisamides are a unique pair of cyclic depsipeptides that are epimeric at C-2 of the β-amino acid, 3-amino-2-methyloctanoic acid.
Keywords: Cyanobacteria, Lyngbya majuscula, depsipeptides, porpoisamide
1. Introduction
An ever increasing number of cyclic peptides and depsipeptides are being isolated from marine cyanobacteria of the genus Lyngbya.1 The structures of these secondary metabolites are as diverse as the biological activities they produce. They can range in size from as many as ten or more amino acid units to as few as two units. Many of the peptides and depsipeptides produced by cyanobacteria contain modified amino acid units as well as polyketide portions. These modifications presumably confer resistance to degradation and produce a variety of biological activities, including antimicrobial, antiviral, and cytotoxic effects.2-4
One type of modification seen in cyclic peptides and depsipeptides produced by marine cyanobacteria are β-amino acid units.4 These β-amino acids often contain functionalized polyketide chains of varying lengths. The guineamides A-F are an example of a series of cyclic depsipeptides that contain a variety of β-amino acid units as well as other modifications to the amino and hydroxy acid units found within the molecule.5 Other examples from cyanobacteria include homodolastatin 16,6 lobocyclamide B,7 obyanamide,8 ulongamides A-F,9 grassypeptolides,10, 11 and ulongapeptin.12 These secondary metabolites have shown biological activity in anti-fungal and cytotoxicity assays.6-12 Reported here is the isolation and structure determination of two epimeric cyclodepsipeptides, porpoisamide A (1)and porpoisamide B (2).
2. Results and Discussion
A sample of the cyanobacterium Lyngbya sp. was collected from the seagrass bed adjacent to Porpoise Key in the Florida Keys in July 2008. The freeze-dried sample was extracted with a 1:1 mixture of MeOH and EtOAc and then partitioned between EtOAc and H2O. The EtOAc-soluble portion was fractionated by silica gel column chromatography. The fraction eluting with EtOAc showed proton resonances indicative of two peptides in a 2:1 ratio and was further separated using normal phase column HPLC. All resulting fractions with similar 1H NMR spectra were combined and purified using reversed-phase HPLC to yield two new cyclic depsipeptides, porpoisamide A (1)and porpoisamide B (2).
Porpoisamide A (1) was isolated as a white powder and its molecular formula was determined to be C33H50N4O6 based on the [M + H]+ ion peak at m/z 599.3795 in HRESIMS. Analysis of the 1H, 13C, and edited HSQC NMR data indicated five carbonyl carbons and a phenyl ring to account for nine of the eleven degrees of unsaturation suggested by the molecular formula. The IR spectrum indicated the presence of phenyl, amide, ester, and NH groups with broad bands at 1032, 1622, 1744, and 3332 cm-1, respectively. Based on the IR data and the 1H and 13C NMR spectroscopic data, the four nitrogen atoms in the molecule could be accounted for by one tertiary amide, two secondary amides (δH 6.57, 8.93) and one N-Me amide (δH 2.96). The six oxygen atoms were found to make up one ester and four amide carbonyl groups, implying that porpoisamide A (1) was a depsipeptide consisting of one hydroxy and four amino acid units.
The DQF COSY and HMBC experiments enabled the identification of three of the amino acid units and one α-hydroxy acid group. The COSY correlations observed from the proton resonance at δH 4.35 to both the methyl group at δH 1.25 and the NH group at δH 6.57 along with the HMBC correlation to the carbonyl carbon resonance at δC 172.7 identified the alanine unit. The HMBC and COSY correlations connected the phenyl ring to the methine at δH 5.52 through a methylene (δH 2.90, 3.26; δC 34.3). Further HMBC correlations linked the methine at δH 5.52 to the N-methyl group at δH 2.96 and to the carbonyl carbon resonance at δC 170.5 to form N-Me-phenylalanine. The chemical shifts of the methine at C-24 (δH 4.51;δC 59.6) indicated that it was adjacent to a nitrogen atom. HMBC correlations connected this methine to the carbonyl carbon resonance at δC 175.3, and the COSY correlations attached it to a methylene (δH 1.80, 1.57), which is part of three consecutive methylenes. The chemical shifts of the end methylene (δH 3.70, 3.44) of this chain and its HMBC correlation to the methine (δH 4.51) through the nitrogen established the proline unit. COSY correlations indicated that the oxymethine C-29 at δH 5.23 was connected to two methyl groups through a methine and a methylene to form 2-hydroxy-3-methylpentanoic acid. Therefore, four of the subunits of the peptide were identified as alanine,N-Me-phenylalanine, proline, and 2-hydroxy-3-methylpentanoic acid (Hmpa).
Based on the NMR data, the remaining subunit of this depsipeptide was identified as an unusual amino acid containing two methyl, four methylene, and two methine groups. HMBC correlations from the methine at δH 2.70 and the methyl doublet at δH 1.13 to the carbonyl carbon resonance at δC 174.8 showed the methine was alpha to the carbonyl carbon. The COSY correlation from the methine at δH 2.70 to the methine at δH 3.97 showed the methine at δH 3.97 to be beta to the carbonyl carbon. The methine at δH 3.97 was shown to be connected to the NH group at δH 8.93 through COSY correlations. The four methylene groups were shown to be connected in a chain starting at the methine at δH 3.97 and ending with the methyl group at δH 0.86 to form the 3-amino-2-methyloctanoic acid (Amoa) as the final subunit of porpoisamide A(1).
The five subunits of porpoisamide A(1) were connected together through HMBC correlations. The 3-amino-2-methyloctanoic acid group had to be connected to alanine based on the correlation between the NH group at δH 8.93 and the carbonyl group at δC 172.7. Alanine was connected to the N-Me-phenylalanine because of a correlation between the NH group at δH 6.57 and the carbonyl carbon atδC 170.5. N-Me-phenylalanine was attached to the proline subunit based on the correlation between the N-Me group at δH 2.96 and the proline carbonyl carbon at δC 175.3. Finally, the proline group was connected to the Hmpa subunit through a correlation between the methylene proton H-27a at δH 3.70 and the carbonyl carbon C-28 at δC 170.0. To account for the remaining degree of unsaturation and the molecular formula the Hmpa subunit had to be attached to the Amoa group to form the cyclic depsipeptide structure depicted for 1. This connection was confirmed by the HMBC correlation between the methine proton H-29 at δH 5.23 and the carbonyl carbon C-1 at δC 174.8.
Porpoisamide B (2) was found to have the same molecular formula as porpoisamide A(1)(C33H50N4O6, m/z599.3798, [M+H]+). Analysis of the 1H, 13C, and 2D NMR spectra indicated that 2 was similar in structure to 1 as well. While the 1H and 13C NMR resonance values for the alanine, N-Me-phenylalanine, proline, and Hmpa were consistent with that of porpoisamide A (1), chemical shifts of the proton and carbon resonances of the β-amino acid portion indicated a difference in this segment of the molecule. The 1D TOCSY experiments suggested the only difference between these segments of porpoisamides A (1) and B (2) involved the stereoconfiguration of the molecule. Excitation of the methine proton at C-2 (δH 2.70 for 1; δH 2.67 for 2) in both molecules produced spectra with proton resonances for the methine at C-3, the methyl group at C-9, and the NH group. Excitation of the methine proton at C-3 (δH 3.97 for 1; δH 4.02 for 2) in both molecules produced spectra with proton resonances for the methyl group at C-9, the methine at C-2, the NH group, the methylene groups at C-4 through C-7, and the methyl group at C-8. This data in conjunction with COSY and HMBC correlations indicated that the planar structure of the unusual amino acid was the same in both porpoisamide A (1) and B (2).
Analysis of the NMR spectra of 1 and 2 showed them to have the same planar structure. Each contained three typical amino acids (alanine, N-Me-phenylalanine, and proline), one α-hydroxy acid (2-hydroxy-3-methylpentanoic acid), and the rare β-amino acid (3-amino-2-methyloctanoic acid). Therefore, it was necessary to investigate the configuration of the eight chiral centers found within the molecules in order to differentiate the structures of the two depsipeptides.
The absolute configurations of the alanine, N-Me-phenylalanine, proline, and Hmpa were determined by stereoselective HPLC. Porpoisamides A (1) and B (2) were subjected to acid hydrolysis and the retention times of the amino acids and the hydroxy acid were compared to those of authentic standards run under the same conditions. The results of this analysis showed the absolute configuration of these α-hydroxy and amino acids to be the same for both compounds. The absolute configuration of alanine and proline was found to be l, the configuration of N-Me-phenylalanine was d, and Hmpa was found to have 2S,3S configuration. This analysis determined that six of the eight stereogenic centers within the peptides were identical.
To determine the absolute configuration of the remaining two stereogenic centers within the 3-amino-2-methyloctanoic acid, advanced Marfey's method was used.13 The methods and retention times published by Williams et al. in 200312 were used for comparison with the derivatized hydrolysate of 1 and 2. The published elution order for the l-FDLA derivatized Amoa units using reversed-phase HPLC is (2R,3S)-Amoa, (2S,3S)-Amoa, (2R,3R)-Amoa, and (2S,3R)-Amoa.12, 14 Performing the same analysis with porpoisamides A and B indicated that 1 contained (2S,3R)-Amoa and 2 contained (2R,3R)-Amoa. The results from this analysis were supported by NOE correlations from the NMR spectra. The 2D NOESY spectrum for porpoisamide A(1)showed strong correlations from H-2 to H-3 and H-9. This data indicated that H-2 and H-3 were on the same side of the ring structure. This configuration matches that seen in the 2S,3R isomer of 3-amino-2-methyloctanoic acid. The 2D NOESY spectrum of porpoisamide B (2) showed a strong correlation from H-3 to H-9 and no NOE correlation between H-2 and H-3,indicating that H-3 and H-9 were close in space and therefore on the same side of the ring structure. The resulting configuration is that seen in the 2R,3R isomer of 3-amino-2-methyloctanoic acid.
Many of the linear and cyclic peptides produced by cyanobacteria have potent activity in cytotoxicity assays.2, 4, 15 For this reason, both of the cyclic peptides were tested for their ability to inhibit the growth of two solid tumor cell types: HCT-116 colorectal carcinoma and U2OS osteosarcoma cells. The IC50 values for compound 1 were 25 μM and 28 μM, respectively. Compound 2 gave similar IC50 values of 21 μM and 22 μM, respectively. Thus, these cyclic depsipeptides produce only moderate antiproliferative activity against these cell lines.
3. Experimental
3.1 General experimental procedures
The optical rotation was measured on a Perkin Elmer model 343 polarimeter. IR spectroscopic data were obtained on a Perkin Elmer Spectrum 100 FT-IR spectrometer. 1H, 13C and 2D NMR spectra for 1 and 2 were recorded in CD3CN on a JEOL ECA-600 operating at 600 MHz using residual solvent resonances for reference (δH 1.94, δC 118.7 for acetonitrile). The HRMS data were obtained using an Agilent 6210 LC-TOF mass spectrometer equipped with an APCI/ESI multimode ion source detector at the Mass Spectrometer Facility at the University of California, Riverside, California.
3.2 Biological material, collection and identification
The marine cyanobacterium, Lyngbya sp., was collected by hand from the seagrass bed located adjacent to Porpoise Key of the Florida Keys in July 2008. The sample was drained of seawater, frozen at -20 °C, and subsequently freeze-dried. A voucher specimen (#VJP_PK_7_22_08) was preserved in 5% formalin/seawater and is retained at the Smithsonian Marine Station, Fort Pierce, FL.
3.3 Extraction and Isolation
The freeze-dried sample (298.1 g) was exhaustively extracted with 1:1 MeOH-EtOAc to yield 40.3 g of crude extract. This extract was then partitioned between EtOAc and H2O. The EtOAc partition (3.1 g) was further separated on a silica gel column using a hexanes-EtOAc-MeOH step gradient system to give six fractions (90:10:0, 50:50:0, 0:100:0, 0:95:5, 0:90:10, 0:0:100; 200 mL of each). Fraction 3 (118.8 mg), 100% EtOAc, was separated by NP-HPLC [Alltima Silica, 10 μm, 10 × 250 mm, 3 mL/min, detection at 220 and 254 nm] with 50% EtOAc in hexanes. All resulting fractions were examined by NMR. Those with similar 1H NMR spectra were combined into one fraction (26.8 mg). This fraction was then separated by RP-HPLC [Econosil C18, 10 μm, 10 × 250 mm, 3 mL/min, detection at 220 and 254 nm], first with 90% MeOH in H2O with 0.1% acetic acid to give a mixture of porpoisamides (9.4 mg, tR 7.2 min) and then with 70% CH3CN in H2O with 0.1% acetic acid to yield 6.0 mg of 1 (tR 16.7 min, yield 0.002% dry wt) and 3.3 mg of 2 (tR 14.7 min, yield 0.0011% dry wt). The NMR guided-fractionation of this sample provided no evidence that epimerization may have occurred during the isolation process. Both purified porpoisamides were stable and did not interconvert.
3.3.1 Porpoisamide A (1)
Yield 6.0 mg. white powder; [α]20d +77.6 (c 0.45, CH3OH); IR υmax 3332, 2940, 1744, 1622, 1456, 1032 cm-1; UV (MeOH), λmax (log ε) 255 (2.45); 1H NMR (CD3CN, 600 MHz) and 13C NMR (CD3CN, 150 MHz) data see Table 1; HRESI/APCIMS m/z599.3795 [M + H]+ (calcd for C33H51N4O6, 599.3809).
Table 1. NMR data for found compound (1 in CD3CN (600 MHz).
| δH(J in Hz) | δC, mult. | COSYa | HMBCb | ||
|---|---|---|---|---|---|
| Amoa | 1 | 174.8, C | |||
| 2 | 2.70, qd (7.5, 4.1) | 44.9, CH | 3, 9 | 1, 3, 4, 9 | |
| 3 | 3.97, m | 52.8, CH | 2, 4b, 9, NH | 1, 2, 4 | |
| 4a | 1.60, m | 29.7, CH2 | 4b, 5a, 5b | 3, 5, 6 | |
| 4b | 1.11, m | 4a, 5a | 3, 5, 6 | ||
| 5a | 1.28, m | 32.9, CH2 | 4a, 4b, 5b | 4, 7 | |
| 5b | 1.13, m | 4a, 5a | 4, 7 | ||
| 6a | 1.28, m | 27.1, CH2 | 6b | ||
| 6b | 1.12, m | 6a | |||
| 7 | 1.27, m | 23.8, CH2 | 8 | 5, 6 | |
| 8 | 0.86, d (6.2) | 14.8, CH3 | 7 | ||
| 9 | 1.13, d (6.9) | 15.7, CH3 | 2 | 1, 2 | |
| NH | 8.93, d (8.9) | 3 | 10 | ||
| Ala | 10 | 172.7, C | |||
| 11 | 4.35, dq (8.3, 6.9) | 52.4, CH | 12, NH | 10, 12, 13 | |
| 12 | 1.25, d (6.8) | 20.8, CH3 | 11 | 11, 10 | |
| NH2 | 6.57, d (8.3) | 11 | 11, 12, 13 | ||
| N-Me-Phe | 13 | 170.5, C | |||
| 14 | 5.52, dd (10.3, 5.5) | 58.4, CH | 15a, 15b | 13, 15, 16, 22, 23 | |
| 15a | 3.26, dd (-15.0, 5.5) | 34.3, CH2 | 14, 15b | 13, 14, 16, 17, 21 | |
| 15b | 2.90, dd (-15.0, 10.3) | 14, 15a | 13, 14, 16, 17, 21 | ||
| 16 | 139.6, C | ||||
| 17 | 7.18, d (6.9) | 130.2, CH | 19, 21 | 15, 18, 19 | |
| 18 | 7.24, m | 129.5, CH | 17, 19, 21 | 16, 17 | |
| 19 | 7.18, t (6.9) | 127.6, CH | 18, 20 | 17, 18, 20, 21 | |
| 20 | 7.24, m | 129.5, CH | 17, 19, 21 | 16, 21 | |
| 21 | 7.18, d (6.9) | 130.2, CH | 18, 20 | 15, 19, 20 | |
| 22 | 2.96, s | 32.4, CH3 | 14, 23 | ||
| Pro | 23 | 175.3, C | |||
| 24 | 4.51,dd (6.9, 3.4) | 59.6, CH | 25a, 25b | 23, 25 | |
| 25a | 1.80, m | 28.8, CH2 | 24, 25b, 26b | 26, 27 | |
| 25b | 1.57, m | 24, 25a, 26a, 26b | 23, 24, 26 | ||
| 26a | 2.06, m | 27.6, CH2 | 25b, 26b, 27a, 27b | 24, 25 | |
| 26b | 1.87, m | 25a, 25b, 26a, 27a, 27b | 25, 27 | ||
| 27a | 3.70, dd (9.0, 9.0) | 48.2, CH2 | 26a, 26b, 27b | 24, 25, 28 | |
| 27b | 3.44, m | 26a, 26b, 27a | 26 | ||
| Hmpa | 28 | 170.0, C | |||
| 29 | 5.23, d (2.7) | 78.3, CH | 30 | 1, 28, 30, 31, 33 | |
| 30 | 2.15, m | 35.4, CH | 29, 31a, 31b, 33 | 31, 32, 33 | |
| 31a | 1.38, m | 24.3, CH2 | 31b, 32 | 29, 30, 32, 33 | |
| 31b | 1.17, m | 30, 31a, 32 | 30, 32, 33 | ||
| 32 | 0.87, t (6.9) | 12.5, CH3 | 31a, 31b | 30, 31 | |
| 33 | 1.01, d (6.9) | 16.4, CH3 | 30 | 29, 30, 31 |
COSY correlations are from proton stated to indicated proton.
HMBC correlations are from proton stated to the indicated carbon.
3.3.2 Porpoisamide B (2)
Yield 3.3 mg. white powder; [α]20d +22.7 (c0.24, CH3OH); IR υmax 3437, 2962, 2344, 1576, 1415, 1046 cm-1; UV (MeOH), λmax (log ε) 253 (2.38); 1H NMR (CD3CN, 600 MHz) and 13C NMR (CD3CN, 150 MHz) data see Table 2; HRESI/APCIMS m/z599.3798 [M + H] + (calcd C33H51N4O6, 599.3809).
Table 2. NMR data for compound(2) in CD3CN (600 MHz).
| δH(J in Hz) | δC, mult. | COSYa | HMBCb | ||
|---|---|---|---|---|---|
| Amoa | 1 | 173.4, C | |||
| 2 | 2.67, qd (6.9, 3.5) | 45.9, CH | 3, 9 | 1, 3, 9 | |
| 3 | 4.02, m | 53.1, CH | 2, 4a, 4b, NH | ||
| 4a | 1.47, m | 35.3, CH2 | 3, 4b, 5a | 3, 5, 6 | |
| 4b | 1.39, m | 3, 4a, 5a, 5b | 3, 5, 6 | ||
| 5a | 1.26, m | 32.8, CH2 | 4a, 4b, 5b | 6, 7 | |
| 5b | 1.18, m | 4a, 5a | 4, 6, 7 | ||
| 6 | 1.26, m | 27.2, CH2 | 5, 7 | ||
| 7 | 1.26, m | 23.7, CH2 | 8 | 5, 6 | |
| 8 | 0.86, t (6.9) | 14.8, CH3 | 7 | 7 | |
| 9 | 1.13, d (7.5) | 14.8, CH3 | 2 | 1, 2, 3 | |
| NH | 8.74, d (9.6) | 3.00 | 10 | ||
| Ala | 10 | 173.8, C | |||
| 11 | 4.37, dq (8.3, 7.3) | 52.8, CH | 12, NH | 10, 12, 13 | |
| 12 | 1.28, d (7.3) | 21.3, CH3 | 11 | 10, 11 | |
| NH | 6.67, d (8.3) | 11 | 13 | ||
| N-Me-Phe | 13 | 170.4, C | |||
| 14 | 5.52, dd (10.9, 5.2) | 58.6, CH | 15a, 15b | 13, 15, 16, 22, 23 | |
| 15a | 3.27, dd (-14.4, 5.5) | 34.3, CH2 | 14, 15b | 14, 16, 17 | |
| 15b | 2.89, dd (-14.4, 10.9) | 14, 15a | 13, 14, 16, 17 | ||
| 16 | 139.6, C | ||||
| 17 | 7.18, d (7.2) | 130.2, CH | 18 | 15, 19 | |
| 18 | 7.24, m | 129.5, CH | 17 | 16, 17, 19 | |
| 19 | 7.18, t (6.8) | 127.6, CH | 18, 20 | 17, 21 | |
| 20 | 7.24, m | 129.5, CH | 19, 21 | 16, 19, 21 | |
| 21 | 7.18, d (7.2) | 130.2, CH | 20 | 15, 19 | |
| 22 | 2.95, s | 32.3, CH3 | 14, 23 | ||
| Pro | 23 | 174.6, C | |||
| 24 | 4.49, dd (6.8, 3.5) | 59.5, CH | 25a, 25b | 23, 25 | |
| 25a | 1.80, m | 28.8, CH2 | 24, 25b, 26b | 26, 27 | |
| 25b | 1.56, m | 24, 25a, 26a, 26b | 24, 26 | ||
| 26a | 2.06, m | 27.5, CH2 | 25b, 26b, 27a, 27b | 24, 25 | |
| 26b | 1.88, m | 25a, 25b, 26a, 27a, 27b | 25, 27 | ||
| 27a | 3.70, t (9.6) | 48.2, CH2 | 26a, 26b, 27b | 24, 25 | |
| 27b | 3.44, dt (10.4, 6.1) | 26a, 26b, 27a | 26 | ||
| Hmpa | 28 | 169.9, C | |||
| 29 | 5.20, d (2.1) | 78.1, CH | 30 | 1, 28, 30, 31, 33 | |
| 30 | 2.15, m | 35.5, CH | 29, 31a, 31b, 33 | 31 | |
| 31a | 1.41, m | 24.5, CH2 | 30, 32 | 30, 32, 33 | |
| 31b | 1.20, m | 30 | 32, 33 | ||
| 32 | 0.88, t (7.6) | 12.5, CH3 | 31a | 30, 31 | |
| 33 | 1.01, d (6.8) | 16.5, CH3 | 30 | 29, 30, 31 |
COSY correlations are from proton stated to indicated proton.
HMBC correlations are from proton stated to the indicated carbon.
3.4 Enantioselective HPLC analysis
2-hydroxy-3-methylpentanoic acid standards were synthesized according to literature methods.16 Compounds 1 and 2 were hydrolyzed (∼ 0.2 mg each) in 6 N HCl at 115 °C for 18 h in a sealed tube. The hydrolysis productswere then concentrated to dryness and re-dissolved in 2.0 mM CuSO4in H2O. Analysis by enantioselective HPLC was carried out using three separate conditions and the retention times were compared to those of authentic standards. Condition 1: 5% CH3CN in 2.0 mM CuSO4 in H2O, 1.0 mL/min, Phenomenex Chirex 3126 (4.6 × 250 mm) column; elution times (tR, min) for standards: l -alanine (6.2), d-alanine (7.3), l-proline (9.2), and d-proline (17.9). Condition 2: 15% CH3CN in 2.0 mM CuSO4 in H2O, 1.0 mL/min, Phenomenex Chirex 3126 (4.6 × 250 mm) column; elution times (tR, min) for standards: l-N-Me-phenylalanine (29.2), d-N-Me-phenylalanine (32.8). Condition 3: 15% CH3CN in 2.0 mM CuSO4 in H2O, 1.0 mL/min, Diacel Chemical Laboratories, Ltd. Chiralpak MA (+) (4.6 × 50 mm) column; elution times (tR, min) for standards: (2R,3S)-Hmpa (16.8), (2R,3R)-Hmpa (19.7), (2S,3R)-Hmpa (25.9), (2S,3S)-Hmpa (31.7). The hydrolysate for 1 was analyzed by the above methods to reveal the presence of l-alanine (6.1 min), l-proline (8.9 min), d-N-Me-phenylalanine (32.5 min), and (2S,3S)-Hmpa (31.8 min). The hydrolysate for 2 was analyzed by the above methods to reveal the presence of l-alanine (6.2 min), l-proline (8.9 min), d-N-Me-phenylalanine (32.0 min), and (2S,3S)-Hmpa (31.8 min). The presence of d-N-Me-phenylalanine in both compounds was confirmed by co-injection with the standard.
3.5 Analysis of 3-amino-2-methyloctanoic acid by advanced Marfey's method
Compounds 1 and 2 were hydrolyzed (∼0.8 mg each) in 6 N HCl at 115 °C for 18 h in a sealed tube. The hydrolysates were dried under air and then re-dissolved in 400 μL of 50:50 acetonitrile:water. Then 100 μL of each hydrolysate product was derivatized with l-FDLA and 100 μL of each was derivatized with dl-FDLA using standard procedures.13, 17 Standard samples of l-valine, l-alanine, l-proline, and N-Me-d-phenylalanine were also derivatized with l-FDLA. The analysis was carried out by RP-HPLC using the same column as Williams et al.12 (YMC-Pack AQ-ODS, 10 × 250 mm, 57% acetonitrile in water with 0.01 N TFA, flow rate 2.5 mL/min, absorbance detection at 340 nm). The published retention times of l-FDLA derivatized Amoa standards were 25.7 min (2R,3S), 26.7 min (2S,3S), 45.6 min (2R,3R), and 54.1 min (2S,3R).12, 14 The retention time of the Amoa portion for 1 was 48.5 and 23.5 min for l-FDLA and d-FDLA, respectively. The retention time of the Amoa portion for 2 was 40.5 and 24.5 min for l-FDLA and d-FDLA, respectively. The previously identified amino acids appeared at 10.1 min (l-Ala), 9.6 min (l-Pro), and 12.6 min (N-Me-d-Phe).
3.6 Activity
Cell viability was assessed using a 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) (MTT) colorimetric assay. Cancer cells were plated in 96-well plates (HCT-116, 10,000; U2OS, 4,000), and after 24 h were treated with various concentrations of compounds 1, 2 (purity >95%) or solvent control (1% EtOH). After 48 h of incubation, cell viability was measured using MTT according to the manufacturer's instructions (Promega). Assays were run in quadruplicate and dose-response curves were generated using XLfit Excel (ID Business Solutions Ltd.).
Supplementary Material
Figure 1. Structures of Porpoisamides A (1) and B (2).
Acknowledgments
This research was supported by the National Institutes of Health, NIGMS grant P41M806210. We are grateful for the use of the 600 MHz NMR spectrometer at Harbor Branch Oceanographic Institute at Florida Atlantic University. We also thank the Florida Atlantic University, Jupiter Campus, for use of their polarimeter and infrared spectrometer. HRMS analyses were performed at the UCR Mass Spectrometry Facility, Department of Chemistry, University of California at Riverside. This is contribution # 849 of the Smithsonian Marine Station at Fort Pierce.
Footnotes
Supporting information available: 1H, 13C, DQF COSY, HSQC, HMBC, and NOESY NMR spectra in CD3CN for compounds(1) and (2).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Liu L, Rein KS. Mar Drugs. 2010;8:1817. doi: 10.3390/md8061817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burja AM, Banaigs B, Abou-Mansour E, Burgess JG, Wright PC. Tetrahedron. 2001;57:9347. [Google Scholar]
- 3.Gademann K, Portmann C. Curr Org Chem. 2008;12:326. [Google Scholar]
- 4.Tan LT. Phytochemistry. 2007;68:954. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Tan LT, Sitachitta N, Gerwick WH. J Nat Prod. 2003;66:764. doi: 10.1021/np020492o. [DOI] [PubMed] [Google Scholar]
- 6.Davies-Coleman MT, Dzeha TM, Gray CA, Hess S, Pannell LK, Hendricks DT, Arendse CE. J Nat Prod. 2003;66:712. doi: 10.1021/np030014t. [DOI] [PubMed] [Google Scholar]
- 7.MacMillan JB, Molinski TF. Org Lett. 2002;4:1883. doi: 10.1021/ol025876k. [DOI] [PubMed] [Google Scholar]
- 8.Williams PG, Yoshida WY, Moore RE, Paul VJ. J Nat Prod. 2002;65:29. doi: 10.1021/np0102253. [DOI] [PubMed] [Google Scholar]
- 9.Luesch H, Williams PG, Yoshida WY, Moore RE, Paul VJ. J Nat Prod. 2002;65:996. doi: 10.1021/np0200461. [DOI] [PubMed] [Google Scholar]
- 10.Kwan JC, Rocca JR, Abboud KA, Paul VJ, Luesch H. Org Lett. 2008;10:789. doi: 10.1021/ol702946d. [DOI] [PubMed] [Google Scholar]
- 11.Kwan JC, Ratnayake R, Abboud KA, Paul VJ, Luesch H. J Org Chem. 2010;75:8012. doi: 10.1021/jo1013564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams PG, Yoshida WY, Quon MK, Moore RE, Paul VJ. J Nat Prod. 2003;66:651. doi: 10.1021/np030050s. [DOI] [PubMed] [Google Scholar]
- 13.Fujii K, Ikai Y, Oka H, Suzuki M, Harada Ki. Anal Chem. 2003;69:5146. [Google Scholar]
- 14.(2R,3S)-Amoa was inferred from (2S,3R)-Amoa derivatized with D-FDLA and (2S,3S)-Amoa was inferred from (2R,3R)-Amoa derivatized with D-FDLA.
- 15.Singh S, Kate BN, Banerjee UC. Crit Rev Biotechnol. 2005;25:73. doi: 10.1080/07388550500248498. [DOI] [PubMed] [Google Scholar]
- 16.Mamer OA. Methods Enzymol. 2000;324:3. doi: 10.1016/s0076-6879(00)24213-4. [DOI] [PubMed] [Google Scholar]
- 17.Marfey P. Carlsberg Res Commun. 1984;49:591. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.