Abstract
Class switch recombination of antibody isotype is mediated by a recombinational DNA deletion event, and must be robustly upregulated during antigen-driven differentiation of B cells. The enhancer region 3′ of the Cα gene is important for the upregulation of switch recombination. Using a transgene of the entire heavy chain constant region locus, we now demonstrate that it is the four 3′ enhancer elements themselves (a total of 4.7 kb) that are responsible for the upregulation, rather than the 24 kb of DNA in between them. Neither allelic exclusion nor transgenic μ expression is reduced by deletion of the four 3′ enhancers. We also test deletions of two or three of the 3′ enhancers, and show that deletion of more 3′ enhancers results in a progressive reduction in both switch recombination and germline transcription of all heavy chain genes. Nevertheless, we find evidence for special roles for some 3′ enhancers--different heavy chain genes are affected by different 3′ enhancer deletions. Thus, we find that the dramatic induction of class switch recombination during antigen-driven differentiation is the result of an interaction among four separated regulatory elements.
Introduction
Class switch recombination (CSR2) changes the type of antibody produced from IgM to IgE, IgA, or IgG. CSR is mediated by a recombinational deletion event that begins in the intron between the V region-encoding exon and the μ C region exon and ends upstream of the Cε, Cα, or one of the four Cγ heavy chain genes (1, 2). CSR is inactive in resting, mature B cells, but must be robustly upregulated during antigen-driven B cell differentiation so that many progeny B cells express an isotype that is better suited for the clearance of the specific type of infection that initiated the immune response. The rate of CSR has been estimated to be as high as 10% per cell per generation (3). Four enhancers that lie 3′ of the heavy chain constant region genes have been proposed to be responsible for this antigen-driven upregulation of CSR (2). The region 3′ of Cα was first implicated in the control of heavy chain gene expression by examination of a spontaneous mutant (4), and later by a directed deletion/replacement (5). DNase I hypersensitive sites (HS) were identified in the region 3′ to Cα, and were named (from Cα proximal to Cα distal): HS3A, HS1,2, HS3B, and HS4 (6–8). The DNase I hypersensitive sites were also shown to be B cell specific, highly synergistic enhancers of transcription (7, 9–13). Replacement of some of these single enhancers by an promoter:neo expression cassette has a negative effect on CSR of some heavy chain genes (14, 15). The deletion of the two most 3′ enhancers (HS3B and HS4) leads to a dramatic reduction in CSR to γ3 and γ2b, a moderate reduction in CSR to γ2a, ε, and α, but little change in expression of μ or γ1 (16). We have found that a 230 kb transgene of the heavy chain C region locus acts very similarly to the endogenous heavy chain locus. CSR, and the germline transcription that precedes it, to all six heavy chain genes is regulated by cytokines like CSR and germline transcription of the endogenous heavy chain genes (17). We demonstrated that a 28 kb deletion of the four 3′ enhancers within the transgene reduced CSR to ε, α, and all four γ heavy chain genes to about 1% of wild type transgenes (17). A similar deletion of the 3′ enhancers from the endogenous locus has the same effect on CSR (18).
The upregulation of CSR would be best understood if the various interactions among cis-acting elements and trans-acting factors could be defined. As expected, many of the transcription factors active in B cells bind the various HS sites 3′ of Cα: Pax5/BSAP, octamer binding proteins, PU.1, NF-κB family members, ets family members, Bach2, and BATF (2, 19). In this study, we address two additional questions to define which cis-acting elements are important for CSR (and implicate the transacting factors that bind those elements). First, is it the 4.7 kb that include the four 3′ enhancers that are important for CSR, or do the 24 kb of DNA in between them have a role? Second, what is the effect of deletion of two or three of the 3′ enhancers, compared to the deletion of all four?
Materials and Methods
Construction and characterization of transgenic mice
The starting bacterial artificial chromosome (BAC) for these constructs included two copies of the chicken β-globin insulator about 3 kb 5′ of a VDJH2 exon located in its normal location. The VDJH2 exon encoded anti-arsonate activity. Both Iγ1 and Iγ2a had 4 bp insertions, and a Flag tag was inserted near the carboxy terminus of the secreted version of γ2a. The starting BAC had a loxP site inserted just 3′ of HS4. The construction of these modifications has been described (17). Two new BACs, with HS4 flanked by loxP sites and deletions of HS1,2 (deletion of residues 11236 to 12572 in Genbank Accession AF450245 http://www.ncbi.nlm.nih.gov/genbank/) and HS3B (deletion of 24040 to 25202), or HS3A (deletion of 1093 to 2129), HS3B, and HS1,2, were constructed for this study. Homology fragments both 5′ and 3′ of either deletions or loxP sites were constructed using appropriate PCR primers.. Targeting vectors in pSV1.RecA (20) included one 5′ and one 3′ homology region of ca. 700 bp each. The 230 kb NotI fragment from these BACs was prepared, purified from the vector sequences, and injected into fertilized SJL × C57BL/6 F2 eggs. By breeding mice with these versions of ARS/Igh transgenes to EIIa Cre-expressing mice (21), we prepared two more sets of transgenic mice, with additional 1.1 kb deletions of HS4 (deletion of 27250 to 28360). All lines of transgenic mice were analyzed for the V exon, JH, Cγ3, Cγ2b, Iγ2a, Cε, HS3A, HS1,2, HS3B, HS4, and sequences 5 kb from the 3′ end of the transgene by PCR and restriction enzyme digestion and for the Sγ1 and Sα regions by Southern hybridization (17, 22). Transgenic lines were tested for Cγ1 sequences using primers CTGACTCCTAAGGTCACGTGTG (Genbank Accession D78344 12993–13014) and GCAGGTCAGACTGACTTTATCC (complement of 13431–13452). Digestion of the PCR product with MboI yielded fragments of 413 bp (transgenic), 336 and 77 bp (endogenous) and 49 bp (both). Transgenic lines were tested for the HS3A deletion using primers GCTAAGTGTCAATCCTCGTCAC (Genbank Accession AF450245 901–922) and TCCACTGGGGTCTCCAGCAGAC (complement of 2271–2292), which resulted in a 353 bp PCR fragment. Transgenic lines were tested for the HS1,2 deletion using primers GGCCAAGATGTAGTAGAGAGACC (11156–11175) and ACAGAGGTAGGAGATGTGCACC (complement of 12737–12748), which resulted in a 254 bp PCR fragment. Transgenic lines were tested for the HS3B deletion using primers GACCATACCCTGCTATGAGTCC (23982–24003) and TGGTGCTCAGCACTGAGTTCTG (complement of 25374–25395), which resulted in a 249 bp PCR fragment. Transgenic lines were tested for the HS4 deletion using primers GAAACTCATATCAGCCTCTGCAC (27053–27075) and GTTTCTGGGTGTCTCTGTGTCTGTTC (complement of 28869–28894), which resulted in a 709 bp PCR fragment. All work with mice has been approved by the University of Michigan Committee on Care and Use of Animals, protocol 08147.
Characterization of B cell development and class switch recombination
Expression of μa (PE) and μb (biotin and PER-CP streptavidin) on splenocytes was analyzed by flow cytometry. For the preparation of RNA samples, splenocytes depleted of erythrocytes and T cells were cultured at 1.5 million per ml for three days with various combinations of murine recombinant IL-4 (35 ng/ml, Biosource, Camarillo, CA), murine recombinant IFN-γ (10 ng/ml, Biosource), human recombinant TFG-β (4 ng/ml, Preprotech, Rocky Hill, NJ), LPS (25 μg/ml, Sigma, St. Louis, MO) or CD40L expressed by Sf21 cells (150,000 cells) (23). The culture medium was RPMI supplemented with penicillin, streptomycin, glutamine, 20 μM 2-ME, and 10% FBS. Germline transcripts for ε, α, and the four γ heavy chain genes were analyzed by RT-PCR and restriction enzyme digestion (17, 22). Post-switch IμCH transcripts and VDJCH transcripts (using a 5′ primer specific for the transgenic VD junction) were analyzed by RT-PCR (17, 22). All PCR products were visualized by incorporation of 32P-dATP during the amplification, followed by Phosphorimager analysis of polyacrylamide gels. CSR was also estimated by quantification of the secreted Ig after culture of splenic B cells (from a single mouse or a pool of two mice) for seven days with B cell activators and cytokines. Analysis of secreted transgenic IgG1a employed ELISA with anti-IgG1a coating antibodies and anti-IgG1 developing antibodies. Analysis of secreted transgenic IgG2a employed anti-Flag coating antibodies and anti-IgG2a developing antibodies. Analysis of secreted transgenic IgA employed anti-IgA coating antibodies, an intermediate AD8 monoclonal antibody (to the transgenic idiotype), and an anti-rat IgG developing antibody. All ELISA developing antibodies were conjugated to alkaline phosphatase.
Results
Generation of ARS/Igh transgenes with deletion of two, three, or four 3′ enhancers
The starting point for this study was a BAC of the heavy chain constant region locus, which included a VDJH2 exon placed in the JH region by homologous recombination, all eight constant region genes, the four known 3′ enhancers, and an additional 15 kb 3′ of the enhancers (17). We have called this BAC “ARS/Igh”. Since the BAC is derived from an Igha locus, and we breed the transgene onto an Ighb background (C57BL/6), there are numerous allotypic and nucleotide polymorphic differences between the transgene and endogenous genes. Using gene targeting in E. coli (20), we made 1.1–1.3 kb deletions of HS1,2 and HS3B in one BAC, and 1.1–1.3 kb deletions of HS3A, HS3B and HS1,2 in a second BAC. In both BACs, HS4 was flanked by loxP sites (Fig. 1A). Generation of female transgenic mice with either of these BACs and an EIIa Cre transgene (21) resulted in offspring with an additional 1.1 kb deletion of HS4. Three to six lines of mice were generated with each of these four types of ARS/Igh transgenes. Each line of transgenic mice was tested for fourteen DNA segments spanning the ARS/Igh transgene. All lines had at least one complete copy of the ARS/Igh transgene. In the majority of lines the copy number was constant across the entire transgene, indicating that all copies were complete (Fig. S1). In two cases, after Cre-mediated deletion, the transgene copy number was reduced by one: 842 to 842Δ and 923 to 923Δ.
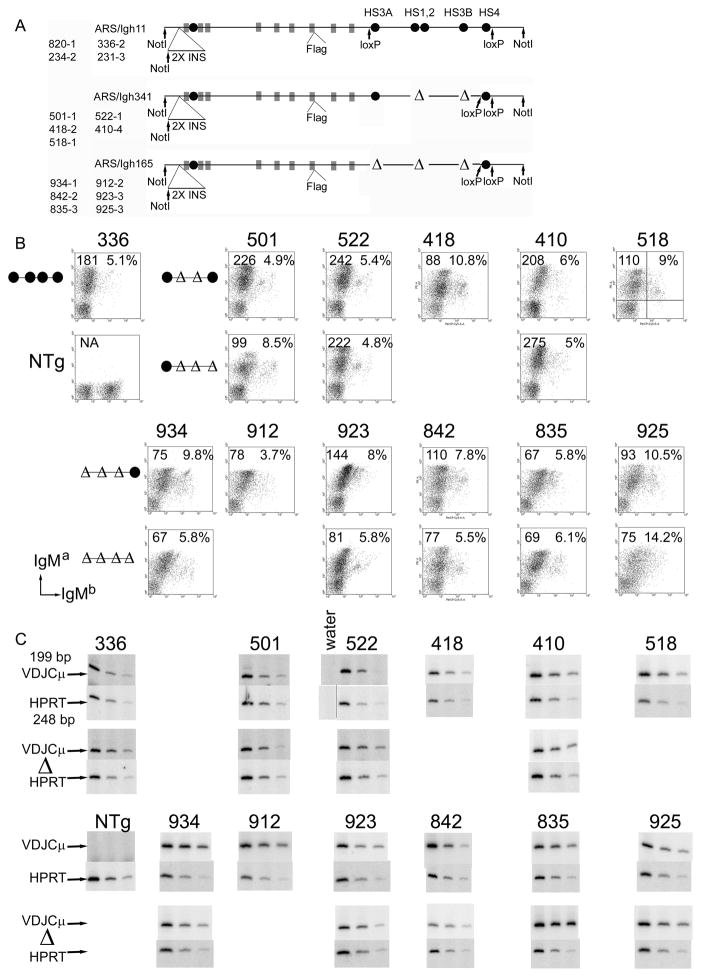
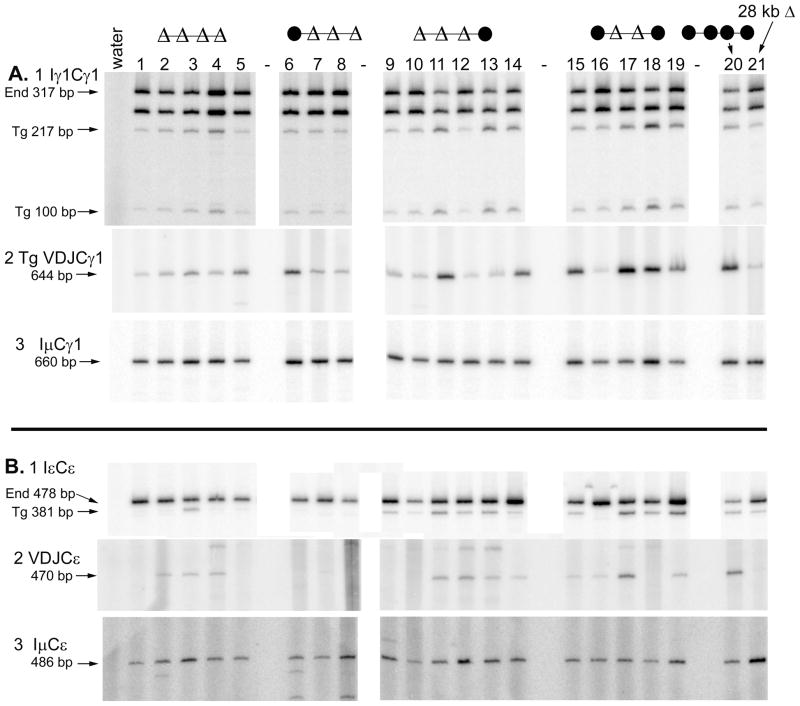
FIGURE 1.
Deletions of 3′ enhancers from a BAC containing the heavy chain constant region locus. A. Transgenic lines are listed to the left of schematics by their founder number, followed by a dash and the transgenic JH copy number of the line. Coding exons are depicted as grey boxes; enhancer elements are depicted as black circles. Insertion of two copies of the chicken β-globin insulator 3 kb 5′ of the VDJH2 exon and insertion of a 24 bp Flag tag-encoding element at the carboxy terminus of secreted γ2a are noted. Four bp insertions in the Iγ1 and Iγ2a regions are not shown (22). 1.1 to 1.3 kb deletions of enhancer elements are depicted as “Δ”s. HS4 is flanked by loxP sites, allowing its deletion by breeding with Cre-expressing transgenic mice. After Cre-mediated deletion, transgenic lines are designated by their founder number with a Δ. B. Analysis of cell surface transgenic IgMa and allelic exclusion by flow cytometry. Founder line numbers are shown above each panel. The composition of the 3′ enhancer region is shown schematically to the left of each set of transgenic lines. The mean fluorescence intensity of the IgMa positive cells is shown in the upper left hand corner of each panel; the percentage of all IgMa positive cells that are also IgMb positive is shown in the upper right hand corner of each panel. The percentage of all IgM positive cells that are IgMb only positive ranged from 0 to 1.9%. An example of how the quadrants were defined is shown for the 518 sample. Part B is derived from 8 experiments: (1) 501, 501Δ, 522, and 522Δ. (2) 923 and 923Δ. (3) 925 and 925Δ. (4) 336 and NTg. (5) 410 and 410Δ. (6) 934, 934Δ, 835, 835Δ, and 912. (7) 518. (8) 418, 842 and 842Δ. C. Expression of LPS-induced (3 day cultures) VDJCμ transcripts and HPRT transcripts was tested by two or three five-fold dilutions of cDNA by semi-quantitative RT-PCR. These cDNA samples were tested in three (HPRT) or four (VDJCμ) experiments, and the gel images were re-organized to construct this figure. For each transgenic line (indicated above the gel images, the version before Cre-mediated deletion is shown in upper set of VDJCμ and HPRT samples, and the version after Cre-mediated deletion (“Δ”) is shown in the lower set. The grey line between the “water” and left-most “522” HPRT sample indicates that the “water” lane was moved from a different location on the same gel as the “522” samples with which it was originally tested.
Splenic B cells were tested for allelic exclusion by analysis of surface IgMa (transgenic) and IgMb (endogenous, Fig. 1B). Mice with an intact ARS/Igh transgene (for example, line 336) demonstrate good allelic exclusion of heavy chain expression; about 95% of the splenic B cells express transgenic IgMa only and the remaining splenic B cells express both transgenic and endogenous IgM (Fig. 1B). All of the transgenic lines with deletion of two, three, or four of the 3′ enhancers retained similar levels of allelic exclusion as line 336 with an intact ARS/Igh transgene, although a few lines showed higher percentages of IgMa/IgMb double positive cells (Fig. 1B), which we did not investigate further. Although there was some variation in the level of surface IgMa expression (as measured by mean fluorescence intensity), this variation was primarily due to differences in one experiment to another; there was no consistent change in surface IgMa expression that correlated with number of enhancers deleted. We also tested RNA from transgenic B cells cultured in LPS for expression of transgenic μ mRNA by semi-quantitative RT-PCR (Fig. 1C). Although there were small changes in μ mRNA expression (both increased and reduced), on average, transgenes with deletion of two, three, or four of the 3′ enhancers expressed about the same amount of μ mRNA as did intact transgenes.
Class switch recombination to γ2a
Transgenic B cells were cultured in vitro for seven days, and the amount of Flag-tagged IgG2a in the culture supernatant was measured, as an estimate of CSR to the transgenic γ2a gene (Fig. 2A). In B cells activated with CD40L, with CSR to γ2a directed by IFN-γ, the deletion of HS1,2 and HS3B led to a reduction in the amount of transgenic IgG2a compared to intact ARS/Igh transgenes, but the levels of transgenic IgG2a were greater than the background level detected in supernatants of nontransgenic cells. Further deletion of HS3A or HS4 resulted in a further reduction in secreted transgenic IgG2a. Deletion of all four 3′ enhancers resulted in transgenic IgG2a levels that were very similar to the nontransgenic control (Fig. 2A). We also measured the amount of total IgM secreted into the supernatants of many of the cultures tested for IgG2a secretion (Fig. 2B). All B cell cultures secreted reasonable amounts of IgM, indicating that B cell activation was successful. Among cultures with different 3′ enhancer deletions in the transgene, there was some variability in the amount of IgM secreted, with transgenes lacking all four 3′ enhancers resulting in the smaller amounts of IgM secreted (Fig. 2B). The differences in IgM secretion were not statistically significant. The secretion of transgenic IgG2a from B cells cultured in LPS + IFN-γ is not as robust as that from B cells cultured in CD40L + IFN-γ. Nevertheless, a similar relationship between expression of transgenic IgG2a and deletion of various 3′ enhancer elements was observed in B cells activated by LPS + IFN-γ (Fig. 2C).
FIGURE 2.
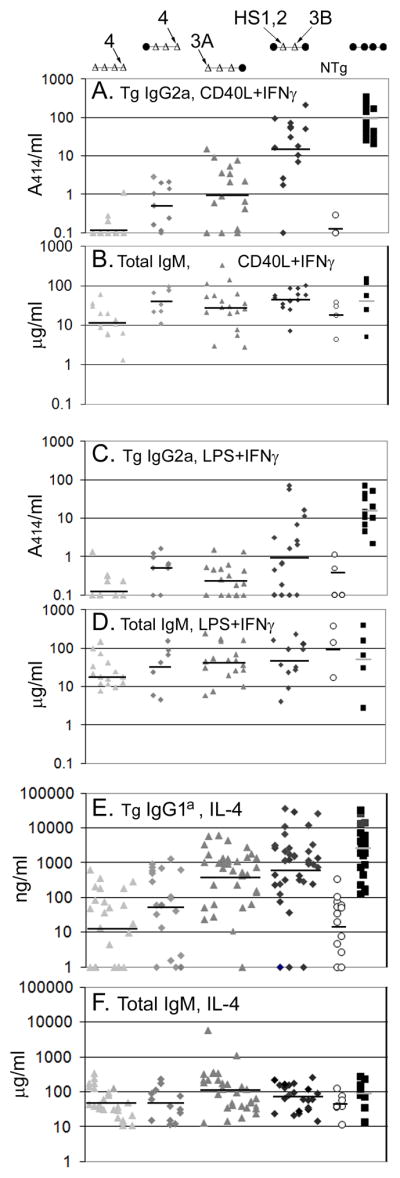
Secretion of transgenic IgG and IgM by transgenic B cells. The composition of the 3′ enhancer region is shown schematically above each set of transgenes. B cells from various transgenic lines were cultured with the indicated B cell activators and cytokines for seven days, and secreted Igs were quantified by ELISA. Results from ARS/Igh transgenes lacking all four 3′ enhancers are depicted by light grey triangles, from transgenes lacking HS3B, HS4, and HS1,2 are depicted by light grey diamonds, from transgenes lacking HS3A, HS3B, and HS1,2 are depicted by dark grey triangles, from transgenes lacking HS1,2 and HS3B are depicted by dark grey diamonds, from non-transgenic B cells are depicted by open circles, and from intact ARS/Igh transgenes are depicted by black squares. Results from multiple cultures of each independent transgenic line are shown in each vertical stack, with transgene copy number increasing from left to right (with some exceptions). This order of presentation of transgenic lines is also used in Figs. 3 and 6. Geometric means of the data from each type of transgene are shown as black horizontal lines (grey horizontal lines for intact ARS/Igh transgenes). A. Secretion of transgenic Flag+ IgG2a from B cells cultured in CD40L+IFN-γ. B. Secretion of total IgM from B cells cultured in CD40L+IFN-γ. C. Secretion of transgenic Flag+ IgG2a from B cells cultured in LPS+IFN-γ. D. Secretion of total IgM from B cells cultured in LPS+IFN-γ. E. Secretion of transgenic IgG1a (or total IgM, part F) with cultures in LPS+IL-4 in the left one-half of each group and cultures in CD40L+IL-4 in the right one-half of each group. Statistical analysis of the data in this Figure is presented in Supplemental Table I.
For ARS/Igh transgenes with deletions of HS1,2 and HS3B or of HS3A, HS3B, and HS1,2, the amounts of secreted transgenic IgG2a can be highly variable, spread out over nearly three logs (Fig. 2A and C). Some of this variation is intrinsic to the culture system and ELISA (the values from both the intact ARS/Igh and nontransgenic B cells vary over a little more than 1.5 logs, Fig. 2A and C). Another component is variation among different lines with the same transgenic construct. For example, line 522 was different from other transgenic lines with the deletions of HS1,2 and HS3B; secreted IgG2a values were always very low (Fig. 2A and C, second stack from the left of five stacks of dark grey diamonds). In fact, for the other four transgenic lines with deletion of HS1,2 and HS3B, IgG2a secretion induced by CD40L + IFN-γ was virtually the same as that from B cells with an intact ARS/Igh transgene (Fig. 2A). We will present in subsequent figures that this poor expression in line 522 extends to ε, α, and all four γ heavy chain genes, including expression of germline transcripts from each of these genes. We suggest that the transgene in line 522 inserted into a chromosomal region that was not favorable for the expression of switched heavy chain genes, in the absence of two of the four 3′ enhancers. In turn, we hypothesize that some of the large variation in expression, when two or three 3′ enhancers are deleted, is due to a role of the 3′ enhancers as a locus control region (see Discussion). Due to this insertion site variation, we have focused our analysis not on individual transgenic lines, but on the “average” expression of all transgenic lines with the same construct. This variation from one transgenic line to another did not extend to the transgenes in which each of the four 3′ enhancers had been removed by 1.1–1.3 kb deletions. Supernatants from these B cells had a distribution of secreted IgG2a values that was virtually identical to nontransgenic mice (Fig. 2A and C).
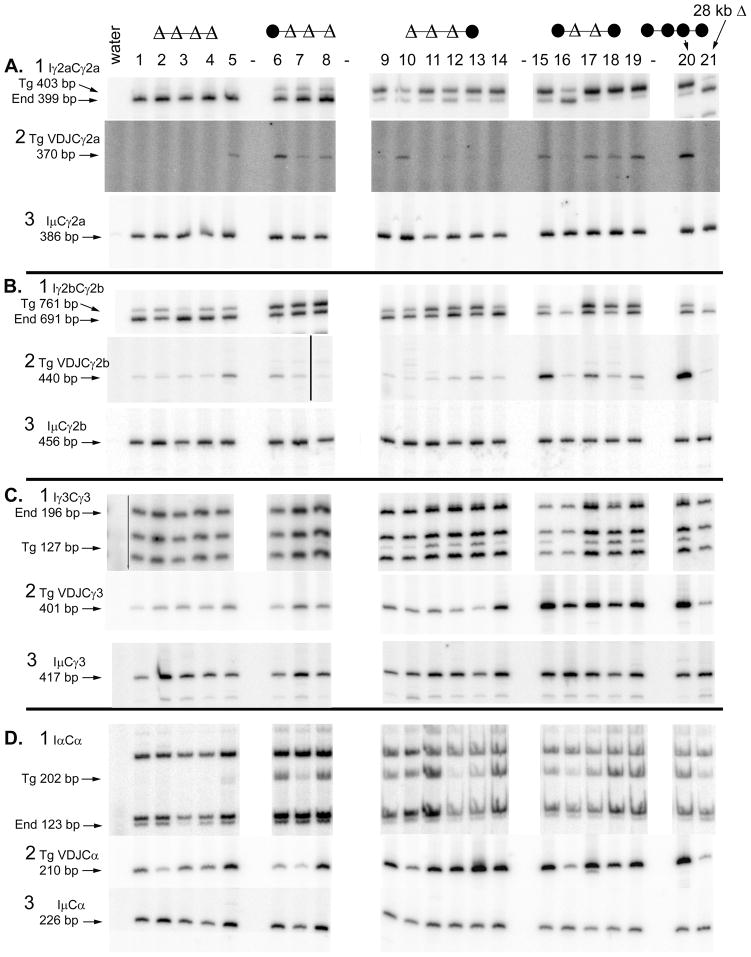
We also estimated CSR by measuring the expression of post-switch VDJCγ2a transcripts from cells cultured in CD40L + IFN-γ. All samples from various transgenic lines expressed IμCγ2a transcripts (24) (Fig. 3A3). IμCγ2a transcripts can be derived from either the transgene or the endogenous genes, as the endogenous genes will undergo CSR even if they lack an in-frame VDJ exon. To the extent possible, we attempted to equalize the amount of the IμCγ2a transcripts in each sample, so that the amount of B cell activation and total cytokine-induced switch recombination would be more or less equal in all samples. The IμCγ2a (and other IμCH transcripts presented subsequently) are a normalization control for these experiments. Expression of transgenic VDJCγ2a post-switch transcripts paralleled the data on secreted transgenic IgG2a. Whereas B cells with a single copy of an intact ARS/Igh transgene expressed VDJCγ2a transcripts, B cells with transgenes with a 28 kb deletion of the 3′ enhancer region did not express detectable levels of these transcripts (Fig. 3A2, lanes 20 and 21). Some post-switch VDJCγ2a transcripts were expressed from transgenes with a deletion of HS1,2 and HS3B (Fig. 3A2, lanes 15 and 17–19). Very small or undetectable levels of these transcripts were expressed from transgenes with further deletions of HS3A, HS4, or both (Fig. 3A2, lanes 1–14). Two exceptions were the samples from line 501Δ B cells (Fig. 3A2, lane 6) and line 912 (lane 10).
FIGURE 3.
Expression of germline and post-switch transcripts. The composition of the 3′ enhancer region is shown schematically above the lane numbers for each set of transgenes. Heavy chain mRNA expression was analyzed after three days of culture with the indicated B cell activators and cytokines. A. γ2a B. γ2b C. γ3 D. α. In each panel, B cells from the following transgenic lines were tested: lane 1, 934Δ; lane 2, 842Δ; lane 3, 923Δ; lane 4, 835Δ; lane 5, 925Δ; lane 6, 501Δ; lane 7, 522Δ; lane 8, 410Δ; lane 9, 934; lane 10, 912; lane 11, 842; lane 12, 923; lane 13, 835; lane 14, 925; lane 15, 501; lane 16, 522; lane 17, 418; lane 18, 410; lane 19, 518; lane 20, 820; and lane 21, 820Δ. Within each type of transgene, this arrangement places single copy transgene to the left and higher copy numbers to the right, with some exceptions (for example, lines 410 and 518 are reversed relative to their JH copy number). The same lane designations are used in subsequent figures. In subpart 1 of each Part, total germline transcripts were amplified by RT-PCR and digested with a restriction enzyme that allows transcripts of the endogenous and transgenes to be distinguished. In each subpart 1, the germline transcripts for lanes 1–8 and 9–21 were tested in separate experiments. In subpart 3 of each Part, the concentration of each cDNA was adjusted so that the amount of Iμ transcripts would be approximately equal, and then the same concentration of cDNA was tested for transgenic VDJ transcripts in subpart 2 of each Part. In Fig. S2, it is demonstrated that these RT-PCR are in the semi-quantitative range, and more complete quantification of some of the samples is shown.
Quantification of VDJCH transcripts
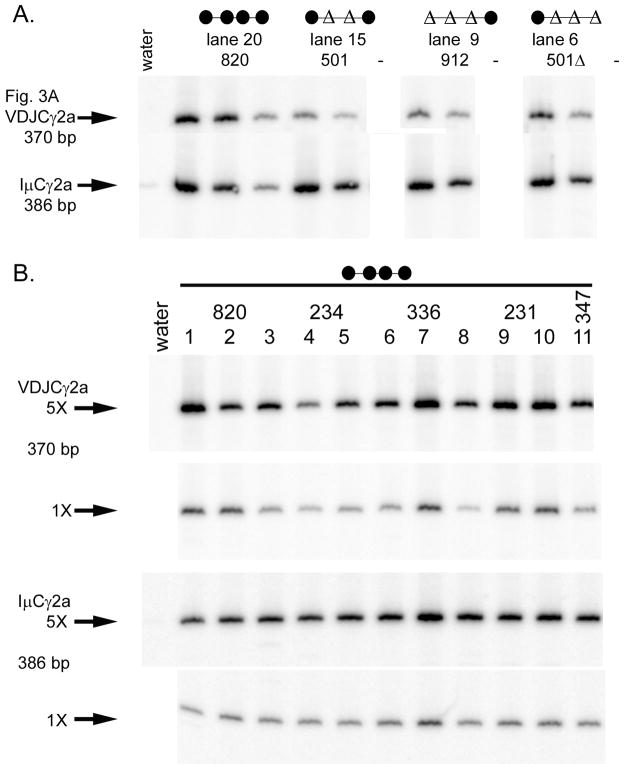
To quantify the amounts of VDJCγ2a transcripts in Lines 501Δ and 912, as well as a few other B cell samples from other transgenic lines, we performed semi-quantitative RT-PCR (Fig. 4A). We first completed a dilution series of IμCγ2a transcripts, confirming that the cDNA samples tested in Fig. 3A3 are more or less equalized. We then determined the dilution at which a given sample matched the dilution of a cDNA from B cells with an intact transgene. As an example of the quantification, we found that the VDJCγ2a signal for the 1:1 dilution of the line 501 (deletion of HS1,2 and HS3B) is a little greater than the 1:25 dilution of the line 820 (intact) sample (Fig. 4A). Hence, we conclude that the expression of VDJCγ2a transcripts in line 501 is approximately 8% that of line 820. Since line 501 (Fig. 3A1, lane 15) is near the middle of the expression level of all mice with a transgene with deletion of HS1,2 and HS3A, we estimate that these transgene expressed approximately 10% the amount of VDJCγ2a transcripts of intact transgenes.
FIGURE 4.
Semi-quantitative RT-PCR analysis of γ2a post-switch transcripts. A. cDNA samples from Fig. 3A, subparts 2 and 3, were amplified using the same cDNA concentration used in Fig. 3A and two additional five-fold dilutions (line 820 samples) or one additional five-fold dilution (other three samples). Lane numbers corresponding to Fig. 3A and transgenic line founder numbers are listed at the top. Empty lanes are indicated by dashes. B. cDNAs from wild type ARS/Igh transgenes (line 820, 1 copy, lanes 1–3; line 234, 2 copies, lanes 4–5; line 336, 2 copies, lanes 6–8; line 231, 3 copies, lanes 9–10; and line 347, 3 copies, lane 11) were first balanced for IμCγ2a expression. Then both VDJCγ2a and IμCγ2a transcripts were tested at identical “5x” and “1X” concentrations of the cDNA. Different lanes from the same transgenic line represent cDNAs from independent B cell cultures.
Our previous studies with wild type ARS/Igh transgenes have demonstrated the amount of VDJCH transcripts (relative to IμCH transcripts) from a given heavy chain gene is reasonably consistent among different lines with a wild type ARS/Igh transgene (17, 22). To confirm this consistency, we measured the relative expression levels of VDJCγ2a transcripts among one to three independent samples from five different lines with wild type transgenes (Fig. 4B). The majority of signals are consistent, within about a two-fold range. We did occasionally observe unusually high or low signals compared to other samples from the same line of transgenic mice; the sample in lane 1 has a markedly greater signal than the other two line 820 samples, and the sample in lane 4 has a markedly lower signal than the other line 234 sample (Fig. 4B). We also observed a few unusually high or low signals for samples from transgenic mice with deletions of two or three enhancers. The samples we present in Fig. 3A, part 2 are neither the highest nor the lowest relative VDJCγ2a signals for a given transgenic line, but rather are in the middle of the observed range. We also present representative samples for other heavy chain genes in subsequent Figures. Given the intrinsic errors in cell culture and the RT-PCR reaction, and the variation among transgenic lines discussed above, we have not emphasized differences in average VDJCH mRNA expression between wild type and transgenes with deletions of 3′ enhancers unless they are 10-fold or more.
Although 4% (1:25 dilution) of the VDJCγ2a expression level of an intact transgene is easily detectable (Fig. 4A, line 820), VDJCγ2a transcripts are not detectable in samples from most transgenes with deletion of three or four 3′ enhancers Fig. 3A3), and therefore we conclude that transgenes lacking three or four 3′ enhancers expressed less than 1% the level of VDJCγ2a transcripts of intact transgenes. From an examination of the semi-quantitative RT-PCR in Fig. 4A, we infer that B cells from line 501Δ expressed about 20% and B cells from line 912 expressed about 15% the amount of VDJCγ2a transcripts of intact transgenes. In subsequent Figures, we present data demonstrating that, in general, the exceptionally greater amounts of VDJCH transcripts have joined the transgenic VDJ exon to an endogenous CH gene. This would suggest that VDJCH transcripts do not result from CSR within transgenes lacking three or more 3′ enhancers.
Expression of γ2a germline transcripts
Expression of γ2a germline transcripts had some parallels to the expression of post-switch VDJCγ2a transcripts. Using the amount of endogenous germline transcripts as a normalization factor, transgenes with deletions of HS1,2 and HS3B expressed, on average, the same amount of germline transcripts as intact ARS/Igh transgenes (Fig. 3A1, compare lanes 15–19 to 20 and 21 and Supplementary Table II). (As noted above, line 522 in lane 16 is the exception, expressing very small amounts of γ2a germline transcripts.) Transgenes with an additional deletion of HS4 expressed 2% or less of the amount of germline transcripts of intact transgenes, similar to transgenes with the 28 kb enhancer region deleted (Fig. 3A1, compare lanes 1–8 to 20 and 21. See also Supplementary Table II). On the other hand, transgenes with deletions of HS3A, HS3B, and HS1,2 (lanes 9–14) expressed 61% the amount of germline transcripts of an intact transgene, (compare lanes 9–14 to 20 and 21 and Supplementary Table II). Nevertheless, these significant amounts of germline transcripts did not, in general, lead to CSR; transgenes with deletion of HS3A, HS3B, and HS1,2 expressed post-switch transcripts at 1% or less compared to the level of intact transgenes.
Class switch recombination to and germline transcription of γ2b, γ3, and α
The expression of γ2b germline transcripts revealed a different sensitivity to deletion of the 3′ enhancer elements. Only transgenes with deletions all four enhancers expressed γ2b germline transcripts at reduced levels, similar to deletion of the same four elements by a 28 kb deletion (Fig. 3B1, lanes 1–5 and 21, Supplementary Table II). All other combinations of enhancer deletions expressed γ2b germline transcripts as well as intact transgenes (Fig. 3B1, lanes 6–20). The expression of post-switch (VDJCγ2b) transcripts was different from that of germline transcripts. Transgenes with deletion of HS1,2 and HS3B expressed reduced (but significant) amounts of post-switch (VDJCγ2b) transcripts (Fig. 3B, lanes 15–19). Further deletion of HS4, HS3A, or both, resulted, in most transgenic lines, in dramatically reduced levels of post-switch transcripts very similar to that of the 28 kb enhancer region deletion (Fig. 3B2, lanes 1–14 and 21). Quantification of VDJCγ2b transcripts, as well as VDJCH transcripts from other heavy chain genes, is presented in Fig. S2.
Overall, 3′ enhancer-dependent expression of the γ3 gene was similar to γ2a or γ2b. Germline transcripts of the transgenic γ3 gene were expressed in transgenic B cells from most of the transgenic mice with deletions of HS1,2 and HS3B or with deletion of HS3A, HS3B, and HS1,2 (Fig. 3C1, lanes 9–19), but not in transgenic B cells with a further deletion of HS4 (Fig. 3C1, lanes 1–8, Supplementary Table II). Abundant post-switch VDJCγ3 transcripts were expressed by transgenes with deletion of two 3′ enhancers (Fig. 3C2, lanes 15–19), but not by transgenes with three or four enhancers deleted (Fig. 3C2, lanes 1–14). Remarkably, taking into account copy number, there are two transgenic lines with deletion of HS3A, HS3B, and HS1,2 that express reasonable amounts of germline transcripts, but switch to the γ3 gene poorly (Fig. 3C, lanes 11 and 13).
When normalized to the level of endogenous (123 bp fragment), transgenic germline transcripts from the α heavy chain gene are reduced by deletion of HS4, with transgenes lacking all four 3′ enhancers expressing no α germline transcripts (Fig. 3D1, lanes 1–5 and Supplementary Table II). We note the difference between transgenes with four 1.1–1.3 kb deletions and those with a 28 kb deletion of the 3′ enhancer region, which express significant α germline transcripts (17) and Fig. 3D1, lane 21). Many heavy chain transgenes with deletions of three or four 3′ enhancers expressed reduced amounts of VDJCα post-switch transcripts, but significantly more VDJCα transcripts than transgenes with a single copy of the 28 kb deletion (Fig. 3D2, compare lanes 1–12 to 20 and 21; see also Fig. S2D). In parallel, B cells with transgenes with three or four 3′ enhancers deleted secreted the same amounts of IgA with the transgenic V region idiotype as did B cells with an intact ARS/Igh transgene (not shown). Thus, we considered the possibility that CSR to α is less dependent on the 3′ enhancer region than are other heavy chain genes.
Transcripts with a transgenic VDJ exon joined to an endogenous CH region are commonly found in RNA from B cells with deletions of three or four 3′ enhancers from the transgene
We had previously reported that B cells with a 28 kb deletion of the 3′ enhancer expressed mRNAs with the VDJ exon joined to an endogenous Cγ1 or Cγ2a coding region (17). In various samples reported in this study, occurring in B cells with three or four of the 3′ enhancers deleted from their transgene, we observed amounts of VDJCγ or VDJCα transcripts that were increased relative to other B cells from lines with the same transgene (Fig. 3). We tested if these transcripts had endogenous CH regions by digesting them with restriction enzymes that distinguished the transgene and endogenous genes. For example, VDJCγ3 transcripts from a heavy chain transgene with deletion of HS1,2 and HS3B were almost entirely from the transgene, as they included the PstI site found in the transgene, but not in the endogenous genes (Fig. 5C, lane 18). On the other hand, transcripts from a transgenic sample with deletions of HS3A, HS3B, and HS1,2 were joined, almost entirely, to an endogenous Cγ3 gene, with a small amount of transgenic Cγ3 gene (Fig. 5C, lane 14). All transcripts from a transgenic sample with deletion of HS3B, HS4, and HS1,2 were joined to an endogenous Cγ3 region (Fig. 5C, lane 7). Similar results were obtained for RNA samples (from B cells with a transgene with deletions of all four 3′ enhancer elements) with exceptionally high levels of VDJCγ2a or VDJCγ2b transcripts (Fig. 5A and 5B, lane 5).
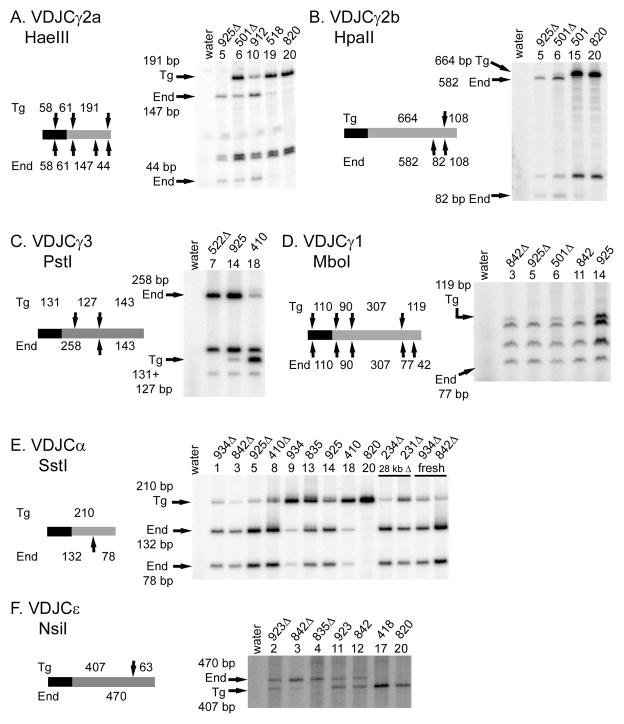
FIGURE 5.
Expression of transgenic VDJ transcripts with endogenous CH regions. A. γ2a. B. γ2b. C. γ3. D. α. E. γ1 F. ε. In each Part, lane numbers correspond to the lane numbers in the relevant parts of Figures 3 and 6. Transgenic line numbers are noted. Refer to Figs. 3 and 6 for the composition of the 3′ enhancers for each sample. Total VDJCH transcripts were amplified by RT-PCR and digested with a restriction enzyme, as indicated at the top of each Part. As shown in the schematic for each Part, the restriction enzymes distinguish transgenic and endogenous CH regions (shown in grey rectangles). Part of the VDJH2 exon is shown as a black rectangle.)
Line 501Δ was unusual in that it expressed significant VDJ transcripts with a transgenic Cγ2a region (Fig. 5A, lane 6). Using a PCR-based strategy, we cloned the chromosomal sequences next to the 3′ end of the ARS/Igh transgene in line 501, and found that the cloned sequences matched mouse chromosome 12 at 502 of 506 bp. Thus, the chromosomal insertion site for lines 501 and 501Δ is the middle of chromosome 12, about 45 Mb centromeric of the endogenous Igh locus. For the single-copy transgenic lines 501 and 501Δ, this insertion site correlates with elevated expression of transgenic γ, ε, and α heavy chain genes (Figs. 2, 3, and subsequent Figures). Perhaps in line 501Δ, deletion of three 3′ enhancers can be compensated by some cis-acting interaction with the endogenous heavy chain regulatory elements. This putative interaction would take place on the same DNA molecule, but at a distance of 45 Mb! Perhaps due to the distances involved in this interaction, CSR within the 501Δ transgene was sporadic. It did not occur to the γ2a gene in all samples, nor did it occur to the transgenic γ2b gene in the sample in Fig. 5B (lane 6).
Since virtually all transgenic B cells expressed significant amounts of VDJCα transcripts, we tested several samples for endogenous Cα regions. An obvious trend was observed: as more 3′ enhancers were deleted from the heavy chain transgene, VDJ transcripts shifted from predominant or complete use of the transgenic Cα region to almost complete use of an endogenous Cα region (with an extra SstI site, Fig. 5E). Although B cells from some lines with a transgene with deletions of each of the four 3′ enhancers express high levels of VDJCα transcripts (Fig. 3D2), virtually all of the transcripts result from VDJ joining to an endogenous Cα region (Fig. 5E). We tested if VDJCα transcripts were present in B cells before culture in vitro. These transcripts were found in B cells freshly isolated from the spleens of transgenic mice, and, like the VDJCα transcripts after culture, were characterized by an endogenous Cα region (Fig. 5E, “fresh” samples at the right end). Thus, some of these transcripts could arise from rare recombination events in vivo that are subject to strong selection. We had also observed some VDJCα transcripts from lines with two or three copies of a transgene with a 28 kb deletion of the 3′ enhancer region (17). We have now determined that these transcripts are also characterized by an endogenous Cα region (Fig. 5E, 234Δ and 231Δ). With four 1.1–1.3 kb deletions of the 3′ enhancers, CSR within the transgene to the α gene is so deficient that an aberrant recombination event to the endogenous Cα gene is more efficient. Alternatively, the VDJCα transcripts may arise from a transplicing event between transgenic VDJCμ transcripts and endogenous α germline transcripts (25, 26). Expression of VDJ exons from other heavy chain transgenes with endogenous CH regions have been shown to be the result of recombination events between the transgene and endogenous heavy chain genes (27, 28). CSR to the α gene is as dependent on the 3′ enhancers as is CSR to γ2a, γ2b, and γ3—heavy chain transgenes lacking all four 3′ enhancers switch very poorly within the transgene. In those B cells where CSR to α within the transgene is greatly impaired, it is replaced by trans-recombination or trans-splicing events to the endogenous CH gene.
Expression of the γ1 heavy chain gene is less dependent on the 3′ enhancer elements
Deletion of HS1,2 and HS3B resulted in a small reduction in the amount of transgenic IgG1 secreted into cultures of transgenic B cells in IL–4 plus B cell activators (Fig. 2E). Deletion of HS3A, HS3B, and HS1,2 led to an apparent further reduction in secreted IgG1. However, the transgenic IgG1 expression in lines with deletion of HS3A, HS3B, and HS1,2 remained significantly greater than that from nontransgenic mice. Further deletion of HS4 or HS4 and HS3A resulted in amounts of IgG1 secretion that are similar to nontransgenic controls. Expression of post-switch VDJCγ1 transcripts followed a similar pattern. Excepting line 522 (Fig. 6A2, lane 16), transgenes with deletions of HS1,2 and HS3B expressed large amounts of VDJCγ1 transcripts (Fig. 6A2, compare lanes 15 and 17–19 to 20). Transgenes with deletions of HS3A, HS3B, and HS1,2 expressed reduced amounts of post-switch γ1 transcripts, which in two lines could be distinguished from the expression from a transgene with the 28 kb deletion of the 3′ enhancer region (Fig. 6A2, compare lanes 11 and 14 to 21). Most lines with transgenes that additionally lack HS4 expressed VDJCγ1 transcripts at about the same level as the transgene with the 28 kb deletion (compare lanes 1–8 to 21). Similar to other γ heavy chain genes, in transgenes with deletions of three or four 3′ enhancers, expression of significant amounts of VDJCγ1 transcripts was associated with expression of an endogenous Cγ1 region (Fig. 5D). Regardless of 3′ enhancer deletions, all transgenes expressed significant amounts of γ1 germline transcripts from the transgene. Like the transgene with the 28 kb 3′ enhancer region deletion, transgenes with deletions of four 3′ enhancers expressed, on average, approximately 20% the amount of γ1 germline transcripts of an intact ARS/Igh transgene (Fig. 6A1, compare lanes 1–5 to 20 and 21, and Supplementary Table II).
FIGURE 6.
Expression of transgenic germline and post-switch γ1 (A) and ε (B) transcripts. This Figure is organized like Fig. 3.
Expression of post-switch VDJCε transcripts (also induced by IL-4) had many similarities to the expression of other heavy chain transcripts. Many transgenes with deletions of HS1,2 and HS3B or of HS3A, HS3B, and HS1,2 expressed reduced amounts of post-switch ε transcripts compared to an intact transgene, but greater amounts than expressed by a transgene with the 28 kb 3′ enhancer region deletion (Fig. 6B2, lanes 9–21). Transgenes with further deletion of HS4 did not express detectable VDJCε transcripts or expressed transcripts with the VDJ exon joined to endogenous Cε regions (Fig. 6B2, lanes 1–8 and Fig. 5F, lanes 2–4). Transgenic germline ε transcripts were expressed in reduced amounts in transgenes with deletions of HS3B and HS1,2, or deletions of HS3A, HS3B, and HS1,2. Germline transcripts from the transgenic ε gene were essentially not expressed in transgenes lacking HS4 (Fig. 6B1).
Discussion
Transgene expression, as measured by expression of the transgenic μ heavy chain gene, is not altered significantly by deletion of the 3′ HS sites
Transgenes with deletion of two, three, or four of the 3′ HS elements excluded the endogenous alleles to about the same extent, expressed about the same amount of surface IgM as intact ARS/Igh transgenes, and expressed about the same amount of LPS-induced VDJCμ transcripts (Fig. 1B and 1C). Also, there was no statistically significant effect on the amount of IgM secreted in tissue culture, although the two heavy chain transgenes lacking HS4 often secrete nominally smaller amounts of IgM (Fig. 2B, 2D, 2F). Our previous results revealed that some transgenic lines had a reduction in surface IgM expression upon deletion of the 28 kb enhancer region (17). Expression of the μ gene from our transgene also differs slightly from the endogenous locus. For the endogenous locus, a 30 kb deletion of the 3′ enhancer region resulted in a reduction to 7% of the amount of secreted IgM of a wild type heavy chain locus (18). However, like our heavy chain transgenes with four deletions of the 3′ enhancer elements, the 30 kb deletion from the endogenous locus had no measurable effect on surface IgM expression (18). On the other hand, deletion of HS4 only from the endogenous locus results in a small reduction in the amount of surface IgM (29). These apparent contradictions for both transgenic loci and the endogenous locus are probably a function of the small magnitude of the changes in μ expression. Any reduction in transgenic IgM expression is small compared to the changes in CSR.
There is a progressive loss in both germline transcription and CSR with additional deletion of 3′ HS sites
It is well-established that deletion of single 3′ enhancer elements from the endogenous locus had little effect on germline transcription or on CSR (29–31), whereas deletion of both HS3B and HS4 had a dramatic effect (16). Hence, we focused on deletion of two or more elements. Regardless of the specific heavy chain gene examined, deletion of more 3′ enhancers resulted in progressive loss of both germline transcription and CSR (Figs. 2, 3, and 6). Deletion of HS1,2 and HS3B led to a modest reduction (if any) in germline transcription for all heavy chain genes and a modest reduction in CSR to γ3 and γ1, but a measurable reduction in CSR to γ2a, γ2b, ε, and α (Figs. 2, 3, 6 and S2). For most heavy chain genes, deletion of HS3A, HS3B, and HS1,2 resulted in a reduction in germline transcription ranging from 16 to 61% of wild type levels (γ1 and γ2b being the exceptions) and in CSR. Likewise, for most heavy chain genes, deletion of HS3B, HS4, and HS1,2 or of all four 3′ HS elements resulted in almost complete loss of CSR and germline transcription that was 2 to 42% of wild type levels (Supplementary Table II).
The pattern of germline transcription, CSR, and deletion of 3′ HS elements indicated a special function for HS4. The most dramatic reduction on both germline transcription and CSR, for all heavy chain genes, was found in the two heavy chain transgenes that lacked HS4, in the context of other 3′ enhancer deletions. With our limited analysis, the best comparison is with HS3A. The transgene that retained only HS3A demonstrated levels of both germline transcription (3 to 10% of wild type levels) and CSR that were near that of non-transgenic mice, but the transgene that retained only HS4 had significant germline transcription (16 to 142% of wild type levels) and some CSR for some heavy chain genes. Differences in heavy chain gene expression from the heavy chain transgene with a deletion of HS1,2 and HS3B (this study) and the endogenous locus with a deletion of HS3B and HS4 (16) are also consistent with a special role for HS4. From the deleted transgene (retaining HS4), expression of γ3 was not reduced and expression of γ2b was partially reduced, whereas from the deleted endogenous locus (lacking HS4), the expression of both was reduced to near background levels (16). The modest (if any) reduction in germline transcription and CSR from a heavy chain transgene lacking both HS3A and HS3B is also consistent with a special role for HS4 (32).
The 3′ enhancers are a locus control region
Madisen and Groudine suggested that the four 3′ enhancers together form a locus control region, as they conferred upon genes inserted into the DNA of a cell line copy number-dependent and insertion site-independent expression (6). The classic definition of a locus control region is insertion site-independent, copy number-dependent expression in transgenic mice. We have found that CSR within intact ARS/Igh transgenes was independent of insertion site for thirteen different transgenic lines (all of the lines we examined) (17, 33). It has been difficult to test copy number dependence of post-switch transcripts, because the majority of heavy chain transgenes have one or two copies. RT-PCR is not well-suited to detection of these two-fold differences. However, DC-PCR tests of CSR to the γ2a gene showed some copy number dependence (17), perhaps because the endogenous DC-PCR product provides an internal normalization control. The wide variation in expression levels of transgenes with deletions of HS3B and HS1,2 or of HS3A, HS3B, and HS1,2 (Fig. 2) is completely consistent with a role for the four 3′ enhancers in forming a locus control region. In the absence of two or three of the 3′ enhancers, the putative locus control region would be disabled, and CSR of the heavy chain transgene would become dependent on insertion site. Not only do different transgenic lines express different amounts of CSR, but within a single transgenic line, the amount of CSR in B cells from different mice can vary widely (for example, Fig. 2E, transgenes with deletion of HS1,2 and HS3B). This suggests that the locus control region may also prevent epigenetic modifications that are not uniform, but effected in a subset of mice. We note that the four 3′ enhancers are a locus control region for only germline transcription and CSR. Transgenic VDJCμ expression is more or less independent of the four 1.1–1.3 kb segments that include the DNase I hypersensitive sites themselves, although the 24 kb in between them may play some role in μ expression (17, 18).
The γ1 heavy chain gene is uniquely resistant to deletion of the 3′ enhancers
In almost all studies of deletion or substitution of the 3′ enhancer elements, expression of the murine γ1 gene has been reduced less than the expression of the α, ε, and other three γ heavy chain genes (14–16). Germline transcription of γ1 heavy chain transgenes, in the absence of any 3′ enhancer elements, appears to be more robust than germline transcripts from similar transgenes of the γ2a or ε heavy chain genes (34–36). Expression of the γ1 gene is often less susceptible to deletion of trans-acting factors (37). We also found that CSR to the γ1 gene is well supported by HS3A and HS4 only, and is supported to some extent by HS4 alone (Figs. 2E and 6A).
Substantial germline transcripts are expressed without subsequent CSR
The 3′ enhancer region physically associates with the promoters for germline transcription and/or the switch regions of various heavy chain genes (38). Is the function of this physical association to merely enhance germline transcription, opening the switch region to AID activity, which in turn results in CSR? Bottaro and colleagues found that germline transcription through a switch region, in conditions where AID should have been induced, was not sufficient for CSR (39). We report herein substantial germline transcripts without CSR of the γ2a gene in transgenes that retain only HS4 (Fig. 3A1), of the γ2b gene in transgenes with three or four of the 3′ enhancers deleted (Fig. 3B1), of the γ3 gene in transgenes that retain only HS4 (Fig. 3C1), of the α gene in the transgenes that retain only HS3A (Fig. 3D1), and of the γ1 gene in transgenes with deletion of HS4 (Fig. 6A1). In each case, the B cells expressed AID, as the IμCH post-switch transcripts demonstrate CSR of the endogenous genes. In the absence of some 3′ enhancers, large amounts of properly spliced germline transcripts can be produced, but these are not sufficient to promote CSR. Therefore, the 3′ enhancers must have a function to enhance recombination that is in addition to their function for enhancing germline transcription (17, 39). We did observe some post-switch transcripts in the absence of germline transcription. In every case, this did not result from CSR within the heavy chain transgene, but was due to expression of a transgenic VDJ joined to an endogenous CH gene. If anything, these transcripts emphasize how dramatically the transgene is disabled for CSR. Expression of transgenic VDJ transcripts joined to an endogenous CH region was sporadic, not consistently observed in independent RNA samples from the same line of transgenic mice. We detected them in B cells before induction of CSR in tissue culture (Fig. 5E). Thus, it is very difficult to estimate the rate at which these “trans” transcripts are produced. Nevertheless, it is clear that the rate of CSR within transgenes not expressing germline transcripts of a given heavy chain gene is much lower.
Deletion of the four 3′ enhancers results in a reduction to less than 1% CSR for all heavy chain genes
The most robust result from these studies is that transgenes lacking the 3′ enhancers, after four 1.1–1.3 kb deletions, were reduced to very low levels of CSR. In parallel, germline transcription was dramatically reduced after the four deletions. These results are similar to those of the heavy chain transgene (28 kb deletion) or the endogenous locus (30 kb deletion) lacking the entire 3′ enhancer region (17, 18). Hence, the 24 kb of DNA in between the four DNase I hypersensitive sites play, at most, a minor role in enhancement of CSR. As we discuss above, other sequences in the 3′ enhancer region may have a positive role in μ heavy chain gene expression. They might also have a negative effect on expression of germline transcripts of the α heavy chain gene (Fig. 3D1, compare lanes 1–5 to 21). Alternatively, the 28 kb deletion may bring positive control elements 3′ of HS4 (40) close enough to the α gene so that they are able to enhance germline transcripts (Fig. 3D1, lane 21). The promoter regions for germline transcripts have a major role in determining the cytokine specificity of CSR (1, 41).
However, the sum of the evidence presented here indicates that other elements in the heavy chain constant region locus do not influence the absolute level of CSR. It is the four enhancer elements/DNase I hypersensitive sites 1–4 that enhance CSR to all heavy chain genes. Effective immune responses in mammals require that large amounts of the optimal type of antibody be produced, as quickly as possible. The primary goal of all vaccinations currently in use is to produce the right isotype of high affinity antibody. We have demonstrated that this ability to produce the optimal type of antibody is the result of an interaction among the four 3′ enhancer elements.
Supplementary Material
Acknowledgments
We thank Drs. Marilia Cascalho, Douglas Engel, and Jeffery Platt for discussions during the execution of these studies and their comments on the manuscript. We acknowledge Wanda Filipiak and Thom Saunders for preparation of transgenic mice and the Transgenic Animal Model Core of the University of Michigan’s Biomedical Research Core Facilities.
Footnotes
This work was supported by the National Institutes of Health (AI076057 to WD). Transgenic core support was provided by The University of Michigan Cancer Center (CA46592).
Abbreviations: BAC, bacterial artificial chromosome; CSR, class switch recombination; End, endogenous; I, 5-most exon of heavy chain germline transcripts; HS, (DNase I) hypersensitive site; NTg, non-transgenic; Tg, transgenic
References
- 1.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. In: Paul WE, Littman DR, editors. Annual Reviews. Palo Alto, CA: 2008. pp. 261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cogne M, Birshtein BK. Regulation of class switch recombination. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B Cells. Academic Press; London: 2004. pp. 289–305. [Google Scholar]
- 3.Coutinho A, Forni L. Intraclonal diversification in immunoglobulin isotype secretion: an analysis of switch probabilities. EMBO J. 1982;1:1251–1257. doi: 10.1002/j.1460-2075.1982.tb00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregor PD, Morrison SL. Myeloma mutant with a novel 3′ flanking region: loss of normal sequence. Mol Cell Biol. 1986;6:1903–1916. doi: 10.1128/mcb.6.6.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberson R, Ong J, Shi X, Eckhardt LA. Immunoglobulin gene transcription ceases upon deletion of a distant enhancer. EMBO J. 1995;24:6229–6238. doi: 10.1002/j.1460-2075.1995.tb00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madisen L, Groudine M. Identification of a locus control region (LCR) in the immunoglobulin heavy chain locus that deregulates c-myc expression in plasmacytoma and Burkitt’s lymphoma cells. Genes & Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- 7.Michaelson JS, Giannini SL, Birshtein BK. Identification of 3′ α-HS4, a novel Ig heavy chain enhancer element regulated at multiple stages of B cell differentiation. Nuc Acids Res. 1995;23:975–981. doi: 10.1093/nar/23.6.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannini SL, Singh M, Calvo CF, Ding G, Birshtein BK. DNA regions flanking the mouse Ig 3′ α enhancer are differentially methylated and DNase I hypersensitive during B cell differentiation. J Immunol. 1993;150:1772–1780. [PubMed] [Google Scholar]
- 9.Pettersson S, Cook GP, Bruggemann M, Williams G, Neuberger MS. A second B cell specific enhancer 3′ of the immunoglobulin heavy-chain locus. Nature. 1990;344:165–168. doi: 10.1038/344165a0. [DOI] [PubMed] [Google Scholar]
- 10.Dariavach P, Williams GT, Campbell K, Pettersson S, Neuberger MS. The mouse IgH 3′ enhancer. Eur J Immunol. 1991;21:1499–1504. doi: 10.1002/eji.1830210625. [DOI] [PubMed] [Google Scholar]
- 11.Lieberson R, Giannini SL, Birshtein BK, Eckhardt LA. An enhancer at the 3′ end of the mouse immunoglobulin heavy chain locus. Nuc Acids Res. 1991;19:933–937. doi: 10.1093/nar/19.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattias P, Baltimore D. The immunoglobulin heavy chain locus contains another B-cell specific 3′ enhancer close to the α constant region. Mol Cell Biol. 1993;13:1547–1553. doi: 10.1128/mcb.13.3.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ong J, Stevens S, Roeder RG, Eckhardt LA. 3′ IgH enhancer elements shift synergistic interactions during B-cell development. J Immunol. 1998;160:4896–4903. [PubMed] [Google Scholar]
- 14.Cogne M, Lansford R, Bottaro A, Zhang J, Gorman J, Young F, Cheng HL, Alt FW. A class switch control region at the 3′ end of the immunoglobulin heavy chain locus. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Seidl KJ, Manis JP, Bottaro A, Zhang J, Davidson L, Kisselgof A, Oettgen H, Alt FW. Position-dependent inhibition of class-switch recombination by PGK-neor cassettes inserted into the immunoglobulin heavy chain constant region locus. Proc Natl Acad Sci USA. 1999;96:3000–3005. doi: 10.1073/pnas.96.6.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogne M. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- 17.Dunnick WA, Collins JT, Shi J, Westfield G, Fontaine C, Hakimpour P, Papavasilliou FN. Switch recombination and somatic hypermutation are controlled by the heavy chain 3′ enhancer region. J Expt Med. 2009;206:2613–2623. doi: 10.1084/jem.20091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogne N, Cogne M, Denizot Y. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010;116:1895–1898. doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- 19.Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, Murphy KM. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature Imm. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XW, Model P, Heintz N. Homologous recombination based modification in Esherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nature Biotech. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 21.Lasko M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunnick WA, Shi J, Graves KA, Collins JT. The 3′ end of the heavy chain constant region locus enhances germline transcription and switch recombination of the four γ genes. J Expt Med. 2005;201:1459–1466. doi: 10.1084/jem.20041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop GA, Warren WD, Berton MT. Signaling via major histocompatibility complex class II molecules and antigen receptors enhances the B cell receptor response to gp39/CD40 ligand. Eur J Immunol. 1995;25:1230–1238. doi: 10.1002/eji.1830250515. [DOI] [PubMed] [Google Scholar]
- 24.Li SC, Rothman PB, Zhang J, Chan C, Hirsh D, Alt FW. Expression of Iμ-Cγ hybrid germline transcripts subsequent to immunoglobulin heavy chain class switching. Int Immunol. 1994;6:491–497. doi: 10.1093/intimm/6.4.491. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu A, Nussenzweig MC, Han H, Sanchez M, Honjo T. Trans-splicing as a possible molecular mechanism for multiple isotype expression of the immunoglobulin gene. J Expt Med. 1991;173:1385–1393. doi: 10.1084/jem.173.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YW, Word CJ, Dev V, Uhr JW, Vitetta ES, Tucker PW. Double isotype production by a neoplastic B cell line. II. Allelically excluded production of μ and γ1 chains without CH gene rearrangement. J Expt Med. 1986;164:562–579. doi: 10.1084/jem.164.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerstein RM, Frankel WN, Hsieh CL, Durdik JM, Rath S, Coffin JM, Nisonoff A, Selsing E. Isotype switching of an immunoglobulin heavy chain transgene occurs by DNA recombination between different chromosomes. Cell. 1990;63:537–548. doi: 10.1016/0092-8674(90)90450-s. [DOI] [PubMed] [Google Scholar]
- 28.Giusti AM, Coffee R, Manser T. Somatic recombination of heavy chain variable region transgenes with the endogenous heavy chain locus in mice. Proc Natl Acad Sci USA. 1992;89:10321–10325. doi: 10.1073/pnas.89.21.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent-Fabert C, Truffinet V, Fiancette R, Cogne N, Cogne M, Denizot Y. Ig synthesis and class switching do not require the presence of the hs4 enhancer in the 3′ IgH regulatory region. J Immunol. 2009;182:6926–6932. doi: 10.4049/jimmunol.0900214. [DOI] [PubMed] [Google Scholar]
- 30.Manis JP, van der Stoep N, Tian M, Ferrini R, Davidson L, Bottaro A, Alt FW. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J Expt Med. 1998;188:1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bebin AG, Carrion C, Marquet M, Cogne N, Lecardeur S, Cogne M, Pinaud E. In vivo redundant function of the 3′ IgH regulatory element HS3b in the mouse. J Immunol. 2010;184:3710–3717. doi: 10.4049/jimmunol.0901978. [DOI] [PubMed] [Google Scholar]
- 32.Yan Y, Pleretti J, Ju Z, Wei S, Christin JR, Bah F, Birshtein BK, Eckhardt LA. Homologus elements hs3a and hs3b in the 3′ regulatory region of the murine immunoglobulin heavy chain (Igh) locus are both dispensable for class-switch recombination. J Biol Chem. 2011;286:27123–27231. doi: 10.1074/jbc.M111.230995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunnick WA, Shi J, Graves KA, Collins JT. Germline transcription and switch recombination of a transgene containing the entire heavy chain constant region locus: Effect of a mutation in a STAT6 binding site in the γ1 promoter. J Immunol. 2004;173:5531–5539. doi: 10.4049/jimmunol.173.9.5531. [DOI] [PubMed] [Google Scholar]
- 34.Elenich LA, Ford CS, Dunnick WA. The γ1 heavy chain gene includes all the cis-acting elements necessary for expression of properly regulated germline transcripts. J Immunol. 1996;157:176–182. [PubMed] [Google Scholar]
- 35.Collins JT, Dunnick WA. Interferon-γ regulated germline transcripts are expressed from γ2a transgenes independently of the heavy chain 3′ enhancers. J Immunol. 1999;163:5758–5762. [PubMed] [Google Scholar]
- 36.Laurencikiene J, Tamosiunas V, Severinson E. Regulation of ε germline transcription and switch region mutations by IgH locus 3′ enhancers in transgenic mice. Blood. 2007;109:159–167. doi: 10.1182/blood-2006-02-005355. [DOI] [PubMed] [Google Scholar]
- 37.Snapper CM, Zelazowski P, Rosas FR, Kehry MR, Tian M, Baltimore D, Sha WC. B cells from p50/NF-κB knockout mice have selective defects in proliferation, differentiation, germ-line CH transcription, and Ig class switching. J Immunol. 1996;156:183–191. [PubMed] [Google Scholar]
- 38.Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2008;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bottaro A, Lansford R, Xu L, Zhang J, Rothman P, Alt FW. S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO J. 1994;13:665–674. doi: 10.1002/j.1460-2075.1994.tb06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. Chromatin architecture near a potential 3′ end of the Igh locus involves modular regulation of histone modifications during B-cell development and in vivo occupancy at CTCF sites. Mol Cell Biol. 2005;25:1511–1525. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunnick WA, Shi J, Holden V, Fontaine C, Collins JT. The role of germline promoters and I exons in cytokine-induced gene-specific class switch recombination. J Immunol. 2011;186:350–358. doi: 10.4049/jimmunol.1003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.