Abstract
Within the testis, each Sertoli cell can support a finite number of developing germ cells. During development, the cessation of Sertoli cell proliferation and the onset of differentiation establish the final number of Sertoli cells and, thus, the total number of sperm that can be produced. The upstream stimulatory factors 1 and 2 (USF1 and USF2, respectively) differentially regulate numerous Sertoli cell genes during differentiation. To identify genes that are activated by USF proteins during differentiation, studies were conducted in Sertoli cells isolated from 5- and 11-day-old rats, representing proliferating and differentiating cells, respectively. Usf1 mRNA and USF1 protein levels were increased between 5 and 11 days after birth. In vitro studies revealed that USF1 and USF2 DNA-binding activity also increased at 11 days for the promoters of four potential target genes, Fshr, Gata4, Nr5a1, and Shbg. Chromatin immunoprecipitation assays confirmed that USF recruitment increased in vivo between 5 and 11 days after birth at the Fshr, Gata4, and Nr5a1 promoters. Expression of Nr5a1 and Shbg, but not of Fshr or Gata4, mRNAs was elevated in 11-day-old Sertoli cells compared with 5-day-old Sertoli cells. Transient transfection of USF1 and USF2 expression vectors up-regulated Nr5a1 and Shbg promoter activity. RNA interference assays demonstrated that USF1 and USF2 contribute to Nr5a1 and Shbg expression in differentiating cells. Together, these data indicate that increased USF levels induce the expression of Nr5a1 and Shbg during the differentiation of Sertoli cells, whereas Fshr and Gata4 expression is not altered by USF proteins during differentiation.
Keywords: developmental biology, differentiation, gene expression, gene regulation, male reproductive tract, Sertoli cell, testis, transcriptional regulation
The increased expression of upstream stimulatory factors 1 and 2 (USF1 and USF2) occurs when differentiation of rat Sertoli cells is sufficient to up-regulate Nr5a1 and Shbg gene expression.
INTRODUCTION
In the adult testis, Sertoli cells are required to provide the physical environment and factors necessary for germ cells to develop into mature sperm. Because Sertoli cells do not divide after puberty and Sertoli cell death is rare, the number of Sertoli cells is fixed in the adult; thus, the number of germ cells that can be supported is determined by the duration of the proliferation period before differentiation [1–5]. Therefore, the timing of Sertoli cell differentiation is critical for determining the final number of Sertoli cells and the upper limit of male fertility. Thus far, however, little is known about the gene regulatory events that occur during Sertoli cell differentiation, including the transcription factors that are required and the downstream target genes that are activated.
Upstream stimulatory factors 1 and 2 (USF1 and USF2, respectively) have been identified as potential mediators of Sertoli cell differentiation [6]. The USF proteins are members of the basic helix-loop-helix (bHLH) family of transcription factors that bind to specific DNA sequence motifs known as E-boxes (CACGTG) [7–9]. The results of studies with Usf knockout mice suggest that the Usf1 and Usf2 genes are partially redundant and that reduction of total USF protein levels below a specific threshold interferes with male reproductive function [10]. Specifically, in Usf2 knockout mice, Usf1 levels are also decreased, and a severe reduction in male reproductive capability is found. In contrast, Usf1 knockout mice have higher Usf2 levels and near-normal levels of USF DNA-binding activity that may compensate for the loss of Usf1 and permit the retention of fertility [10]. It is not yet known whether USF proteins are required for male fertility or Sertoli cell differentiation, because Usf1/Usf2 double-knockout mice are an embryonic lethal mutation [10] and, to our knowledge, cell-specific USF knockout studies have not been performed.
Previously, we found that the expression of Usf1 mRNA and levels of USF binding to E-box motifs increased during the differentiation of Sertoli cells [6]. To determine whether the increased DNA-binding activity of USF during differentiation results in the alteration of E-box-regulated gene expression in Sertoli cells, we investigated USF regulation of four target genes in Sertoli cells that contribute to maintaining fertility. Two of the target genes, follicle-stimulating hormone receptor (Fshr) and GATA binding protein (Gata4), are expressed at relatively constant levels throughout Sertoli cell differentiation and are not expected to be induced by USF proteins in vivo. The expression of steroid hormone-binding globulin (Shbg) increases during differentiation and, thus, is a candidate for regulation by USF proteins, whereas the expression of the other target gene, nuclear receptor subfamily 5, group A, member 1 (Nr5a1), during Sertoli cell differentiation is less well characterized.
The FSHR is required for FSH-mediated Sertoli cell proliferation, pubertal initiation of spermatogenesis, and maintenance of normal sperm production [11]. GATA4 is a transcription factor expressed throughout Sertoli cell development [12–14] and is required for proper sex determination and testicular organogenesis [12, 15]. Knockout of Gata4 specifically in Sertoli cells results in disrupted spermatogenesis due to the premature release of spermatocytes and spermatids [16]. The promoters of the Fshr and Gata4 genes are activated through E-box motifs by overexpression of USF proteins in cell-culture transfection studies [17–20]. However, the expression of these two genes is either stable during Sertoli cell differentiation (Fshr) [11, 21] or reportedly decreases (Gata4) [12–14]; thus, elevated USF levels are not expected to activate the genes during differentiation in vivo.
The Nr5a1 gene (also known as SF-1) encodes a transcription factor that activates genes essential for testicular organogenesis as well as the steroidogenic enzyme cytochrome P450, family 19, subfamily a, polypeptide 1 (Cyp19a1; commonly known as aromatase) [22–27]. The transcription of Nr5a1 requires USF1 and USF2 in cultured Sertoli cells [28], but the expression of Nr5a1 during Sertoli cell differentiation has not been fully characterized [22]. The Shbg gene product, also known as androgen-binding protein (ABP), functions to carry testosterone into the lumen of the seminiferous tubules and farther downstream through the male reproductive tract [29, 30]. Shbg is expressed by mature and maturing Sertoli cells and is therefore a marker of Sertoli cell differentiation. Expression of Shbg mRNA increases during Sertoli cell differentiation between 5 and 20 days after birth [31]. Studies in fully differentiated Sertoli cells revealed that two E-box motifs within the promoter are necessary for Shbg expression [32]. Thus far, the pattern of Shbg expression during Sertoli cell differentiation has not been well characterized, and to our knowledge, USF regulation of Shbg has not been investigated.
Because the factors that cause Sertoli cells to differentiate are not well characterized, we tested the hypothesis that USF1 and USF2 are required to increase gene expression in postnatal proliferating and differentiating Sertoli cells isolated directly from testes and in culture. USF protein levels and activities as well as the expression of Fshr, Nr5a1, Gata4, and Shbg target genes were assayed. DNA-protein interactions were examined at E-box motifs within the promoters of the Fshr, Nr5a1, Gata4, and Shbg genes before and during differentiation both in vitro, through the use of electrophoretic mobility shift assays (EMSAs), and in vivo, through the use of chromatin immunoprecipitation (ChIP) assays. USF1 and USF2 regulation of promoter activity was assessed for genes found to be induced during differentiation. Finally, RNA interference assays were performed to determine the contribution of USF1 and USF2 to the expression of genes essential for Sertoli cell-mediated support of fertility.
MATERIALS AND METHODS
Animal Care and Use
Male Sprague Dawley rat pups were obtained from Charles River Laboratories. Animals used in the present study were maintained and euthanized according to the principles and procedures described in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The present study was approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Isolation of Sertoli Cells for Direct Assay
Rats were euthanized 5 and 11 days after birth, and Sertoli cells were isolated as described by Anway et al. [33] with slight modifications [6]. Briefly, decapsulated testes were digested with collagenase (0.5 mg/ml) in enriched Krebs-Ringer bicarbonate medium (EKRB; 118.5 mM NaCl, 4.7 mM KCl, 2.1 mM CaCl2·H2O, 1.0 mM KH2PO4, 1.2 mM MgSO4·7H2O, 25 mM NaHCO3, 10 mM Hepes [pH 7.4], and 0.1% bovine serum albumin [BSA]) at 34°C for 10–15 min in a shaking water bath (80 oscillations/min), followed by three settlings in EKRB to isolate seminiferous tubules. Tubules were dispersed through digestion with trypsin (0.5 mg/ml) in the presence of DNase (1 μg/ml) for 5–10 min at 37°C without shaking. After digestion, tubule fragments were washed with soybean trypsin inhibitor (0.3 mg/ml), followed by two washes with EKRB. Sertoli cells were separated from germ cells by incubation with 0.1% collagenase, 0.2% hyaluronidase, 0.04% DNase I, and 0.03% trypsin inhibitor in a shaking water bath for 40 min at 34°C (80 oscillations/min). Cells were then collected by centrifugation 82 × g for 5 min and washed three times with EKRB. To remove contaminating germ cells, the suspension of single cells and clusters of 5–10 cells were subjected to hypotonic shock by resuspending the pellet in EKRB diluted with water (final concentration, 0.2× EKRB), then gently inverting three times to disperse cells, followed by immediate centrifugation at 700 rpm (63 × g) for 10 min. Cells were next washed three times with serum-free media containing 50% Dulbecco modified Eagle medium (DMEM), 50% Ham F-12, 5 mg/ml of insulin, 5 mg/ml of transferrin, 10 ng/ml of epidermal growth factor, 3 μg/ml of cytosine β-d-arabinofuranosidase, 1 mM sodium pyruvate, 200 U/ml of penicillin, and 200 μg/ml of streptomycin. The resuspended cells were counted and directly aliquoted for preparation of protein extracts or RNA or were plated for analysis of purity. Preparations of isolated cells are routinely 85% enriched for Sertoli cells [6].
Isolation of Sertoli Cells for Cell Culture
Rats were euthanized 5 and 11 days after birth as described previously [34]. Decapsulated testes were digested with collagenase (0.5 mg/ml, 33°C, 12 min) in EKRB, followed by three settlings in EKRB to isolate seminiferous tubules. The tubules were digested with trypsin (0.5 mg/ml, 32°C, 12 min) in the presence of DNase (2 μg/ml), and cell aggregates were passed repeatedly through a Pasteur pipette. An equal volume of DMEM containing 10% fetal bovine serum was added to the Sertoli cells, which were then subjected to centrifugation (500 × g, 5 min), after which the pellet was resuspended in the supplemented serum-free media as described above (Isolation of Sertoli Cells for Direct Assay). Sertoli cells were cultured on Matrigel (Collaborative Research)-coated dishes (33°C, 5% CO2). Sertoli cells were routinely 95% pure as determined by phase microscopy and alkaline phosphatase staining [35]. Cells were cultured for 3 days before transfection or fixation.
Western Blot Analysis
Sertoli cells were prepared for immediate assay in extract lysis buffer (250 mM NaCl, 0.1% Nonidet P40, 50 mM Hepes [pH 7.0], 5 mM ethylenediaminetetra-acetic acid [EDTA], 0.5 mM dithiothreitol [DTT], and a cocktail of protease and phosphatase inhibitors) and then sonicated and subjected to centrifugation (12 000 × g, 10 min). Supernatants containing whole-cell lysates were stored at −80°C. Protein concentrations were determined by the Bradford method (Bio-Rad Protein Assay; Bio-Rad Laboratories, Inc.). Protein was concentrated when necessary using a Microcon Ultracel YM-3 Centrifugal Filter Unit (Millipore). For detection of USF1 and USF2, 50 and 10 μg of protein, respectively, were fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and incubated with primary antisera against USF1 (sc-229; Santa Cruz Biotechnologies), USF2 (sc-862; Santa Cruz Biotechnologies), or β-actin (A1978; Sigma-Aldrich), followed by either goat anti-rabbit antisera (11-035-003; Jackson ImmunoResearch Laboratories, Inc.) or goat anti-mouse antisera (170-6516; Bio-Rad Laboratories, Inc.). The antigen-antibody complex was visualized with Immobilon Western Chemiluminescent HRP Substrate (Millipore Corporation), and the levels of USF1 and USF2 were normalized to β-actin expression levels. Relative signal intensities were measured using the ImageJ software program [36, 37]. The specificities of USF1 and USF2 antisera were confirmed using extracts from COS7 cells transfected with plasmids encoding USF1 or USF2 (Supplemental Fig. S1A, all Supplemental Data are available online at www.biolreprod.org).
Nuclear Protein Extracts
Sertoli cells for direct assay (1.5 × 106 cells/sample) were collected in a total of 1 ml of PBS and used for the preparation of nuclear extracts. Nuclear and cytoplasmic extracts were prepared by detergent lysis [38]. Briefly, after centrifugation (82 × g for 5 min), the cell pellet was resuspended and incubated in buffer A (10 mM Hepes [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM ethyleneglycoltetra-acetic acid [EGTA], 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail) for 15 min on ice, followed by the addition of 0.06% Nonidet P40. Cells were vortexed for 10 sec, and nuclei were collected by centrifugation (12 000 × g, 30 sec). The nuclei pellet was then washed once with buffer A and resuspended in buffer C (20 mM Hepes [pH 7.9], 0.4 mM NaCl, 1 mM EDTA, 1 mM EGTA, 20% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail). Nuclei were incubated for 15 min on a shaking vortex platform at 50% power at 4°C. The cellular debris was cleared by centrifugation (12 000 × g, 5 min), and the supernatant containing nuclear proteins was stored at −80°C. Protein concentrations were determined by the Bradford method.
DNA-Protein Binding Studies
The 32P-radiolabeled DNA probes were generated by annealing 24- to 42-nucleotide templates containing protein-binding motifs plus flanking promoter sequences to 10-nucleotide primers complementary to the 5′ end of the templates (Table 1). The overhangs were filled in with the Klenow fragment in the presence of [α-32P]dATP and 5 mM each of dCTP, dGTP, and dTTP at 16°C overnight. EMSAs were performed as described previously [39]. Briefly, 32P-labeled probes were incubated with nuclear extracts (1 μg) for 15 min at room temperature in the presence of 1 μg of poly(deoxyinosine-deoxycytosine), 0.25 mg/ml of BSA, 5 mM DTT, 50–100 mM KCl, 20 mM Hepes (pH 7.9), 20% glycerol, and 0.2 mM EDTA. For supershift assays, 1 μl of antiserum was added to the mixture for 15 min at room temperature before addition of the probe. All antisera were obtained from Santa Cruz Biotechnologies: α-USF1 (sc-229), α-USF2 (sc-862), α-E47 (sc-763), α-E2A.E12 (sc-349), and α-MYC (sc-40). DNA-protein complexes were resolved by 5% PAGE under nondenaturing conditions in a Tris borate/EDTA buffer. The gels were dried at 80°C for 1 h and exposed to Classic x-ray Film, Blue Sensitive (Laboratory Products Sales) at −80°C. The relative intensities of DNA-protein complexes were measured from digitized autoradiograms using ImageJ software, and the intensity of E-box-protein complexes was normalized to that of a control cAMP response element-binding protein (CREB) probe [36, 37]. The specificities of USF1 and USF2 antisera were confirmed using extracts from COS7 cells transfected with plasmids encoding USF1 or USF2 (Supplemental Fig. S1B).
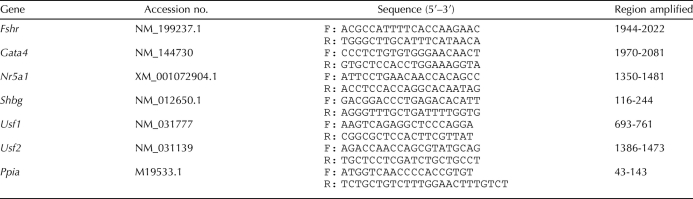
TABLE 1.
Electrophoretic mobility shift assay probes.
ChIP Assays
Primary Sertoli cells were isolated as described above (Isolation of Sertoli Cells for Direct Assay). Resuspended cells were incubated with 1% formaldehyde for 10 min at 37°C. The reaction was halted by the addition of 0.125 m glycine for 5 min at room temperature. ChIP assays were performed following the Upstate Biotechnology ChIP Assay Kit protocol, with modifications. Nuclei were isolated as described above (Nuclear Protein Extracts) with minor changes, washed in buffer A, resuspended in 150 μl of SDS lysis buffer (10 mM EDTA [pH 8.0], 50 mM Tris [pH 8.0], and 1% SDS), and flash-frozen for storage at −80°C until sonication. Thawed samples were resuspended in a total of 1 ml of SDS lysis buffer and aliquoted into two 500-μl aliquots for sonication to yield 200- to 1000-bp genomic DNA fragments utilizing a Kontes Micro Ultrasonic Cell Disruptor. Nuclei were sonicated on ice at 50% output for 20 pulses of 30 sec each. Samples were allowed to cool at least 2 min between each pulse. Cellular debris was eliminated by centrifugation at 14 000 × g for 10 min at 4°C. From the supernatant, 10-μl aliquots, corresponding to 1% input, were removed, and the remaining supernatant was diluted with 2.5 ml of dilution buffer (16.7 mM Tris [pH 8.1], 167 mM NaCl, 0.01% SDS, 1.2 mM EDTA [pH 8.0], and 1.1% Triton-X 100) containing protease and phosphatase inhibitors and stored at −80°C in 1-ml aliquots. Protein concentrations were determined using the BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific, Inc.). Fragmentation was verified using 10 μl of sample, reversing the cross-link (see below), and resolving the purified chromatin in 0.05% glycerol on a 2% agarose gel. The lysate (35 μg in total) of 1 ml of dilution buffer was precleared by incubation with 40 μl of protein A sepharose (Invitrogen) in the presence of 10 μl of preimmune normal rabbit serum (Pierce) and 5 μg of sheared salmon sperm DNA (Invitrogen). After centrifugation at 1000 × g for 1 min, aliquots of the supernatant were incubated with 2 μg of antiserum directed against USF1 or USF2 (sc-229 Trans-X or sc-862 Trans-X, respectively; Santa Cruz Biotechnologies) or a control antiserum, WT1 (sc-192; Santa Cruz Biotechnologies). Controls also were incubated with nonimmune serum or without protein. Samples were incubated with antibodies with rocking at 4°C overnight. Immunocomplexes were recovered by incubation with 50 μl of 50% slurry of protein A sepharose for 2 h at 4°C. The beads were washed 5 min each with low-salt buffer (20 mM Tris [pH 8.1], 150 mM NaCl, 2 mM EDTA [pH 8.0], 1% Triton-X 100, and 0.1% SDS), high-salt buffer (20 mM Tris [pH 8.1], 500 mM NaCl, 2 mM EDTA [pH 8.0], 1% Triton-X 100, and 0.1% SDS), and LiCl buffer (240 mM LiCl, 1% IGEPAL CA-630 (Sigma), 1% sodium deoxycholate, 10 mM Tris [pH 8.1], and 1 mM EDTA [pH 8.0]). The beads were then washed two times in a buffer consisting of 10 mM Tris (pH 8.0) and 1 mM EDTA. The chromatin complexes were eluted by incubation with 250 μl of freshly prepared elution buffer (1% SDS and 0.1 m NaHCO3) with rotation at room temperature for 15 min. The supernatant was saved to a new tube, and the elution was repeated. The supernatant from the second elution was pooled with the supernatant from the first elution. Cross-linking was reversed by adding NaCl to a final concentration of 200 mM and heating at 65°C for 4 h. After incubation with 40 μg of proteinase K for 1 h at 45°C, the DNA was purified using the QIAquick PCR Purification Kit (Qiagen, Inc.). Eluates of immunoprecipitated chromatin were stored at −20°C.
Immunoprecipitation of promoter regions was quantified by quantitative PCR (qPCR) with PowerSYBR Green (Applied Biosystems, Inc.) using 2 μl of the purified chromatin fragments described above. Primers are listed in Table 2. For each primer set, the efficiency of amplification [efficiency = 10(1/−slope)] was calculated for each chromatin sample utilizing a nonprecipitated input sample [40]. The relative quantity of immunoprecipitated chromatin fragments that was amplified for each primer set was then determined through the efficiency-corrected ΔCt (cycle threshold) method of qPCR analysis. The relative quantity is derived from the following equation: quantity = (efficiency)−Ct [40]. The calculated quantity of each primer set amplifying a promoter region was normalized to the quantity found for the control primer set (designed to the Nr5a1 coding region) by dividing the values for each promoter region by the values for the control, unbound region. Relative quantities were expressed as fold-induction over the relative quantity obtained with nonimmune immunoglobulin G.
TABLE 2.
Primers for chromatin immunoprecipitation assays.
Quantitative PCR Analysis of Gene Expression
The RNA was isolated from Sertoli cells using the RNeasy Mini Kit (Qiagen, Inc.) according to manufacturer's instructions. RNA was treated with RNase-free RQ1 DNase (Promega) for 2 h at 37°C and purified by phenol/chloroform extraction. The RNA was subjected to reverse transcription using random hexamers [41]. For reverse transcription, 250 ng of RNA were incubated with 100 μl of reaction mix containing 7.5 mM MgCl2, 400 μM deoxynucleotide triphosphates (Promega), 10× PCR II Buffer (Applied Biosystems), 40 U of RNasin Ribonuclease Inhibitor (Promega), 2.25 μM random hexamers (Integrated DNA Technologies), 250 U Superscript II Reverse Transcriptase (Invitrogen Corporation), and nuclease-free water (Applied Biosystems/Ambion). Parallel reactions were performed without reverse transcriptase. The samples were incubated at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min, followed by 4°C for 5 min.
Real-time qPCR amplifications were performed in a 96-well plate using the ABI Prism 7900HT Sequence Detection System v2.3 (Applied Biosystems). Reactions were conducted in a total volume of 20 μl, which included 2 μl of cDNA, 10 μl of PerfeCTa SYBR Green Fast Mix ROX (Quanta Biosciences, Inc.), and 2 μM of each primer. Amplification reactions were also performed on samples containing no cDNA as negative controls, either from reverse transcription reactions lacking either reverse transcriptase or mRNA template. Primer pairs (Table 3) were independently validated for use in the ΔΔCt method of gene expression analysis [40] through use of a standard curve derived from serial dilutions of the cDNA obtained from the reverse transcription reactions as previously described [6]. Primers with an efficiency of 2 ± 0.2 were considered to be acceptable. Peptidylprolyl isomerase A (Ppia; commonly known as cyclophilin) was used as an endogenous control. The qPCR analysis initiated with melting of cDNA at 95°C for 15 min, followed by 40 amplification cycles of 15 sec at 95°C and 1 min at 60°C. Dissociation-curve analysis was performed immediately after amplification. Ct values were recorded and analyzed via the ΔΔCt method [40] to characterize relative fold-changes in mRNA expression between treatment groups as previously described [6].
TABLE 3.
Primers for quantitative PCR.
Reporter Plasmid Constructs, Transient Transfections, and Luciferase Assays
The Nr5a1Luc reporter plasmid contains the Nr5a1 promoter region, including the E-box at −32 to −77 bp [42]. The Shbg600Luc reporter plasmid contains 600 bp of the rat Shbg promoter region upstream of the proximal start site [32, 43]. Cultured Sertoli cells from 5- and 11-day-old rats were transfected with 0.5 μg of reporter plasmid in the presence or absence of 0.5 μg of empty expression vector or vectors expressing USF1, USF2, or USF1+USF2 (pSVUSF1 and pSVUSF2 [44]) using FuGENE Transfection Reagent 6 (Roche Applied Science) according to the manufacturer's instructions. Three days after transfection, the Sertoli cells were collected, and total cellular proteins were extracted using Reporter Lysis Buffer (Promega). Luciferase assays were performed using a Victor2 luminometer (PerkinElmer). Luciferase activities were normalized for total protein as determined by the Bradford method.
RNA Interference Assays
The Usf1 (siRNA ID: s136901) and Usf2 (siRNA ID: si35812) siRNAs were obtained from Applied Biosystems. The sequences are as follows: Usf1 sense, 5′-GGA CAA CGC GAG AUG AGA Att-3′; Usf1 antisense, 5′-UUC UCA UCU CGC GUU GUC Ctg-3′; Usf2 sense, 5′-GGA UGU GCU UCA AAC AGG Att-3′; and Usf2 antisense, 5′-UCC UGU UUG AAG CAC AUC Ctg-3′. Control siRNA (directed against androgen receptor [Ar]) used as a negative control was obtained from Dharmacon and had the following sequences: sense, 5′-AGG AGC GUU CCA GAA UCU G dT dT-3′; and anti-sense, 5′-dT dT UCC UCG CAA GGU CUU AGA C-3′. Sertoli cells isolated from 5- and 11-day-old rats were cultured in 60-mm2 plates and transfected with siRNAs using TransIT-TKO Transfection Reagent (Mirus) according to manufacturer's instructions. Cells were transfected with either 40 mM Usf1 or Usf2 siRNA, 20 mM Usf1 plus 20 mM Usf2 siRNA, or 40 mM control siRNA or were mock transfected with no siRNA. Twenty-four hours after transfection, the media were replaced by serum-free media. RNA was isolated as described above (Quantitative PCR Analysis of Gene Expression) three days after transfection, and gene expression was analyzed using qPCR with the primers listed in Table 3.
Statistical Analysis
For EMSAs, ChIP assays, qPCR, and RNA interference studies, the mean ± SEM value or intensity of three individual experiments was determined for each sample group unless otherwise noted. For luciferase assays, the mean ± SEM value for three or four independent experiments of two replicates each was determined for each sample group. Results were analyzed by ANOVA with Fisher PLSD at a 5% significance level utilizing Statview 4.5 (Abacus Concepts, Inc.).
RESULTS
USF1 Protein Levels Increase Between 5 and 11 Days after Birth in Sertoli Cells
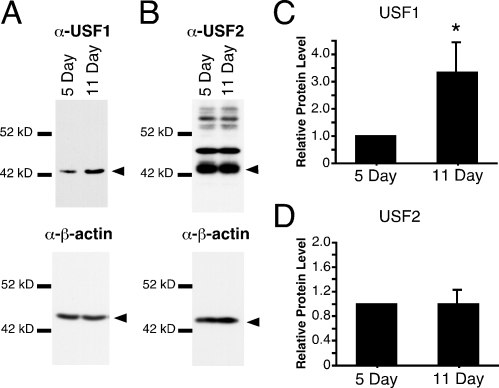
Our previous studies determined that Usf1 mRNA levels increased 1.6-fold during Sertoli cell differentiation [6]. To determine whether USF protein levels similarly increased during Sertoli cell differentiation, Western blot analysis of USF1 and USF2 was performed using whole-cell extracts prepared from Sertoli cells immediately after isolation from rat testes. These studies demonstrated that USF1 protein levels increased 3.3-fold between 5 and 11 days after birth (Fig. 1, A and C). USF2 protein levels did not change (Fig. 1, B and D). Detection of USF1 required 5-fold more protein extract than the amount needed for USF2 protein detection. Although the efficiency of the USF1 and USF2 antibodies may vary, the need for less protein to detect USF2 is consistent with our previous qPCR results demonstrating that Usf2 mRNA levels are greater than those of Usf1 [6].
FIG. 1.
USF1 protein levels increase during differentiation in Sertoli cells. A and B) Western immunoblotting of whole-cell extracts from 5- and 11-day-old Sertoli cells immediately after isolation was performed sequentially with antisera specifically recognizing USF1 (A) or USF2 (B) followed by β-actin. Arrowheads denote the band of interest. C and D) Levels of USF1 and USF2 were normalized to β-actin levels, and levels for 5-day-old Sertoli cells were arbitrarily set equal to one. The fold-induction (mean ± SEM) of 11-day-old Sertoli cells over 5-day-old Sertoli cells from three independent experiments for USF1 and USF2, respectively, is reported. *P < 0.05.
Binding of USF Proteins to Target Gene Promoters Increases During Sertoli Cell Differentiation
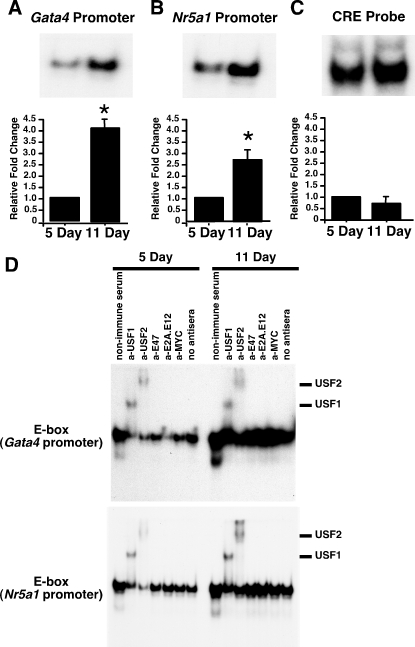
Previously, we found that USF binding to the Fshr promoter E-box increases during Sertoli cell differentiation [6]. To determine whether increased USF binding to target gene promoters is common during Sertoli cell differentiation, we first assayed USF binding to the E-box regions of the Gata4 and Nr5a1 gene promoters. Nuclear extracts prepared from Sertoli cells immediately after isolation from 5- and 11-day-old rats were incubated with radiolabeled, double-stranded oligonucleotide probes containing E-box motifs from the Gata4 and Nr5a1 promoters. Protein binding to the Gata4 and the Nr5a1 E-boxes was low in extracts from 5-day-old Sertoli cells, but binding activity at both promoter regions increased 4.1- and 2.7-fold, respectively, at 11 days (Fig. 2, A and B). In contrast, protein interactions with a consensus cAMP response element (CRE) that binds the CREB transcription factor remained constant between 5 and 11 days (Fig. 2C) [6, 45]. To confirm that USF proteins contribute to forming the DNA-protein complexes with the Gata4 and Nr5a1 E-box motifs, supershift studies were performed. Sertoli cell nuclear extracts from 5- and 11-day-old testes were preincubated with nonimmune serum or antisera against USF1, USF2, E47, E2A, or MYC E-box-binding transcription factors before incubation with probes containing the E-box regions of the Gata4 or Nr5a1 promoters (Fig. 2D). Both the USF1 and USF2 antisera supershifted the DNA-protein complexes, indicating that the complexes contain USF proteins. The E47 and MYC antisera did not cause a supershift of protein complexes at the Gata4 E-box. However, the E2A-E12 antiserum did decrease Gata4 E-box binding with extracts from 5-day-old Sertoli cells, raising the possibility that E2A or E12 proteins may also bind to the Gata4 E-box. The E47, E2A-E12, and MYC antisera had little effect on the binding of proteins to the Nr5a1 promoter E-box. Overall, these data support the hypothesis that elevated levels of USF proteins are available to bind E-box motifs within the Gata4 and Nr5a1 target gene promoters during Sertoli cell differentiation.
FIG. 2.
USF1 and USF2 DNA-binding increases at E-box motifs in the Gata4 and Nr5a1 promoters in differentiating Sertoli cells. A–C) In EMSAs, radiolabeled probes containing E-box regions of the Gata4 promoter (A), the Nr5a1 promoter (B), or a consensus CRE-binding motif for the CREB transcription factor (CRE probe; C) were incubated with nuclear extracts of Sertoli cells prepared immediately after isolation from 5- and 11-day-old testis. Representative images of the resulting DNA-protein complexes are shown, and quantitation of the mean ± SEM of four independent experiments is provided. The relative binding to the Gata4 and Nr5a1 promoters for each condition was normalized to CREB binding for each experiment, and the value of binding activity from 5-day-old Sertoli cell extracts was set equal to one. The relative CREB-binding activity represents the mean ± SEM of the fold-inductions derived directly from the intensities of the protein-DNA complexes. Representative full-length gel images for A–C are provided in Supplemental Figure S2. *P < 0.05. D) DNA-protein complexes from nuclear extracts isolated from 5- and 11-day-old Sertoli cells were incubated immediately after isolation with nonimmune sera or antisera against USF1, USF2, E47, E2A, or MYC or with no antisera. Supershifted DNA-protein complexes containing USF1 or USF2 are indicated. Exposure times for all complexes are less than 48 h. Free probe was run off the gel.
Shbg Promoter Proximal E-box Is a Target of USF1 and USF2
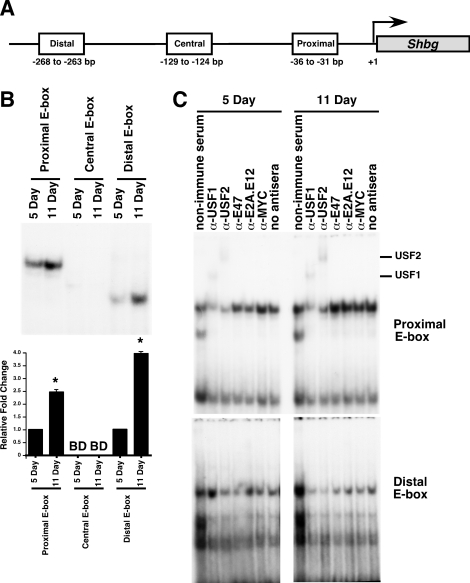
The Shbg mRNA levels are known to increase during Sertoli cell differentiation. Shbg mRNA levels first increase between 10 and 15 days after birth and then increase more dramatically between 15 and 40 days after birth [31]. To determine whether USF1 and USF2 bind to any of the three E-box regions within the Shbg promoter (Fig. 3A), EMSAs of nuclear extracts from 5- and 11-day-old rat Sertoli cells were conducted. For the probes containing the proximal (−36 to −31 bp) and distal (−268 to −263 bp) E-box motifs, protein-binding activity was low in extracts from 5-day-old Sertoli cells but increased in extracts from 11-day-old Sertoli cells (2.5-fold and 3.9-fold for binding to the proximal and distal sites, respectively) (Fig. 3B). DNA binding was not detected at the central E-box (−129 to −124 bp) at either 5 or 11 days after birth.
FIG. 3.
USF1 and USF2 exhibit increasing DNA binding at the proximal E-box of the Shbg promoter in differentiating Sertoli cells. A) Schematic of the Shbg promoter showing the presence of three E-box motifs: the proximal E-box motif from −36 to −31 bp, the central E-box motif from −129 to −124 bp, and the distal E-box from −268 to −263 bp [32]. Promoter region is not drawn to scale. B) In EMSAs, radiolabeled probes containing a region of the Shbg promoter including the proximal E-box, the central E-box, or the distal E-box (top) were incubated with nuclear extracts prepared immediately after isolation from 5- and 11-day-old Sertoli cells. Representative images of the resulting DNA-protein complexes are shown, and quantitation of the mean ± SEM of three independent experiments is provided (bottom). The relative binding for each condition is normalized to that of CREB binding, and the value of binding from 5-day-old Sertoli cell extracts was set equal to one. BD, below detection limit of the assay. *P < 0.05. C) DNA-protein complexes from nuclear extracts isolated from cultured 5- and 11-day-old Sertoli cells were incubated with nonimmune sera or antisera against USF1, USF2, E47, E2A, or MYC or with no antisera. Supershifted DNA-protein complexes containing USF1 or USF2 are indicated. Exposure times for all complexes are less than 48 h. Free probe was run off of the gel.
To determine whether USF1 or USF2 contributed to the DNA-protein complexes at either the proximal or distal Shbg E-box motifs, supershift studies were performed. DNA-protein complexes produced with Sertoli cell extracts from 5- and 11-day-old rats incubated with the proximal E-box motif could be supershifted with either USF antisera, whereas the E47, E2A, and MYC antisera had little effect (Fig. 3C). In contrast, none of the antibodies tested was able to shift or disrupt the DNA-protein complexes detected for the distal E-box. Additionally, the DNA-protein complexes formed at the distal E-box motif migrated faster than usually observed for USF-bound probes, further suggesting that an alternative protein or protein complex is bound at the site (Fig. 3B). These data suggest that USF1 and USF2 could be involved in the transcriptional activation of Shbg through the proximal E-box region, whereas a different protein complex forms at the distal E-box during differentiation.
Recruitment of USF Proteins to Fshr, Nr5a1, and Gata4 Target Gene Promoters In Vivo Increases During Sertoli Cell Differentiation
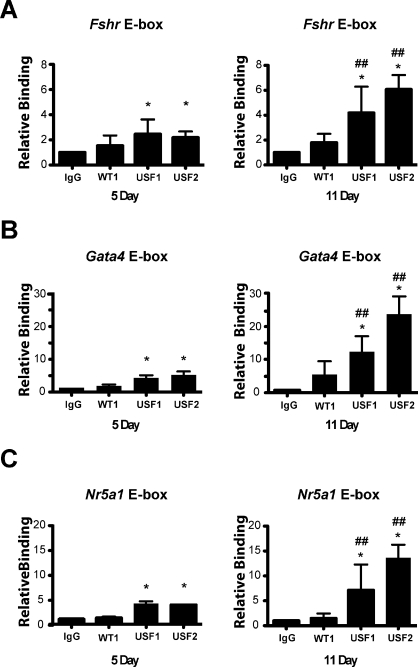
Chromatin immunoprecipitation analysis was employed to assess the recruitment of USF1 and USF2 to the promoters of USF target genes during Sertoli cell differentiation in vivo. USF1 and USF2 were recruited to the E-box regions of the Fshr, Gata4, and Nr5a1 promoters at both 5 and 11 days of age (Fig. 4). However, we were unable to detect USF1 or USF2 recruitment to the Shbg promoter in chromatin samples from 5- or 11-day-old Sertoli cells by ChIP analysis (data not shown). For all three promoters at which USF proteins were detected, the levels of USF1 and USF2 recruitment in the 11-day-old Sertoli cells were greater than those in the 5-day-old cells. USF1 and USF2 recruitment, respectively, increased 1.7- and 2.7-fold at the Fshr promoter (Fig. 4A), 2.9- and 4.8-fold at the Gata4 promoter (Fig. 4B), and 2.0- and 3.7-fold at the Nr5a1 promoter (Fig. 4C). These data indicate that increased levels of USF protein expression and in vitro DNA-binding activity correlate with increased recruitment to E-box motifs in vivo.
FIG. 4.
USF1 and USF2 binding to the Fshr, Gata4, and Nr5a1 gene promoters in vivo during differentiation. Chromatin-protein complexes were immunoprecipitated with nonimmune serum (IgG) or antiserum against WT1, USF1, or USF2. The relative quantities of PCR amplified E-box regions from the Fshr (A), Gata4 (B), or Nr5a1 (C) promoters are shown for 5- and 11-day-old Sertoli cells. Recruitment to E-box regions was normalized to the relative quantity of complexes recruited by IgG for the same age cells (relative binding set equal to one). Results shown are the mean ± SEM of three experiments performed in triplicate. Experiments employing cells from 5- and 11-day-old rats were run concurrently. *P < 0.05 vs. the IgG value of the same age, ##P < 0.05 vs. the same condition in 5-day-old rats.
Expression of USF Target Genes Nr5a1 and Shbg Increases During Sertoli Cell Differentiation
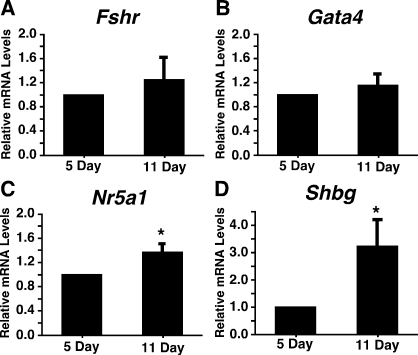
To determine whether increased USF recruitment to E-box motifs during Sertoli cell differentiation corresponded with increased USF target gene expression, the expression of mRNAs encoding Fshr, Gata4, Nr5a1, and Shbg was assayed during Sertoli development using qPCR. Fshr and Gata4 mRNA levels did not increase between 5 and 11 days after birth (Fig. 5, A and B). In contrast, the mRNA levels of Nr5a1 increased 1.4-fold from 5 to 11 days after birth (Fig. 5C), and mRNA levels of Shbg in Sertoli cells increased 3.2-fold (Fig. 5D). For all studies, the mRNA levels of the housekeeping gene Ppia did not change over the course of differentiation (data not shown).
FIG. 5.
Nr5a1 and Shbg mRNA levels increase during differentiation in Sertoli cells. The mRNAs isolated from 5- and 11-day-old Sertoli cells assayed immediately after isolation were analyzed by qPCR using primers for Fshr (A), Gata4 (B), Nr5a1 (C), and Shbg (D). Data were analyzed using the ΔΔCt method, and quantitation of the mean ± SEM of three individual experiments for each condition is provided for each primer set. The relative mRNA levels were normalized to Ppia levels and made relative to 5-day-old Sertoli cells (set equal to one). *P < 0.05.
USF Proteins Induce Nr5a1 and Shbg Promoter Activity in 11-Day-Old Rat Sertoli Cells
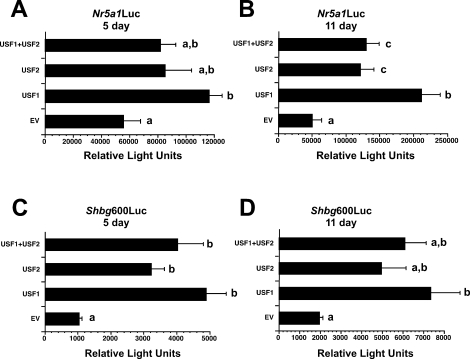
Because only Nr5a1 and Shbg mRNA levels increased in differentiating Sertoli cells, we next focused on USF regulation of the promoters of these genes in proliferating and differentiating Sertoli cells. To test the hypothesis that USF proteins mediate increased Nr5a1 promoter activity, the luciferase reporter construct Nr5a1Luc, containing the Nr5a1 promoter including the E-box at −82 to −77 bp, was used in transient transfection assays [42]. Cultured Sertoli cells, isolated from 5- and 11-day-old rats, were cotransfected with the luciferase reporter constructs and with plasmids encoding Usf1, Usf2, or Usf1+Usf2. Overexpression of USF1, USF2, and USF1+USF2 increased Nr5a1Luc activity in Sertoli cells from 5-day-old rats by 2.1-, 1.5-, and 1.5-fold, respectively (Fig. 6A), and in Sertoli cells from 11-day-old rats by 4.2-, 2.4-, and 2.6-fold, respectively (Fig. 6B).
FIG. 6.
Overexpression of USF proteins up-regulates Nr5a1 and Shbg promoter activity in cultured Sertoli cells. Sertoli cells isolated from 5-day-old rats (A and C) or 11-day-old rats (B and D) were cotransfected with Nr5a1Luc (A and B) or Shbg600Luc (C and D) reporter plasmids and empty expression plasmid (EV) or plasmids encoding USF1, USF2, or USF1+USF2 proteins as noted. Promoter activities of the reporter plasmids are expressed in relative light units normalized to 1 μg of protein. The results shown are the mean ± SEM luciferase activities for three (Shbg600Luc) or four (Nr5a1Luc) experiments performed in duplicate. Values with different lowercase letters differ significantly (P < 0.05).
Additional transient transfection assays were carried out with the Shbg600 luciferase reporter construct, containing 600 bp of the Shbg proximal promoter [32, 43, 46]. Addition of USF1, USF2, or USF1+USF2 induced Shbg600Luc activity in Sertoli cells 4.7-, 3.1-, and 3.9-fold, respectively, in Sertoli cells isolated from 5-day-old rats (Fig. 6C) and 3.7-, 2.5-, and 3.1-fold, respectively, in 11-day-old Sertoli cells (Fig. 6D). Together, the transient transfection studies indicate that elevated USF expression results in increased Nr5a1 and Shbg promoter activity in cultured Sertoli cells isolated from both 5- and 11-day-old rats.
Knockdown of Usf1 Plus Usf2 Inhibits the Expression of Fshr and Shbg in 11-Day-Old Sertoli Cells
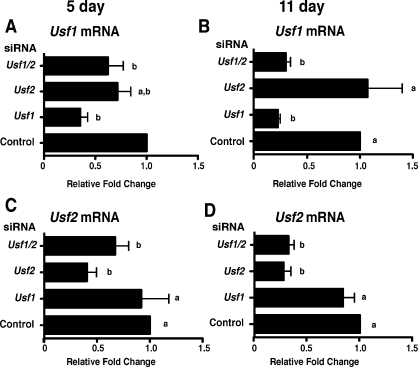
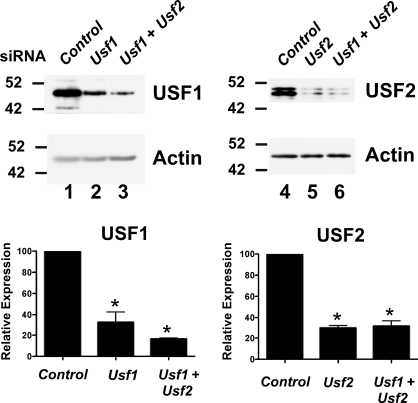
To investigate whether USF proteins are required to regulate endogenous target gene expression in proliferating and differentiating Sertoli cells, RNA interference assays were performed in cultured Sertoli cells. Evaluation of the efficiency of mRNA knockdown revealed that Usf1 mRNA levels were reduced 58.0% by Usf1 siRNA in 5-day-old Sertoli cells (Fig. 7A) and by 78% in 11-day-old Sertoli cells (Fig. 7B). Usf2 siRNA reduced Usf2 mRNA levels 59% and 69% at 5 and 11 days, respectively, after birth (Fig. 7, C and D). The actions of the siRNAs were specific as Usf1 mRNA levels in the presence of Usf2 siRNA and Usf2 mRNA after Usf1 siRNA transfection were not significantly altered in 5- or 11-day-old Sertoli cells. In Sertoli cells cultured from 5-day-old rats, the combination of Usf1 and Usf2 siRNAs decreased Usf1 and Usf2 mRNAs 48% and 33%, respectively. In 11-day-old Sertoli cells, the combination of siRNAs resulted in 70% and 65% decreases in Usf1 and Usf2 mRNA levels, respectively. Further evaluation of USF protein expression after transfection of siRNA determined that Usf1 and Usf2 siRNA reduced expression of their respective proteins by 67% and 70%, respectively (Fig. 8). Protein levels of USF1 and USF2 were similarly reduced when the combination of siRNAs was used. Together, these results indicate that Usf1 and Usf2 siRNAs effectively and specifically inhibit their respective mRNA targets.
FIG. 7.
Usf1 and Usf2 siRNAs specifically knock down their target mRNAs. Sertoli cells cultured from 5- and 11-day-old rats were transfected with siRNAs against no siRNA (Control), Usf1, Usf2, or Usf1 and Usf2 (Usf1/2). The mRNA was isolated for analysis by qPCR using primers for Usf1 (A and B) or Usf2 (C and D). The USF siRNA-induced fold-changes in Usf1 and Usf2 mRNA levels relative to that of Control (set equal to one) are provided. The results shown are the mean ± SEM for three transfection experiments. Values with different lowercase letters differ significantly (P < 0.05).
FIG. 8.
Usf1 and Usf2 siRNAs specifically knock down their target proteins. Sertoli cells cultured from 20-day-old rats were transfected with siRNAs against Ar (Control), Usf1, Usf2, or Usf1, and Usf2. Whole-cell extracts were assayed by Western blot analysis using antisera against USF1 (lanes 1–3) or USF2 (lanes 4–6), followed by reprobing with β-actin. Quantitation of the mean ± SEM for two experiments is shown. Values were made relative to Ar controls (set equal to 100%). *P < 0.05 vs. Ar controls.
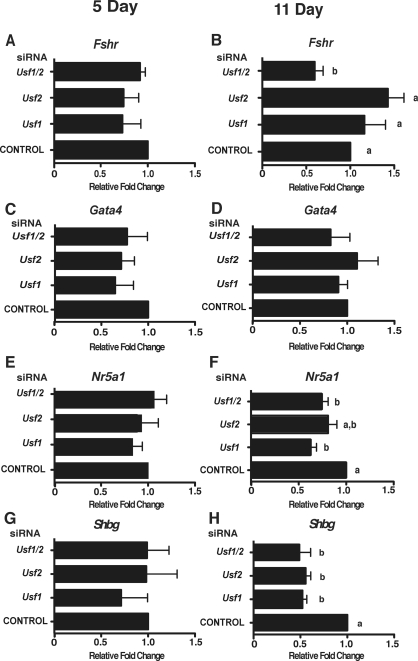
To determine whether USF proteins contribute to the expression of the Fshr and Gata4 genes during proliferation or differentiation, Sertoli cells were treated with Usf siRNAs. The use of either Usf siRNA alone or the combination of siRNAs did not result in significant reduction of Fshr or Gata4 mRNA levels in 5-day-old Sertoli cells (Fig. 9, A and C). At 11 days, treatment with either Usf1 or Usf2 siRNA alone did not alter Fshr and Gata4 mRNA levels (Fig. 9, B and D). The combination of Usf1 and Usf2 siRNAs did decrease Fshr mRNA expression by 40%, but Gata4 mRNA levels were not significantly altered (Fig. 9, B and D).
FIG. 9.
Knock down of Usf mRNAs results in decreased expression of Fshr and Shbg target genes. Sertoli cells cultured from 5- and 11-day-old rats were transfected with no siRNA (CONTROL) or with siRNAs against Usf1, Usf2, or Usf1 and Usf2 (Usf1/2). The mRNA was isolated for analysis by qPCR using primers for the USF target genes Fshr (A and B), Gata4 (C and D), Nr5a1 (E and F), or Shbg (G and H). The Usf siRNA-induced changes in target gene mRNA levels relative to that after transfection with control (set equal to one) are provided. The results are shown as the mean ± SEM for three to five transfection experiments. Values with different lowercase letters differ significantly (P < 0.05).
In addition, RNA interference studies were conducted to determine whether USF proteins are required for the expression of Nr5a1 and Shbg. Expression of Nr5a1 and Shbg at five days was not altered by Usf1 or Usf2 knockdown (Fig. 9, E and G). In Sertoli cells isolated from 11-day-old rats, transfection of Usf1 or Usf2 siRNA alone as well as the combination of Usf1 and Usf2 siRNA caused a 37%, 20%, and 26% decrease, respectively, in Nr5a1 mRNA levels. These data suggest that USF proteins contribute to Nr5a1 expression during Sertoli cell differentiation. In 11-day-old Sertoli cells, Usf1 siRNA resulted in a 48% decrease in Shbg levels, whereas Usf2 siRNA decreased Shbg mRNA 44% (Fig. 9H). Usf1 and Usf2 siRNA together produced a 51% decrease in Shbg mRNA levels. These results indicate that USF1 and USF2 similarly activate Shbg gene expression during the differentiation of Sertoli cells.
DISCUSSION
We tested the hypothesis that elevated USF levels contribute to the activation of genes supporting male fertility that are expressed during the differentiation of Sertoli cells. We focused on three classes of genes characterized by their USF-mediated regulation during Sertoli cell differentiation. These genes included 1) Fshr and Gata4, the expression of which is not greatly altered during Sertoli cell differentiation; 2) Shbg, which represented a potential target of USF, because Shbg expression increases during differentiation; and 3) the USF-regulated Nr5a1 gene, which has not been well characterized during Sertoli cell differentiation.
The USF proteins were identified as potential regulators of gene expression during the differentiation of Sertoli cells, because USF protein binding to the E-box in the Fshr promoter in EMSAs increased 7-fold from 5 to 11 days after birth [6]. In the present study, we similarly found that USF binding to E-boxes in the promoters of the Gata4, Nr5a1, and Shbg increased 2.5- to 4-fold for Sertoli cells from 11-day-old rats. The increase in E-box binding is likely due to the corresponding 3.3-fold increase in USF1 protein expression, because levels of Usf2 mRNA [6] and protein remain relatively constant during the transition from the proliferating to the differentiating state.
In vivo ChIP assay results also indicated that USF1 recruitment to the Gata4, Nr5a1, and Fshr E-box motifs increased in 11-day-old Sertoli cells. Interestingly, USF2 recruitment to the target promoters also increased in vivo, although USF2 protein expression did not increase. One possible explanation for the increased recruitment of USF2 is that the increase in USF1 expression that occurs in 11-day-old Sertoli cells may drive the formation of heterodimers that preferentially bind to the E-box motifs studied. A similar increase in USF heterodimer formation is proposed for endometriotic stroma cells, except that an increase in USF2 expression drives heterodimer formation and increased target gene expression [47]. The E-box motifs assayed in the present study may specify recruitment of USF1/USF2 heterodimers over USF2 homodimers. This idea is consistent with results from Hermann et al. [18], who found USF1/USF2 heterodimers to be the most abundant dimer bound to the Fshr promoter E-box in Sertoli cells (58.9% of dimers), followed by USF1 homodimers (34.5%) and USF2 homodimers (6.6%). Alternatively, it is possible that differentiation of Sertoli cells results in chromatin alterations that facilitate the binding of all USF dimers to E-box motifs.
In vitro DNA-protein interaction studies identified two complexes that form with Shbg promoter E-box motifs, a USF-containing complex at the proximal E-box and a complex at the distal E-box that contains unidentified proteins. Protein binding to both the proximal and distal E-box motif increased between 5 and 11 days after birth. Both E-box motifs are important for promoter activity, because previous DNase protection assays identified binding complexes essential for promoter activity within the proximal promoter in MSC-1 cells [46] and mutation of either the proximal or the distal E-box prevented Shbg promoter activity in 20-day-old Sertoli cells [32]. In fully differentiated Sertoli cells, the central E-box (−129 to −124 bp upstream of the transcription start site) was reported to bind transcription factor E2A (TCFE2A; also known as E47) [32]). However, we did not detect protein binding to the central E-box using extracts from 5- or 11-day-old Sertoli cells, which may indicate an alternative mechanism of regulation for Shbg in the fully differentiated Sertoli cells [32].
An additional E-box motif is found in the human SHBG promoter that is not found in rodents. The unique human SHBG E-box binds USF proteins but also acts to inhibit SHBG expression in Sertoli cells and likely is responsible for the differing expression patterns for SHBG/Shbg in humans versus rodents [48]. In addition to E-box motifs, a CRE motif has also been identified within the Shbg promoter (−125 to −118 bp) and overlaps with the central E-box, but the regulation of the CRE motif has not been investigated in undifferentiated Sertoli cells [32]. Together, these data suggest that multiple proteins bind to the Shbg promoter and that the regulation of Shbg is complex.
We were unable to detect either USF1 or USF2 recruitment to the Shbg promoter by ChIP assay. It is possible that USF proteins are not recruited to the Shbg promoter in vivo. However, such a result is difficult to reconcile with our findings that 1) in 11-day-old Sertoli cells, USF binding to the proximal E-box in EMSAs increased; 2) in vivo Shbg mRNA levels increased coincident with elevated USF levels; and 3) Shbg mRNA levels decreased after knockdown of Usf. One explanation for the ChIP result is that USF proteins are recruited to the Shbg promoter but cannot be accessed or recognized by the USF antisera used for the immunoprecipitations due to the presence of complex, multiprotein structures. Further studies are required to identify the proteins in the transcriptional complex at the Shbg promoter in vivo and to determine whether the USF proteins are components of a multiprotein structure.
The increase in USF recruitment to target gene promoters in Sertoli cells between 5 and 11 days after birth correlated with an increase in gene expression for Nr5a1 and Shbg during differentiation in vivo. Furthermore, the Nr5a1 and Shbg gene promoters could be activated by the overexpression of USF1 and/or USF2 in Sertoli cells cultured from either 5- or 11-day-old rats. These findings are consistent with earlier studies showing that E-box motifs in both target gene promoters regulated promoter activity [28, 32, 49–51] and that overexpression of USF proteins induced the Nr5a1 promoter in the MSC-1 mouse Sertoli cell line [28, 52]. Together, the USF-mediated induction of Nr5a1 and Shbg promoter activity and the increase in mRNA expression that occurs with elevated USF levels in vivo support the hypothesis that increased USF protein levels up-regulate Nr5a1 and Shbg target genes during the differentiation of Sertoli cells.
The USF regulation of Nr5a1 and Shbg in differentiating Sertoli cells was confirmed by RNA interference studies. Although knockdown of USF1 or USF2 did not affect the expression of any target gene in 5-day-old Sertoli cells, siRNA-mediated knockdown of USF1 and/or USF2 inhibited Shbg target gene expression in 11-day-old Sertoli cells by 44%–51%. Knockdown of USF1 also reduced the expression of the Nr5a1 gene by 37%. The siRNA-mediated change in Nr5a1 mRNA levels corresponded closely to the 40% increase in gene expression observed in 11-day-old Sertoli cells, in which USF levels were elevated. Together, these results suggest that USF proteins contribute to the increase in expression that is observed for the Nr5a1 and Shbg genes in Sertoli cells in vivo between 5 and 11 days after birth.
In contrast to the activation of the Nr5a1 and Shbg genes by USF proteins, the present results indicate that elevated USF expression and increased USF recruitment to E-box motifs was not sufficient to increase expression of the Fshr and Gata4 target genes in differentiating Sertoli cells in vivo. Although USF1 and USF2 have each been shown in transient transfection studies of cultured cells to induce a segment of the Fshr promoter that contains an E-box [19, 53], our studies indicate that in vivo mRNA levels remain constant during the period in which proliferating cells mature to differentiating cells [6]. This result agrees with previous studies showing that Fshr mRNA levels in Sertoli cells increase until Day seven and remain constant during Days 10–20 after birth before decreasing into adulthood [11, 21]. The finding that elevated USF levels alone are unable to activate Fshr expression in vivo suggests that additional factors are required to support expression of the Fshr and is consistent with the previous determination that extensive regulatory sequences (>413 kb) were required for expression of the Fshr gene in transgenic mice [54]. Our RNA interference studies indicate that only after knockdown of both USF1 and USF2 were Fshr mRNA levels reduced in cultured 11-day-old Sertoli cells. These data suggest that a minimal level of USF is necessary to maintain Fshr expression in cultured Sertoli cells. Together, the available data suggest that elevated USF levels alone are unable to activate expression of the Fshr gene during differentiation in vivo. However, USF has been implicated in the cyclical increase and decrease in Fshr expression that occurs in adult Sertoli cells [19].
Previous reports showed that Gata4 expression decreases in the whole testis during puberty. However, these studies, as mentioned, assayed mRNA levels from whole testis, and the decreased Gata4 mRNA levels could be accounted for by the dilution of Sertoli cell-specific mRNAs with mRNA from the increasing numbers of germ cells that arise during puberty [12–14]. Our data indicate that Gata4 mRNA levels remain constant during postnatal Sertoli cell development, despite an increase in USF1 and USF2 recruitment to the Gata4 promoter during Sertoli cell differentiation. These data, coupled with the finding that knockdown of USF expression does not decrease Gata4 mRNA levels in cultured Sertoli cells at either age, indicates that increased USF recruitment to the Gata4 promoter is not sufficient to up-regulate Gata4 expression in vivo at these stages of development.
Our studies support the following working model. During proliferation, low levels of USF1 expression in Sertoli cells limit USF recruitment to E-box motifs within the promoter regions of select genes required for differentiated Sertoli cell functions. With the onset of differentiation, Usf1 mRNA and USF1 protein production is increased, USF1 recruitment to E-box motifs in the promoters of target genes increases, and expression of selected USF target genes, including Nr5a1 and Shbg, is induced. This mechanism does not apply to all USF target genes, because additional factors may be required to activate expression (as proposed for the Fshr and Gata4 genes). The proposed model is limited to USF protein actions and does not include other members of the E-box protein family that may be regulated differently during differentiation and alter target gene activity with different characteristics. Future studies of the physiological role of USF proteins in Sertoli cell differentiation and identification of the transcription complexes with which USF proteins are involved will contribute to our understanding of transcriptional control of Sertoli cell differentiation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following people for reporter constructs: Keith Parker and Nancy Stallings for SF1Luc, Michael Skinner and David Joseph for ABP600Luc, and Reza Zarneger for pSVUSF1 and pSVUSF2. We thank the laboratories of Donald Scott (University of Pittsburgh) and Gary Hammer (University of Michigan) for their expertise in ChIP assays, Dr. Anthony Zeleznik for the Ppia primers and for his continued advice and mentoring, and John Shupe for excellent technical assistance. This work is part of the requirements for M.A.W. to obtain the Ph.D. degree in Cell Biology and Molecular Physiology at the University of Pittsburgh.
Footnotes
Supported by the National Institute of Child Health and Human Development/National Institutes of Health through cooperative U54 (HD008610) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
REFERENCES
- Orth JM, Gunsalus GL, Lamperti AA. Evidence from Sertoli cell-depleted rats indicates that spermatid number in adults depends on numbers of Sertoli cells produced during perinatal development. Endocrinology 1988; 122: 787 794. [DOI] [PubMed] [Google Scholar]
- Sharpe RM. Regulation of spermatogenesis. : Knobil E, Neil JD. (eds.), The Physiology of Reproduction. New York: Raven Press; 1994: 1363 1434.
- Sharpe RM, McKinnell C, Kivlin C, Fisher JS. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reproduction 2003; 125: 769 784. [DOI] [PubMed] [Google Scholar]
- Orth JM. Proliferation of Sertoli cells in fetal and postnatal rats: a quantitative autoradiographic study. Anat Rec 1982; 203: 485 492. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Wreford NG, De Kretser DM. Determination of Sertoli cell numbers in the developing rat testis by stereological methods. Int J Androl 1989; 12: 58 64. [DOI] [PubMed] [Google Scholar]
- Wood MA, Walker WH. USF1/2 transcription factor DNA-binding activity is induced during rat Sertoli cell differentiation. Biol Reprod 2009; 80: 24 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell 2003; 3: 525 530. [DOI] [PubMed] [Google Scholar]
- Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene 2001; 20: 8317 8325. [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol 2003; 13: 410 418. [DOI] [PubMed] [Google Scholar]
- Sirito M, Lin Q, Deng JM, Behringer RR, Sawadogo M. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc Natl Acad Sci U S A 1998; 95: 3758 3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni M, Gromoll J, Nieschlag E. The follicle-stimulating hormone receptor: biochemistry, molecular biology, physiology, and pathophysiology. Endocr Rev 1997; 18: 739 773. [DOI] [PubMed] [Google Scholar]
- Viger RS, Mertineit C, Trasler JM, Nemer M. Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the mullerian-inhibiting substance promoter. Development 1998; 125: 2665 2675. [DOI] [PubMed] [Google Scholar]
- Ketola I, Rahman N, Toppari J, Bielinska M, Porter-Tinge SB, Tapanainen JS, Huhtaniemi IT, Wilson DB, Heikinheimo M. Expression and regulation of transcription factors GATA-4 and GATA-6 in developing mouse testis. Endocrinology 1999; 140: 1470 1480. [DOI] [PubMed] [Google Scholar]
- Imai T, Kawai Y, Tadokoro Y, Yamamoto M, Nishimune Y, Yomogida K. In vivo and in vitro constant expression of GATA-4 in mouse postnatal Sertoli cells. Mol Cell Endocrinol 2004; 214: 107 115. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Albrecht KH, Crispino JD, Fujiwara Y, Eicher EM, Orkin SH. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development 2002; 129: 4627 4634. [DOI] [PubMed] [Google Scholar]
- Kyronlahti A, Euler R, Bielinska M, Schoeller EL, Moley KH, Toppari J, Heikinheimo M, Wilson DB. GATA4 regulates Sertoli cell function and fertility in adult male mice. Mol Cell Endocrinol 2011; 333: 85 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Heckert LL. Transcriptional regulation of the FSH receptor: new perspectives. Mol Cell Endocrinol 2007; 260–262: 100 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Hornbaker K, Rice DA, Sawadogo M, Heckert LL. In vivo regulation of follicle-stimulating hormone receptor by the transcription factors upstream stimulatory factor 1 and upstream stimulatory factor 2 is cell specific. Endocrinology 2008; 149: 5297 5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan P, Wood MA, Walker WH. Follicle-stimulating hormone (FSH) transiently blocks FSH receptor transcription by increasing inhibitor of deoxyribonucleic acid binding/differentiation-2 and decreasing upstream stimulatory factor expression in rat Sertoli cells. Endocrinology 2009; 150: 3783 3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaud Guittot S, Tetu A, Legault E, Pilon N, Silversides DW, Viger RS. The proximal Gata4 promoter directs reporter gene expression to Sertoli cells during mouse gonadal development. Biol Reprod 2007; 76: 85 95. [DOI] [PubMed] [Google Scholar]
- Bortolussi M, Zanchetta R, Belvedere P, Colombo L. Sertoli and Leydig cell numbers and gonadotropin receptors in rat testis from birth to puberty. Cell Tissue Res 1990; 260: 185 191. [DOI] [PubMed] [Google Scholar]
- Hatano O, Takayama K, Imai T, Waterman MR, Takakusu A, Omura T, Morohashi K. Sex-dependent expression of a transcription factor, Ad4BP, regulating steroidogenic P-450 genes in the gonads during prenatal and postnatal rat development. Development 1994; 120: 2787 2797. [DOI] [PubMed] [Google Scholar]
- Shen WH, Moore CC, Ikeda Y, Parker KL, Ingraham HA. Nuclear receptor steroidogenic factor 1 regulates the mullerian inhibiting substance gene: a link to the sex determination cascade. Cell 1994; 77: 651 661. [DOI] [PubMed] [Google Scholar]
- Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol 2004; 274: 271 279. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008; 453: 930 934. [DOI] [PubMed] [Google Scholar]
- Gurates B, Amsterdam A, Tamura M, Yang S, Zhou J, Fang Z, Amin S, Sebastian S, Bulun SE. WT1 and DAX-1 regulate SF-1-mediated human P450arom gene expression in gonadal cells. Mol Cell Endocrinol 2003; 208: 61 75. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Clarke TR, Lane AH, Wang X, Donahoe PK. Endogenous expression of mullerian-inhibiting substance in early postnatal rat Sertoli cells requires multiple steroidogenic factor-1 and GATA-4-binding sites. Proc Natl Acad Sci U S A 2000; 97: 1624 1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett MA, Rice DA, Heckert LL. Expression of steroidogenic factor 1 in the testis requires an E-box and CCAAT box in its promoter proximal region. Biol Reprod 2000; 62: 670 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenas L, Ritzen EM, Plooen L, Hansson V, French FS, Nayfeh SN. Sertoli cell origin of testicular androgen-binding protein (ABP). Mol Cell Endocrinol 1975; 2: 339 350. [DOI] [PubMed] [Google Scholar]
- Griswold MD. Protein secretions of Sertoli cells. Int Rev Cytol 1988; 110: 133 156. [DOI] [PubMed] [Google Scholar]
- Bunick D, Kirby J, Hess RA, Cooke PS. Developmental expression of testis messenger ribonucleic acids in the rat following propylthiouracil-induced neonatal hypothyroidism. Biol Reprod 1994; 51: 706 713. [DOI] [PubMed] [Google Scholar]
- Saxlund MA, Sadler-Riggleman I, Skinner MK. Role of basic helix-loop-helix (bHLH) and CREB transcription factors in the regulation of Sertoli cell androgen-binding protein expression. Mol Reprod Dev 2004; 68: 269 278. [DOI] [PubMed] [Google Scholar]
- Anway MD, Folmer J, Wright WW, Zirkin BR. Isolation of Sertoli cells from adult rat testes: an approach to ex vivo studies of Sertoli cell function. Biol Reprod 2003; 68: 996 1002. [DOI] [PubMed] [Google Scholar]
- Walker WH, Fucci L, Habener JF. Expression of the gene encoding transcription factor cyclic adenosine 3′,5′-monophosphate (cAMP) response element-binding protein (CREB): regulation by follicle-stimulating hormone-induced cAMP signaling in primary rat Sertoli cells. Endocrinology 1995; 136: 3534 3545. [DOI] [PubMed] [Google Scholar]
- Chapin RE, Phelps JL, Miller BE, Gray TJ. Alkaline phosphatase histochemistry discriminates peritubular cells in primary rat testicular cell culture. J Androl 1987; 8: 155 161. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International 2004; 11: 36 42. [Google Scholar]
- Rasband WS. ImageJ. U.S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2011. [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts,' prepared from a small number of cells. Nucleic Acids Res 1989; 17: 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard DW, Walker WH, Doerre S, Sista P, Molitor JA, Dixon EP, Peffer NJ, Hannink M, Greene WC. The v-rel oncogene encodes a kappa B enhancer-binding protein that inhibits NF-kappa B function. Cell 1990; 63: 803 814. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol 2006; Chapter 15:Unit 8.1–8.28, Supplement 73. [DOI] [PubMed]
- Innis MA. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. [Google Scholar]
- Cui S, Ross A, Stallings N, Parker KL, Capel B, Quaggin SE. Disrupted gonadogenesis and male-to-female sex reversal in Pod1 knockout mice. Development 2004; 131: 4095 4105. [DOI] [PubMed] [Google Scholar]
- Fenstermacher DA, Joseph DR. Analysis of promoter and androgen regulatory sequences required for optimal transcription of the rat androgen-binding protein gene. J Androl 1998; 19: 81 91. [PubMed] [Google Scholar]
- Meier JL, Luo X, Sawadogo M, Straus SE. The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol Cell Biol 1994; 14: 6896 6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M, Brindle P, Harootunian A, Armstrong R, Rivier J, Vale W, Tsien R, Montminy MR. Coupling of hormonal stimulation and transcription via the cyclic AMP-responsive factor CREB is rate limited by nuclear entry of protein kinase A. Mol Cell Biol 1993; 13: 4852 4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenstermacher DA, Joseph DR. DNA sequences and their binding proteins required for Sertoli cell-specific transcription of the rat androgen-binding protein gene. Mol Endocrinol 1997; 11: 1387 1400. [DOI] [PubMed] [Google Scholar]
- Utsunomiya H, Cheng YH, Lin Z, Reierstad S, Yin P, Attar E, Xue Q, Imir G, Thung S, Trukhacheva E, Suzuki T, Sasano H. et al. Upstream stimulatory factor-2 regulates steroidogenic factor-1 expression in endometriosis. Mol Endocrinol 2008; 22: 904 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selva DM, Hogeveen KN, Hammond GL. Repression of the human sex hormone-binding globulin gene in Sertoli cells by upstream stimulatory transcription factors. J Biol Chem 2005; 280: 4462 4468. [DOI] [PubMed] [Google Scholar]
- Nomura M, Bartsch S, Nawata H, Omura T, Morohashi K. An E-box element is required for the expression of the ad4bp gene, a mammalian homologue of ftz-f1 gene, which is essential for adrenal and gonadal development. J Biol Chem 1995; 270: 7453 7461. [DOI] [PubMed] [Google Scholar]
- Harris AN, Mellon PL. The basic helix-loop-helix, leucine zipper transcription factor, USF (upstream stimulatory factor), is a key regulator of SF-1 (steroidogenic factor-1) gene expression in pituitary gonadotropin and steroidogenic cells. Mol Endocrinol 1998; 12: 714 726. [DOI] [PubMed] [Google Scholar]
- Gao L, Kim Y, Kim B, Lofgren SM, Schultz-Norton JR, Nardulli AM, Heckert LL, Jorgensen JS. Two regions within the proximal steroidogenic factor 1 promoter drive somatic cell-specific activity in developing gonads of the female mouse. Biol Reprod 2011; 84: 422 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer SP, Rice DA, Heckert LL. Expression of steroidogenic factor 1 in the testis requires an interactive array of elements within its proximal promoter. Biol Reprod 2002; 67: 1509 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert LL, Sawadogo M, Daggett MA, Chen JK. The USF proteins regulate transcription of the follicle-stimulating hormone receptor but are insufficient for cell-specific expression. Mol Endocrinol 2000; 14: 1836 1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Hornbaker KI, Maran RR, Heckert LL. Distal regulatory elements are required for Fshr expression in vivo. Mol Cell Endocrinol 2007; 260–262: 49 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.