Abstract
NNRTI drug resistance mutations (DRM) are increasingly reported in Africans failing their first antiretroviral regimen. The Phidisa II trial randomized treatment-naïve participants to LPV/r or EFV with d4T+3TC or ZDV+ddI. We report the prevalence of DRM in subjects who achieved HIV RNA < 400 copies/mL at 6-months but subsequently had 2 consecutive HIV RNA >1000 copies/mL. Sixty-eight participants fulfilled the inclusion criteria. NNRTI-DRM were found in 17/36 (47.2%) EFV-recipients, and M184V/I mutation in 14/40 (35.0%) 3TC-recipients. No PI mutation was identified in 38 LPV/r-recipients. This is one of the first studies in Africa confirming the paucity of PI-associated DRM despite virologic failure.
Keywords: genotypic resistance, non-nucleoside reverse transcriptase inhibitors, protease inhibitors, HIV, South Africa, antiretroviral naïve
INTRODUCTION
Combination antiretroviral therapy (cART) has been effective in reducing morbidity and mortality of HIV infection in resource-limited countries. In South Africa, cART became available through the national ART roll-out since April 20041. Current South African HIV treatment guidelines recommend a non-nucleoside reverse transcriptase inhibitor (NNRTI), combined with two nucleos(t)ide reverse transcriptase inhibitors (NRTI) one of which should be lamivudine (3TC) or emtricitabine, as first-line cART2. Although these regimens are highly effective, there are increasing reports of HIV-1 drug resistance compromising treatment. Studies from sub-Saharan Africa have reported drug resistance mutation frequencies of 66% to 91% in those failing first cART. Resistance to 3TC and the NNRTIs were the most commonly identified3–9.
Ritonavir-boosted protease inhibitor (PI/r)-based regimens are commonly used as first-line cART in developed countries where many additional second-line cART options exist10, but are seldom available as initial cART regimens in Africa. PI/r have a greater genetic barrier to development of resistance than NNRTI- or NRTI, and presence of PI-associated resistance mutations at virologic failure are uncommon11.
The Phidisa II trial compared the efficacy and safety of efavirenz (EFV) to lopinavir/ritonavir (LPV/r)-based regimens in South Africa.12 It was designed in 2003 and started enrolment before the national ART roll-out. This study represents one of the largest cohorts initiating PI/r-based first-line cART in Africa, and a unique opportunity to describe genotypic resistance patterns in those failing EFV vs. LPV/r- based cART. We conducted a descriptive substudy to investigate the prevalence and type of genotypic resistance in participants who had viral rebound after viral suppression at 6 months.
METHODS
Phidisa II was a randomized, open-label 2×2 factorial study which enrolled members of the South African National Defence Force (SANDF) and/or their dependents ≥14 years of age, with advanced HIV disease and/or CD4+ cell counts < 200 cells/mm3, who were treatment-naïve or had <7 days of prior therapy. It compared the efficacy and safety of four regimens: (EFV vs. LPV/r) + (stavudine+lamivudine [d4T+3TC] vs. zidovudine+didanosine [ZDV+ddI]). Patients had follow-up visits at months 1, 2, 3, and then every three months. The primary study was approved by the SANDF and the United States National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Boards. All participants signed written informed consent before enrolment.
This substudy selected participants with confirmed viral rebound (defined as HIV RNA >1,000 copies/mL on two consecutive visits after having documented viral suppression to <400 copies/mL at month 6). Baseline characteristics including age, gender, WHO stage, body mass index (BMI), hemoglobin, CD4 count and HIV RNA level were recorded. All cART regimens and changes were recorded.
Genotypic resistance testing was performed on stored blood samples from the date of second consecutive determination of HIV RNA >1,000 copies/mL, using an in-house genotyping assay previously described13. This assay has been successfully validated through regular participation in the Viral Quality Assurance (VQA) program run by the NIH. Briefly, viral RNA was isolated from plasma using the MagNa Pure LC automated system (Roche Diagnostics, Indianapolis, IN, USA) and the region spanning protease (PR, amino acids 1–99) through reverse transcriptase (RT, amino acids 1–440) was amplified by nested PCR using an Expand Long Template kit (Roche Diagnostics, Germany). The PCR products were sequenced using BigDye Terminators v3.1 on an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Consensus sequences were aligned and manually edited using the Sequencher v4.5 software (GeneCodes, Ann Arbor, MI). Phylogenetic analysis of nucleic acid sequences was performed using MEGA 4 for internal quality purposes and viral subtype was assigned using the REGA HIV subtyping tool. Drug resistance mutations (DRM) were identified using the 2009 IAS-USA list14 and confirmed using the Stanford Sequence Resistance Database15. Prevalence of at least one known HIV-1 RT or PR DRM was reported for samples with successful amplification of the relevant RT or PR region. The participants with and without DRM were compared using the Wilcoxon rank sum test for continuous characteristics and Fisher’s exact test for binary characteristics. A p-value of <0.05 was considered statistically significant. Pre-cART samples from participants with DRM at rebound were sequenced to determine the baseline genotype.
RESULTS
Key Findings from the Primary Phidisa II Trial
1,771 participants enrolled into the primary Phidisa II trial. After a median follow-up of 24.7 months, there was no statistically significant difference in the hazard for the primary endpoint of AIDS/death between the EFV and LPV/r groups, and between the d4T+3TC and ZDV+ddI groups. At 36 month of follow-up, there was no difference in the proportion of participants reaching HIV RNA < 400 copies/mL between the EFV and LPV/r groups (66.7% vs. 68.9%% respectively, p=0.588), but a significantly higher increase in CD4 count (18 cells averaged over all follow-up) in the LPV/r group compared to the EFV group (P <0.001). HIV RNA levels were lower (P < 0.001) and CD4 cell counts were greater (P <0.01) over follow-up for d4T+3TC versus ZDV+ddI.12
Results of the Current Substudy
Seventy-three of 1,771 Phidisa II participants satisfied the inclusion criteria for this substudy, namely, had confirmed rebound HIV viremia (to > 1,000 copies/mL) after suppression at month 6. Five participants were excluded from this analysis: four were known to have discontinued cART 3–6 months prior to genotype testing, and one had an uncertain treatment history. Amongst the remaining 68 participants, 38 were randomized to LPV/r and 30 to EFV; 36 to d4T+3TC and 32 to ZDV+ddI. The median age =35.9 years (IQR:32.9–38.8), 38.2% were female, median hemoglobin = 12.3g/dl (IQR: 10.6–14.1), median CD4 count = 100 cells/mm3(IQR: 53.5–153), median HIV RNA VL = 5.2 log10 copies/ml (IQR: 4.7–5.6), 44% were WHO Stage 3 or 4, and median BMI =22.7kg/m2 (IQR: 20.5–26.7). The median time to confirmed rebound was 17.1 months (15.9 months for patients with RT mutations, and 18.4 months for patients with no RT mutations, p=0.29); median CD4 count = 261 cells/mm3 and HIV RNA=3.6 log10 copies/ml at rebound. Eight patients had EFV or LPV/r changes prior to rebound, six due to the need to commence rifampicin-based tuberculosis treatment, necessitating switches from LPV/r to EFV, and two due to adverse effects related to EFV.
Genotypic Drug Resistance
Sixty-eight participants’ samples were successfully sequenced in the RT and/or PR region. Of the 68 participants, 23 (33.8%, CI: 22.8–46.3%) had at least one known DRM at rebound. The overall frequencies of DRM are shown in Table 1. The specific DRM, CD4 count and HIV-RNA at baseline and change at rebound, and cART for individual participants with DRM are presented in Figure 1. DRM were not detected from pre-cART samples in any participant who had DRM at failure. HIV subtypes were determined for 65 patients: 61 (93.8%) were subtype C, with two A, 1 of each of C/D, and D.
Table 1.
Antiretroviral Drug Exposure and Drug Resistance Mutations Identified at Rebound
| Resistance Mutations | No. of subjects with DRM | n (%) who received EFV (total N=36) | n (%) who received 3TC (total N=40) | n (%) who received EFV + 3TC (total N=16) | n (%) who received ZDV/ddI (total N=28) | n (%) who received LPV/r (total N=38) |
|---|---|---|---|---|---|---|
| Major PI mutations | 0 | 0 (0%) | ||||
| Any major NNRTI mutations | 17 | 17 (47.2%) | ||||
| K103N | 9 | 9 (25.0%) | ||||
| V106M | 6 | 6 (16.7%) | ||||
| Y188C/L +/− G190S/A | 6 | 6 (16.7%) | ||||
| Y181C | 0 | 0 (0%) | ||||
| Major NRTI mutations | ||||||
| M184V | 14 | 14 (35.0%) | 5 (15.6%)* | |||
| K70R +/− D67N | 3 | 3 (10.7%) | 1 (3.1%)* | |||
| N348I | 2 | 2 (14.1%) | ||||
| K103N + M184V | 7 | 7 (43.8%) | ||||
| Any NNRTI mutation + M184V | 9 | 9 (56.3%) | ||||
3TC = lamivudine, ddI = didanosine, DRM = drug resistance mutation, EFV = efavirenz, LPV/r = lopinavir/ritonavir, NNRTI = non-nucleoside reverse transcriptase inhibitor, NRTI = nucleoside reverse transcriptase inhibitor, PI = protease inhibitor, ZDV = zidovudine
6 patients received both LPV/r and EFV before genotype testing, for calculating the percentage of patients who were on LPV/r and had M184V or K70R +/− D67N mutations, only those who received LPV/r without EFV (n=32) were included.
Figure 1. Antiretroviral Therapy, Pre-Treatment CD4 and HIV RNA, Time to Failure, CD4 and HIV RNA at Failure, and Major Genotypic Resistance Mutations for Individual Participants with Detected Mutations at Failure.
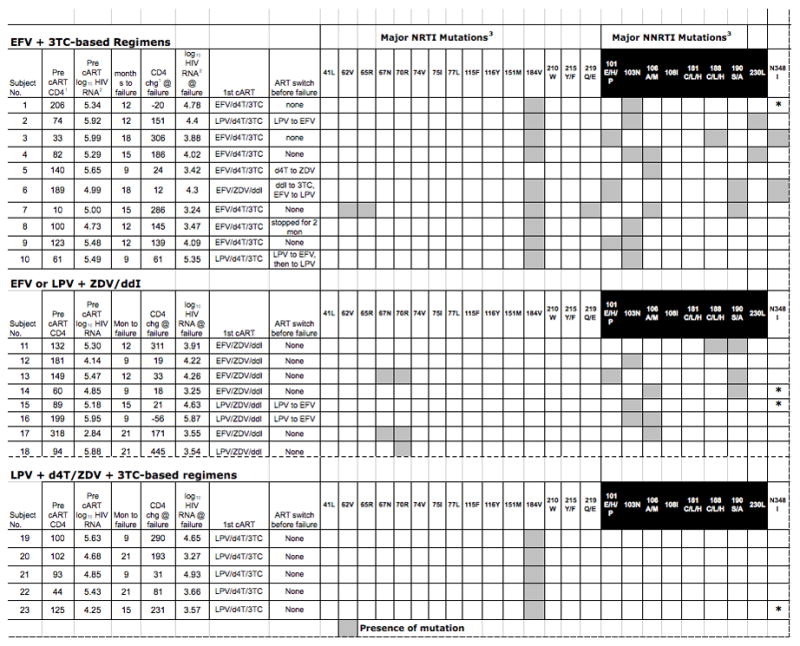
1 (cells/mm3)
2 (copies/mL)
3 Major reverse transcriptase mutations according to the International AIDS Society-USA Drug Resustance Mutations in HIV-1, December 2009[14]
3TC = lamivudine, ART = antiretroviral therapy, cART = combination antiretroviral therapy, chg = change, d4T = stavudine, ddI = didanosine, EFV = efavirenz, LPV = lopinavir/ritonavir, mon = months
NRTI-resistance mutations
All patients received ZDV+ddI, d4T+3TC, or ZDV+3TC (after drug substitution for toxicity) as the 2-NRTI backbone. NRTI-resistance mutations were detected in 17 participants. The most common was M184V, found in 14 of 40 (35.0%) 3TC-recipients. Four participants had at least one thymidine-associated mutation (TAM) – two recipients of ZDV+ddI had D67N + K70R, another ZDV+ddI recipient had K70R alone. One participant who received EFV+d4T+3TC had four RT mutations (A62V, K65R, M184V, and K219E) in addition to two NNRTI-associated mutations.
LPV/r-based regimens
No PR resistance mutations were detected in the 38 LPV/r recipients. Six of these individuals also received EFV before rebound. Of the 32 individuals who never received EFV, 6 (18.8%, CI: 7.2–36.4%) had NRTI-associated mutations; five had M184V and one with K70R.
EFV-based regimens
Seventeen of 36 EFV recipients (47.2%) had NNRTI resistance mutations. K103N was the most commonly detected in 9 (25%) participants, V106M in 6 (16.7%), and G190S/A +/− Y188C/L in 6 (16.7%). No Y181C mutation was detected. DRM to both NRTI and NNRTI were found in 11/36 (30.5%) of EFV recipients, with the combination of M184V and K103N being most prevalent. The N348I mutation of the connection domain of RT was detected in two participants; both also had NNRTI mutations. There was no statistical difference in the frequency of NRTI mutations between the group exposed to EFV and those never exposed to EFV (p=0.4).
Comparison between participants with and those without RT mutations
There was no statistically significant difference in the following baseline characteristics between the groups with and without RT mutations: age, hemoglobin, CD4 count, HIV RNA, BMI, gender, proportion with WHO Stage 3 or 4 diseases, and the time to viral rebound. Those with RT mutations had a smaller increase in CD4 counts (+139 vs. +155 cells/mm3, p=0.05). The HIV RNA levels at rebound were similar between the 2 groups (3.9 vs. 3.6 log10 copies/mL, p=0.25).
DISCUSSION
Our study is amongst the largest HIV cohort in South Africa who started PI/r as initial cART to date. Our data confirmed those of other studies that PI-associated DRM is much less common than NNRTI- or NRTI-associated DRM at failure. Compared to other reports from sub-Saharan Africa, a smaller proportion of our participants had DRM at confirmed rebound viremia. This could be due to only a quarter of our population being randomized to a regimen containing both 3TC and an NNRTI, the drugs most frequently associated with DRM at failure11; whereas in other cohorts, 94% to 100% of patients were on these RT inhibitors in their first cART5–7. As seen in other studies, M184V and the NNRTI-associated mutations were most prevalent in our cohort5,6.
Since our participants were enrolled in a randomized control trial, they were evaluated on a relatively frequent follow-up schedule of monthly for the first three months, then every three months thereafter. We performed genotype testing at viral rebound confirmed by the second viremia sample, a time point within three months of the first detection of viremia of > 1,000 copies/mL. As a consequence, patients were viremic on therapy for a relatively short period, which may explain the fewer DRM detected. Greater accumulation of DRM, especially NRTI-associated mutations, occurs the longer one remains on a failing regimen5. In a Malawi study where treatment failure was not determined virologically, but clinically or immunologically, 93% of participants with viremia (on stored samples) had NNRTI mutations, 81% had M184V, and 56% harbored at least one TAM along with M184V and NNRTI mutations16.
Two patients in our study developed the connection domain mutation N348I, identified previously as contributing to substantial increase in AZT resistance, including patients with non-subtype B infection17. N348I may emerge on NRTI or NNRTI exposure, both of our patients received EFV. The detection of N348I in patients with DRM resistance suggests that sequencing programs that include this portion of RT will be useful in deriving a comprehensive drug resistance evaluation.
The pre-cART samples of those with DRM at failure all had wild-type virus, suggesting that pre-treatment transmitted drug resistance was not a reason for failure. However, as more patients in South Africa receive first-line NNRTI based regimens and failure with NNRTI-resistance increases, transmitted resistance will likely increase, which has already been reported in several African countries18,19.
Reasons for rebound viremia in our cohort are uncertain, but non-adherence most likely plays a key role. Adherence assessments were solely based on participants’ adherence reports which have limitations. Measures to improve adherence to cART, like: continuous adherence support, using co-formulations and once-daily dosing of cART, may reduce virologic failure with or without DRM.
Our sub-study has a few limitations. First, we only selected patients who demonstrated viral suppression at six month and then had viral rebound, in order to eliminate those who may have had transmitted HIV drug resistance at baseline. This is not representative of all the patients who either did not respond or fail to achieve viral suppression until a later time point. Secondly, as genotype testing was done on the second rebound samples, we cannot prove that PI-resistance mutations did not emerge at a later time point while the patients were viremic and continued to receive LPV/r. Despite these limitations, our findings are important to note as they confirm data from developed countries showing that PI/r has a greater genetic barrier to resistance than NNRTI-based regimens.
In many settings in sub-Saharan Africa, viral load testing is not available and antiretroviral failure is only recognized by CD4 decline or clinical progression. Based on this study and others, virologic failure with DRM precedes clinical or immunologic failure. Phidisa II utilized a relatively frequent viral RNA determination schedule, permitting early detection of rebound viremia. Development of affordable point of care viral load tests in resource-limited setting is critical to allow for early identification and management of treatment failure.
In conclusion, this study confirms the risk of selection of NNRTI and M184V mutations in patients who received EFV and 3TC in first-line cART. Phidisa II represents the largest cohort of adults who received PI/r as first-line cART in South Africa. In this subgroup analysis, no PI-resistance mutations were detected at failure, demonstrating that a PI/r-containing regimen maybe a potential ART option in Africa. The relatively high cost of PI/r and lack of co-formulations with other ARV drug classes, however, precludes their use as first-line regimen in this population. The absence of PI DRM noted here suggests, however, that the increased cost may be offset by durable efficacy of these antivirals. More affordable generic PI/r and co-formulations available for initial cART options may reduce development of DRM and transmitted drug resistant HIV in Africa.
Acknowledgments
We wish to express our gratitude to: Phidisa II study participants, South African Military Health Service leadership and Units, Phidisa Executive Committee and Phidisa staff.
The contents of this publication do not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
FUNDING SUPPORT: The South African National Defence Force through SAMHS, the United States NIH-NIAID, and the United States Department of Defense (US DoD). This project has also been funded in in part with federal funds from the National Cancer Institute, NIH, under Contract No. HHSN261200800001E.
Footnotes
This study was presented at the 17th Conference on Retroviruses and Opportunistic Infection, San Francisco, CA, USA, February 2010.
Conflict of Interest: None
Contributor Information
J. Nomthandazo Dlamini, Project Phidisa, South African Military Health Service (SAMHS), South Africa.
Zonghui Hu, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), USA
Johanna Ledwaba, National Institute of Communicable Diseases (NICD), Johannesburg, South Africa.
Lynn Morris, NICD, Johannesburg, South Africa.
Frank M. Maldarelli, National Cancer Institute (NCI), NIH, USA
Robin L. Dewar, NCI-Frederick, SAIC-Frederick, Inc., USA
Helene C. Highbarger, NCI-Frederick, SAIC-Frederick Inc., USA
Harsha Somaroo, Project Phidisa, SAMHS, South Africa.
Phumelele Sangweni, Project Phidisa, SAMHS, South Africa.
Dean A. Follmann, NIAID, NIH, USA
Alice K. Pau, NIAID, NIH, USA
References
- 1.National Department of Health SA. National Antiretroviral Treatment Guidelines. Pretoria; South Africa: 2004. pp. 1–93. [Google Scholar]
- 2.Department of Health RSA. The South African Antiretroviral Treatment Guidelines. 2010. pp. 1–8. [Google Scholar]
- 3.Barth RE, Wensing AM, Tempelman HA, Moraba R, Schuurman R, Hoepelman AI. Rapid accumulation of nonnucleoside reverse transcriptase inhibitor-associated resistance: evidence of transmitted resistance in rural South Africa. AIDS. 2008 Oct 18;22(16):2210–2212. doi: 10.1097/QAD.0b013e328313bf87. [DOI] [PubMed] [Google Scholar]
- 4.El-Khatib Z, Ekstrom AM, Ledwaba J, et al. Viremia and drug resistance among HIV-1 patients on antiretroviral treatment: a cross-sectional study in Soweto, South Africa. AIDS. 2010 Jul 17;24(11):1679–1687. doi: 10.1097/QAD.0b013e32833a097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann CJ, Charalambous S, Sim J, et al. Viremia, resuppression, and time to resistance in human immunodeficiency virus (HIV) subtype C during first-line antiretroviral therapy in South Africa. Clin Infect Dis. 2009 Dec 15;49(12):1928–1935. doi: 10.1086/648444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallis CL, Mellors JW, Venter WD, Sanne I, Stevens W. Varied patterns of HIV-1 drug resistance on failing first-line antiretroviral therapy in South Africa. J Acquir Immune Defic Syndr. 2010 Apr 1;53(4):480–484. doi: 10.1097/QAI.0b013e3181bc478b. [DOI] [PubMed] [Google Scholar]
- 7.Marconi VC, Sunpath H, Lu Z, et al. Prevalence of HIV-1 drug resistance after failure of a first highly active antiretroviral therapy regimen in KwaZulu Natal, South Africa. Clin Infect Dis. 2008 May 15;46(10):1589–1597. doi: 10.1086/587109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillay V, Pillay C, Kantor R, Venter F, Levin L, Morris L. HIV type 1 subtype C drug resistance among pediatric and adult South African patients failing antiretroviral therapy. AIDS Res Hum Retroviruses. 2008 Nov;24(11):1449–1454. doi: 10.1089/aid.2008.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orrell C, Walensky RP, Losina E, Pitt J, Freedberg KA, Wood R. HIV type-1 clade C resistance genotypes in treatment-naive patients and after first virological failure in a large community antiretroviral therapy programme. Antivir Ther. 2009;14(4):523–531. [PMC free article] [PubMed] [Google Scholar]
- 10.Panel on Antiretroviral Guidelines for Adults and Adolescents. [Accessed February 8, 2011];Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. 2011 http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf.
- 11.Hirsch MS, Gunthard HF, Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008 Jul 15;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 12.Ratsela A, Polis M, Dhlomo S, et al. A randomized factorial trial comparing 4 treatment regimens in treatment-naive HIV-infected persons with AIDS and/or a CD4 cell count <200 cells/muL in South Africa. J Infect Dis. 2010 Nov 15;202(10):1529–1537. doi: 10.1086/656718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillay V, Ledwaba J, Hunt G, et al. Antiretroviral drug resistance surveillance among drug-naive HIV-1-infected individuals in Gauteng Province, South Africa in 2002 and 2004. Antivir Ther. 2008;13( Suppl 2):101–107. [PubMed] [Google Scholar]
- 14.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009 Dec;17(5):138–145. [PubMed] [Google Scholar]
- 15.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006 Jun 1;42(11):1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosseinipour MC, van Oosterhout JJ, Weigel R, et al. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS. 2009 Jun 1;23(9):1127–1134. doi: 10.1097/QAD.0b013e32832ac34e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delviks-Frankenberry KA, Nikolenko GN, Boyer PL, et al. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc Natl Acad Sci U S A. 2008 Aug 5;105(31):10943–10948. doi: 10.1073/pnas.0804660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lihana RW, Khamadi SA, Lubano K, et al. HIV type 1 subtype diversity and drug resistance among HIV type 1-infected Kenyan patients initiating antiretroviral therapy. AIDS Res Hum Retroviruses. 2009 Dec;25(12):1211–1217. doi: 10.1089/aid.2009.0007. [DOI] [PubMed] [Google Scholar]
- 19.Price MA, Wallis CL, Lakhi S, et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses. 2011 Jan;27(1):5–12. doi: 10.1089/aid.2010.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]


