Abstract
Background
A growing proportion of head and neck squamous cell carcinoma (HNSCCs) is caused by the human papillomavirus (HPV). In light of the unique natural history and prognosis of HPV-related HNSCCs, routine HPV testing is being incorporated into diagnostic protocols. Accordingly, there is an escalating demand for an optimal detection strategy that is sensitive and specific, transferrable to the diagnostic laboratory, standardized across laboratories, cost effective, and amenable to broad application across specimen types including cytologic preparations.
Methods
Cytologic preparations (fine needle aspirates and brushes) were obtained from surgically resected HNSCCs and evaluated for the presence of high risk HPV using the hybrid capture 2 assay. HPV analysis was also performed on the corresponding tissue sections using HPV in-situ hybridization and p16 immunohistochemistry. In cases where the immunohistochemical and in-situ hybridization results were discordant, HPV status was determined by real time PCR detection of E7 expression. HPV status in the tissues and corresponding cytologic samples were compared.
Results
Based on benchmark HPV testing of the tissue sections, 14 HNSCCs were classified as HPV positive and 10 as HPV negative. All corresponding cytologic preparations were correctly classified using the hybrid capture 2 assay.
Conclusions
The hybrid capture 2 strategy, already widely used for the detection of high risk HPV in cervical brushes, is readily transferrable to HNSCCs. Consistent accuracy in cytologic preparation suggests its potential application in FNAs from patients who present with lymph node metastases, and may eliminate the need to obtain tissue solely for the purpose of HPV testing.
Introduction
High risk human papillomavirus (HPV), particularly the 16 type, has been established as a causative agent for a significant proportion of head and neck squamous cell carcinomas (HNSCC)(1), and the incidence of these HPV-related carcinomas is on the rise. (2–4) Given the distinctiveness of HPV-related carcinoma as a biological and clinical variant of HNSCC, the need for routine HPV testing of oropharyngeal carcinomas is urgent and compelling. First, HPV status is a powerful indicator of patient prognosis. HPV-positivity correlates with a lower risk of tumor progression and death, reflecting in part an enhanced sensitivity to ionizing radiation with or without chemotherapy.(5–7) Second, knowledge of HPV status is compulsory for meaningful comparison of treatment responses for patients enrolled in clinical trials. The direction of current clinical trials, where patient selection for specific therapies is predicated on HPV tumor status, dramatically heightens the stakes for accurate HPV detection. For example, false negative results would deny access to therapeutic HPV vaccines; and false positive results would wrongly assign high-risk patients to less intensive multimodality therapy designed for low-risk patients with HPV-positive tumors. Third, HPV assessment may play some present or future role in early cancer detection,(8) tumor localization,(9;10) post-treatment tumor surveillance,(11;12) and informed consultation of patients and their partners.
The increasing incidence of HPV-associated HNSCC along with the growing importance of HPV status as a versatile biomarker are prompting a growing demand for HPV testing of clinical samples. There is not yet a standard assay for HPV detection. Current methods include consensus and type-specific PCR techniques, real-time PCR assays to quantify viral load, DNA in situ hybridization, and immunohistochemical detection of surrogate biomarkers (e.g. p16 protein). Preferential use of one method over another must consider not just test sensitivity and specificity, but must also carefully balance a variety of practical concerns related to cost, ease of specimen acquisition and processing, turnaround time, automation and standardization. Most currently used assays depend on the availability of excised tumor tissues obtained by biopsy or surgical resection. The ability to perform liquid phase HPV testing of cytologic preparations could greatly expand the application of HPV testing in the clinical arena.
The hybrid capture 2 assay is a commercially available microplate analysis approved by the US Food and Drug Administration for the detection of HPV DNA as part of cervical cancer screening.(13;14) It is a liquid-phase hybridization assay that uses an RNA probe mixture for the detection of up to thirteen high risk HPV types. Studies have shown that the assay is highly sensitive and specific when it comes to detecting high risk HPV in cytologic brushes from the uterine cervix,(15) but its use as a tool to evaluate the HPV status of HNSCCs is largely unexplored. The purpose of this feasibility study was to test the sensitivity and specificity of the hybrid capture 2 assay in cytologic preparations from excised HNSCCs of known HPV status.
Methods
Cases
Study approval was obtained from the Johns Hopkins Medical Institutions Internal Review Board including a waiver for HIPAA privacy authorization. Twenty-four consecutive patients who had undergone resection of a primary and/or metastatic squamous cell carcinoma of the head and neck (HNSCC) formed the bases of this study. During routine surgical pathology dissection of the specimens, the tumor was identified and aspirated with a 12-gauge needle or brushed using the Rovers® Cervex-Brush® (Therapak® Corporation, Buford, Georgia). Specifically, metastatic implants were aspirated and primary tumors were brushed. The brush was disconnected from the stem and placed into a BD SurePath™ vial (BD Diagnostics-TriPath, Burlington, N.C.). Cells obtained by fine needle aspiration (FNA) were directly injected into the BD SurePath™ vial. For these FNA specimens, a second pass was performed and evaluated cytologically (Diff Quick stained air dried smears) for the presence of tumor cells and overall sample cellularity (Figure 1). Extra passes were made for the collection of additional material if the specimen were judged to be of low tumor cellularity to ensure collection of more than 5000 tumor cells – the quantitative threshold of detection for the hybrid capture 2 assay. Four brushes from non-neoplastic tonsils and 3 FNAs from benign lymph nodes were used as HPV negative controls.
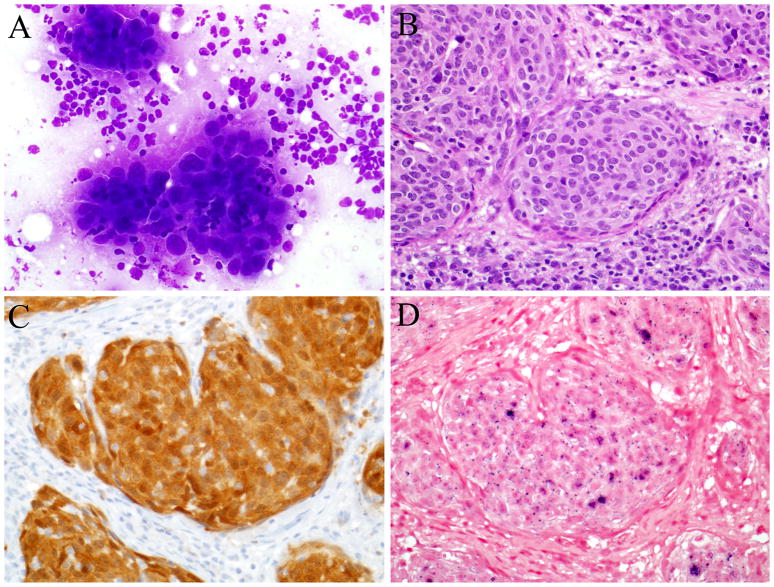
Figure 1.
An HPV-related oropharyngeal squamous cell carcinoma. Cytologic specimens were sent for hybrid capture 2 testing after the presence of tumor was confirmed by a Diff Quick stain (A). Tumor in the corresponding tissue sections (B) was determined to be HPV positive on the basis of strong p16 immunohistochemical staining (C) and the presence of in situ hybridization signals within the tumor nuclei (D).
Hybrid capture 2 analysis
The Digene HPV Test using Hybrid Capture II (hc2) technology is an in vitro nucleic acid hybridization microplate assay with signal amplification using microplate chemiluminescence for the qualitative detection of thirteen high-risk types of HPV (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68). Specimen DNA was denatured into single stranded DNA by the addition of Denaturation Solution, and then hybridized with specific high-risk HPV RNA probes. RNA/ DNA hybrids are captured and immobilized onto the surface of microplate wells coated with antibodies specific for the RNA/ DNA hybrids. The immobilized hybrids were then reacted with alkaline phosphatase conjugated antibodies specific for the RNA/ DNA hybrids, and detected with a chemiluminescent substrate. As the substrate is cleaved by the bound alkaline phosphatase, light is emitted which is measured as relative light units (RLUs) on a luminometer. The intensity of the light emitted denotes the presence or absence of target DNA in the specimen.
Only specimens with RLUs above 250 were deemed of adequate DNA quantity for HPV analysis. Sample scores were calculated as a ratio of the sample RLU divided by the positive control (CO) RLU (1 pg/mol of HPV DNA). Based on our threshold for detection of high risk HPV in cervical brushes, a sample score (RLU/CO) of 3 or greater defined an HPV-related carcinoma. A sample score less than 0.85 defined an HPV-unrelated carcinoma. Specimens with sample scores in the range of 0.85 to 3.0 were interpreted as equivocal for high risk HPV. Negative and positive calibrators were used for quality control. Negative and positive calibrators were used as per manufacturer’s package insert to establish run validity. In addition to the manufacturer’s quality control material, SiHa cells were denatured and tested as positive process controls. In addition to quality control, calibration was performed with each run to ensure that the reagents and furnished calibrator material were functioning properly.
P16 immunohistochemistry
HPV status was confirmed in the formalin-fixed and paraffin-embedded tissue sections of the resected HNSCCs. Immunohistochemical analysis targeted expression of a biomarker of HPV E7 oncoprotein activity, the CDK-inhibitor p16. Five micron sections of formalin-fixed and paraffin-embedded tissues were deparaffinized and subjected to antigen retrieval using 10 mM citrate buffer (92° C for 30 minutes). P16 expression was evaluated by use of a mouse monoclonal antibody against p16 (MTM Laboratories, Heidelberg, Germany) visualized using the Ultra view polymer detection kit (Ventana Medical Systems, Inc. Tucson, AZ) on a Ventana BenchmarkXT autostainer (Ventana). P16 expression was scored as positive if strong and diffuse nuclear and cytoplasmic staining was present in ≥70% of the tumor (Figure 1).(16)
High risk HPV In situ hybridization
Five-micron sections from the tissue microarrays and the formalin-fixed paraffin embedded tumor blocks were evaluated for the presence of HPV DNA by in situ hybridization. Slides were conditioned using Ventana cell conditioner #2 and ISH-protease 3. Hybridization was performed using the HPV III Family16 probe set that captures HPV genotypes 16, 18, 33, 35, 45, 51, 52, 56 and 66. Signals were detected with the ISH iView Blue Plus Detection Kit, which is an indirect biotin-streptavidin system that detects fluorescein-labeled probes. The kit utilizes an alkaline phosphatase enzyme and NBT/BCIP substrate chromogen reaction that provides an intense blue, permanent color and a red counter stain. All reagents are provided pre-diluted and ready-to-use on BenchMark Series automated slide stainers (Ventana Medical Systems, Tucson, AZ). Punctate hybridization signals localized to the tumor cell nuclei defined an HPV-positive tumor (Figure 1). HPV16-positive controls included an HPV16-positive oropharyngeal cancer and the SiHa and CaSki cell lines. The SiHa cell line is known to harbor 1 to 2 copies of integrated HPV-16 per cell, and the CaSki cell line is known to harbor 60 to 600 copies of integrated HPV-16 per cell.
Quantitative PCR detection of E7 expression
To quantify the viral load for cases with discrepant p16 immunostaining and high risk HPV in situ hybridization results, DNA was extracted from fresh tumor and used for real-time PCR with primers and probes sets specific for the E7 region of HPV-16 DNA as previously described.(12) PCR for β-actin was performed in parallel to normalize the input DNA. gDNA from the CaSki cell line was used to develop standard curves for the HPV-16 viral load, as it has been previously characterized to harbor 600 copies of HPV-16 DNA/genome equivalent. gDNA from CaSki cells was serially diluted to the following concentrations: 30ng, 3ng, 0.3ng, 0.03ng, and 0.003ng. A standard curve was developed for β-actin which has 2 copies/genome equivalent, using the same serial dilutions as described previously.(17). HPV16 quantities that were >0.1 (i.e. more than 1 HPV16 viral genome copy/10 cells) were scored as positive. This threshold takes into account background stroma and infiltrating lymphocytes, and has been used to distinguish HPV infections that are clinically relevant from HPV that is incidental and non-causal.(17) All samples were run in triplicate with water control. Taqman Fast Universal PCR Master Mix was used according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA).
Results
Cytologic preparations were obtained from 24 consecutive patients who had undergone a resection of a primary and/or metastatic squamous cell carcinoma of the head and neck (HNSCC). The primary sites of tumor origin were the palatine tonsil (n=9), base of tongue (n=5), oral cavity (n=4), larynx (n=2), skin (n=2) and maxillary sinus (n=1). One patient presented with a lymph node metastasis of unknown primary origin. Cytologic preparations were obtained from the primary tumor (n=11), the lymph node metastasis (n=14) and/or the lung metastasis (n=2). All together, 27 cytologic preparations from 24 patients were evaluated for high risk HPV using the hybrid capture 2 assay.
The results of HPV analysis are summarized in Table 1. Using a normalized ratio of >3 for identification of high risk HPV, 14 of 24 (58%) of the patients had HNSCCs that were HPV-related. HPV was detected in 13 of 14 (93%) carcinomas of oropharyngeal origin, but in only 1 of 9 (11%) carcinomas arising in a non-oropharyngeal site (p < 0.001, Fisher’s exact, two-tail). The non-oropharyngeal HPV-positive tumor arose in the sinonasal tract. The lymph node metastasis from an unknown primary was HPV negative. In the 3 cases (patients 2, 7 and 18) where both the primary tumor and the corresponding lymph node metastasis were evaluated, HPV status was concordant between the paired samples. HPV was not detected in any of the samples obtained from non-neoplastic tonsils (n=4) or non-neoplastic lymph nodes (n=3).
Table 1.
Correlation of hybrid capture 2 classification with tumor origin and HPV status by p16 immunohistochemistry and high risk HPV in-situ hybridization
| Patient | tumor origin | sample site | p16 IHC | HPV ISH | Hybrid capture 2 (score) |
|---|---|---|---|---|---|
| 1 | tonsil | tonsil | positive | positive | positive (18.12) |
| 2 | tonsil | tonsil lymph node |
positive positive |
positive positive |
positive (858.16) positive (1993.00) |
| 3 | tonsil | lymph node | positive | positive | positive (709.16) |
| 4 | tonsil | lung | positive | positive | positive (786.68) |
| 5 | tonsil | lymph node | positive | positive | positive (6.24) |
| 6 | tonsil | tonsil | positive | positive | positive (45.78) |
| 7 | tonsil | tonsil lymph node |
positive positive |
positive positive |
positive(710.91) positive (1789.52) |
| 8 | tonsil | tonsil | positive | positive | positive (1781.85) |
| 9 | tongue base | tongue base | positive | positive | positive (3.21) |
| 10 | tongue base | lymph node | positive | positive | positive (210.09) |
| 11 | tongue base | tongue base | positive | positive | positive (3.47) |
| 12 | tongue base | lung | positive | negative | positive (5.45) |
| 13 | tongue base | tongue base | positive | positive | positive (26.26) |
| 14 | maxillary sinus | maxillary | positive | positive | positive (1860.52) |
| 15 | oral tongue | lymph node | negative | negative | negative (0.08) |
| 16 | oral tongue | oral tongue | negative | negative | negative (0.12) |
| 17 | floor of mouth | lymph node | negative | negative | negative (0.17) |
| 18 | gingiva gingiva |
gingiva lymph node |
negative negative |
negative negative |
negative (0.10) negative (0.11) |
| 19 | tonsil | tonsil | negative | negative | negative (0.14) |
| 20 | larynx | lymph node | negative | negative | negative (0.12) |
| 21 | larynx | lymph node | negative | negative | negative (0.11) |
| 22 | skin | lymph node | negative | negative | negative (0.16) |
| 23 | skin | lymph node | negative | negative | negative (0.12) |
| 24 | unknown | lymph node | negative | negative | negative (0.11) |
| Control | tonsil | not done | not done | negative (0.15) | |
| Control | tonsil | not done | not done | negative (0.13) | |
| Control | tonsil | not done | not done | negative (0.14) | |
| Control | tonsil | not done | not done | negative (0.10) | |
| Control | lymph node | not done | not done | negative (0.29) | |
| Control | lymph node | not done | not done | negative (0.11) |
There was strong agreement for HPV status across the various methods of HPV analysis (Figure 2). All 13 HNSCCs that were p16 positive by immunohistochemistry and HPV positive by in situ hybridization (p16+/HPV+) were HPV positive by hybrid capture 2 analysis. Conversely, the 10 HNSCCs that were p16−/HPV− were negative by hybrid capture 2 analysis. The single discordant case (p16+/HPV−) was found to be HPV positive using the hybrid capture 2 assay. Using real-time PCR amplification of the viral oncoprotein E7, the presence of HPV16 in this case was confirmed at 1.02 copies of HPV16/ genome equivalent (Figure 3).
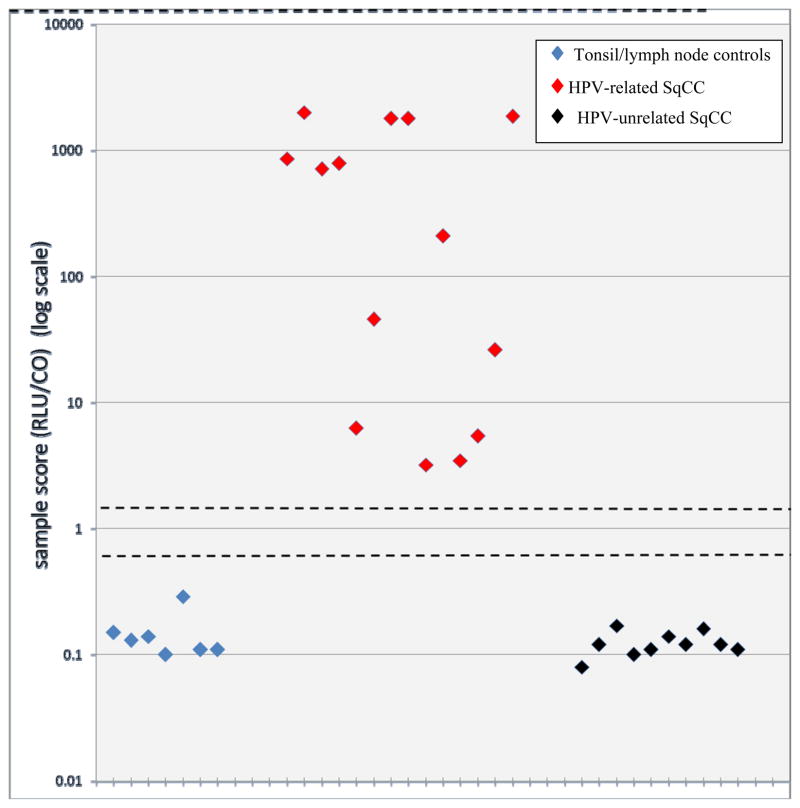
Figure 2.
HPV status by sample scores. For the cytologic preparations from head and neck squamous cell carcinomas, the reference scores of >3.0 and <0.85 (dashed lines) was found to have 100% sensitivity and 100% specificity in differentiating HPV positive (red diamonds) from HPV negative (black diamonds). The benign tonsils and lymph nodes (blue diamonds) were also scored as HPV negative. The y-values are shown on log scale.
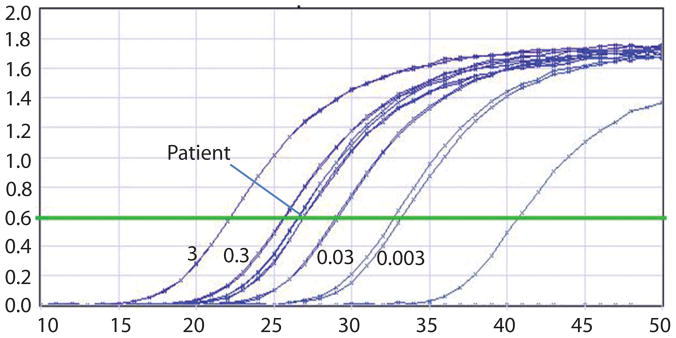
Figure 3.
Representative real-time quantitative PCR curves showed the amplification of E7 of HPV16. Standard curves were generated with series dilution of DNA (ng) from Caski cells. The samples have been normalized for DNA equivalence using actin dilution curves (data not shown). The X axis represents the PCR cycle number as the reaction takes place.
Discussion
HPV status is a powerful prognostic indicator for patients with HNSCC, and it is being used as an essential parameter in the recruitment of patients into various clinical trials. Despite a growing need for the accurate and practical HPV testing of clinical samples, there is not yet a consensus regarding the optimal detection method for universal implementation. Demands for flawless accuracy must be balanced against practical considerations regarding cost, turnaround time, ease of application, and ease of transfer to the clinical diagnostic laboratory. In this feasibility study, we show that the hybrid 2 capture test can detect HPV in cells scraped or aspirated from a squamous cell carcinoma of the head and neck (HNSCC). This assay may offer some advantages that would favor its use over other detection strategies.
In this study, the benchmark HPV status was established in the corresponding tissue sections using a detection strategy that combines the strengths of highly sensitive p16 immunohistochemistry and highly specific HPV in situ hybridization. The utility of this approach, however, is limited by a subset of HPV ambiguous carcinomas with discordant findings (i.e. p16+/HPV−). In one large series of HNSCCs, the p16+/HPV− discrepancy rate was 6%.(16) The basis of these discordant results is not always clear, but may reflect the inability of in situ hybridization to consistently detect HPV when present at low copy numbers. In our single p16+/HPV− carcinoma, real time PCR detection of E7 confirmed the presence of HPV-16 at about 1 copy per tumor cell. Notably, the tumor was scored as HPV positive using the hybrid capture 2 assay suggesting that the assay is highly sensitive even at the low end of viral copy number where in situ hybridization may be most susceptible to failure. The high sensitivity of the hybrid capture 2 assay does not appear to compromise its specificity: HPV was not detected in any of the cytologic preparations from HPV negative controls (non-neoplastic tonsils and lymph nodes) or the HPV negative HNSCCs.
This study used a standard threshold established from a large experience of HPV detection in cervical brushings, and was able to confirm the fidelity of this reference standard across different specimen types. This ability to unambiguously score samples as positive or negative based on where values reside in relation to a standard reference is preferable to “semi”-quantitative assays that does not lend themselves to objective quantitative analysis. Reproducibility of immunohistochemical analysis, for example, is highly vulnerable to staining variation as a function of tissue preservation and processing, to disparities in assay protocol and reagents, and to the interpretation bias inherent with observer-based assessment devoid of quantitative scoring systems.(18) Quantitative separation of HPV-positive from HPV-negative HNSCCs may not only benefit patients individually, but will promote enhanced objectivity and reproducibility among clinical trials where participants are selected on the basis of HPV status.
Various practical factors may also support the preferential use of the hybrid capture 2 assay over other methods of HPV testing. Specimens can be easily collected without the need for tumor micro dissection, formalin-fixation, or specimen processing of any kind. The liquid phase media serves not only as a transport media but also as a storage media. Accordingly, there is no need for a complex workflow to support rapid specimen transport from collection site to laboratory to minimize specimen degradation. Automation ensures reproducibility and provides a capacity for handling increasing clinical volumes. Same day results permits timely treatment decisions including study enrollment by eliminating HPV determination as a rate limiting factor. The hybrid capture 2 assay is cost effective. At our institution, the unit cost associated of the hybrid capture 2 assay is less than the cost of a single immunohistochemical stain. Determination of HPV status using a single and simple assay sharply contrasts with current trends toward complex multi-tiered staged algorithms using multimodality detection assays.(19;20)
Perhaps most importantly, the hybrid capture 2 assay allows for real time HPV testing of cytologic preparations. For HNSCCs, HPV detection strategies have primarily tried to adapt tissue-targeted approaches (e.g. p16 immunohistochemistry and HPV in situ hybridization) to archived cytologic specimens.(10;21–23) In most instances, HPV testing of cytologic specimens is restricted to a small subset of cases where ample cellular material is available for the construction of cell blocks. The hybrid capture 2 assay appears to reliably detect approximately 5,000 viral copies per reaction. In effect, the cellular demands of the assay ranges from 5,0000 tumor cells to less than 20 tumor cells depending on the viral load. Admittedly, our study design facilitated the collection of highly cellular specimens, but the modest cellular demands of the assay suggest that this strategy will be effective and transferrable to the clinics where neck node metastases can be aspirated and oropharyngeal tumors can be brushed.
The development and implementation of HPV detection strategies that are transferrable to clinical cytologic specimens is compelled by factors that restrict tissue access for HPV testing. The first is related to tumor size. HPV-related HNSCCs are often small and deeply concealed within the crypts of the lingual or palatine tonsils. Indeed for those patients with cervical lymph node metastases of unknown primary origin, their small and elusive primary cancers are likely to be HPV-related.(9;10;16;24) A second is related to evolving treatment practices. HPV-related HNSCCs tend to be sensitive to radiation and chemotherapy such that surgical removal is often unwarranted.(7;25) A third is related to current diagnostic practices. The routine use of FNAs for establishing a diagnosis of HNSCC in patients with lymph node metastases renders subsequent tissue acquisition unnecessary and excessive. Limited access to excised tissues argues for the development of other assays that permit the reliable, convenient and cost-effective determination of HPV status of cytological specimens. Although this feasibility study was limited to a relatively small number of samples that were surgically removed, our findings suggest that the hybrid capture 2 assay is a promising method that warrants additional large scale testing of cytologic specimens in the clinical arena.
Acknowledgments
This work was funded by the NIDCR (P50 DE019032)
Footnotes
Financial disclosures: none
Reference List
- 1.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 2.Junor EJ, Kerr GR, Brewster DH. Oropharyngeal cancer. Fastest increasing cancer in Scotland, especially in men. BMJ. 2010;340:c2512. doi: 10.1136/bmj.c2512. [DOI] [PubMed] [Google Scholar]
- 3.Ramqvist T, Dalianis T. Oropharyngeal cancer epidemic and human papillomavirus. Emerg Infect Dis. 2010;16:1671–7. doi: 10.3201/eid1611.100452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010 doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, et al. Improved Survival of Patients With Human Papillomavirus-Positive Head and Neck Squamous Cell Carcinoma in a Prospective Clinical Trial. J Natl Cancer Inst. 2008 doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 6.D’Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 7.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao M, Rosenbaum E, Carvalho AL, Koch W, Jiang W, Sidransky D, et al. Feasibility of quantitative PCR-based saliva rinse screening of HPV for head and neck cancer. Int J Cancer. 2005;117:605–10. doi: 10.1002/ijc.21216. [DOI] [PubMed] [Google Scholar]
- 9.Begum S, Gillison ML, Ansari-Lari MA, Shah K, Westra WH. Detection of human papillomavirus in cervical lymph nodes: a highly effective strategy for localizing site of tumor origin. Clin Cancer Res. 2003;9:6469–75. [PubMed] [Google Scholar]
- 10.Begum S, Gillison ML, Nicol TL, Westra WH. Detection of human papillomavirus-16 in fine-needle aspirates to determine tumor origin in patients with metastatic squamous cell carcinoma of the head and neck. Clin Cancer Res. 2007;13:1186–91. doi: 10.1158/1078-0432.CCR-06-1690. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal Y, Koch WM, Xiao W, Westra WH, Trivett AL, Symer DE, et al. Oral human papillomavirus infection before and after treatment for human papillomavirus 16-positive and human papillomavirus 16-negative head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:7143–50. doi: 10.1158/1078-0432.CCR-08-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuang AY, Chuang TC, Chang S, Zhou S, Begum S, Westra WH, et al. Presence of HPV DNA in convalescent salivary rinses is an adverse prognostic marker in head and neck squamous cell carcinoma. Oral Oncol. 2008;44:915–9. doi: 10.1016/j.oraloncology.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cervix Cancer Screening. IARC Press; 2005. IARC Working Group on the Evaluation of Cancer Preventive Strategies. [Google Scholar]
- 14.Carozzi FM, Del Mistro A, Confortini M, Sani C, Puliti D, Trevisan R, et al. Reproducibility of HPV DNA Testing by Hybrid Capture 2 in a Screening Setting. Am J Clin Pathol. 2005;124:716–21. doi: 10.1309/84E5-WHJQ-HK83-BGQD. [DOI] [PubMed] [Google Scholar]
- 15.Bozzetti M, Nonnenmacher B, Mielzinska II, Villa L, Lorincz A, Breitenbach VV, et al. Comparison between hybrid capture II and polymerase chain reaction results among women at low risk for cervical cancer. Ann Epidemiol. 2000;10:466. doi: 10.1016/s1047-2797(00)00147-2. [DOI] [PubMed] [Google Scholar]
- 16.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116:2166–73. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
- 17.Ha PK, Pai SI, Westra WH, Gillison ML, Tong BC, Sidransky D, et al. Real-time quantitative PCR demonstrates low prevalence of human papillomavirus type 16 in premalignant and malignant lesions of the oral cavity. Clin Cancer Res. 2002;8:1203–9. [PubMed] [Google Scholar]
- 18.Taylor CR, Levenson RM. Quantification of immunohistochemistry--issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411–24. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- 19.Thavaraj S, Stokes A, Guerra E, Bible J, Halligan E, Long A, et al. Evaluation of human papillomavirus testing for squamous cell carcinoma of the tonsil in clinical practice. J Clin Pathol. 2011;64:308–12. doi: 10.1136/jcp.2010.088450. [DOI] [PubMed] [Google Scholar]
- 20.Smeets SJ, Hesselink AT, Speel EJ, Haesevoets A, Snijders PJ, Pawlita M, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121:2465–72. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 21.Jannapureddy S, Cohen C, Lau S, Beitler JJ, Siddiqui MT. Assessing for primary oropharyngeal or nasopharyngeal squamous cell carcinoma from fine needle aspiration of cervical lymph node metastases. Diagn Cytopathol. 2010;38:795–800. doi: 10.1002/dc.21293. [DOI] [PubMed] [Google Scholar]
- 22.Umudum H, Rezanko T, Dag F, Dogruluk T. Human papillomavirus genome detection by in situ hybridization in fine-needle aspirates of metastatic lesions from head and neck squamous cell carcinomas. Cancer. 2005;105:171–7. doi: 10.1002/cncr.21027. [DOI] [PubMed] [Google Scholar]
- 23.Zhang MQ, El Mofty SK, Davila RM. Detection of human papillomavirus-related squamous cell carcinoma cytologically and by in situ hybridization in fine-needle aspiration biopsies of cervical metastasis: a tool for identifying the site of an occult head and neck primary. Cancer. 2008;114:118–23. doi: 10.1002/cncr.23348. [DOI] [PubMed] [Google Scholar]
- 24.Weiss D, Koopmann M, Rudack C. Prevalence and impact on clinicopathological characteristics of human papillomavirus-16 DNA in cervical lymph node metastases of head and neck squamous cell carcinoma. Head Neck. 2010;33:856–62. doi: 10.1002/hed.21548. [DOI] [PubMed] [Google Scholar]
- 25.Marklund L, Hammarstedt L. Impact of HPV in Oropharyngeal Cancer. J Oncol. 2011;2011:509036. doi: 10.1155/2011/509036. [DOI] [PMC free article] [PubMed] [Google Scholar]