Abstract
Model studies in mice indicate that the severity of alcohol withdrawal is associated with polymorphic variation and expression of the MPDZ gene. Current knowledge about variation in the human MPDZ gene is limited; however, our data indicate its potential association with alcohol dependence. The MPDZ protein is an important part of the NMDA-dependent AMPA receptor trafficking cascade that controls glutamate-related excitatory neurotransmission. To investigate association of variation in the NMDA-dependent AMPA trafficking cascade with alcohol dependence, we performed a gene-set (pathway) analysis using single nucleotide polymorphism (SNP) data from the Study of Addiction: Genetic and Environment (SAGE). Rather than testing for association with each SNP individually, which typically has low power to detect small effects of multiple SNPs, gene set analysis applies a single statistical test to evaluate whether variation in a set of genes is associated with the phenotype of interest. Gene-set analysis of 988 SNPs in 13 genes in the pathway demonstrated a significant association with alcohol dependence, with p<0.01 for the global effect of variation in this pathway. The statistically significant association of alcohol dependence with genetic variation in the NMDA-dependent AMPA receptor trafficking cascade indicates a need for further investigation of the role of this pathway in alcohol dependence.
Keywords: alcohol dependence, AMPA, genetics, NMDA, pathway analysis
INTRODUCTION
Alcoholism is a complex disease with a major genetic contribution (Goldman, Oroszi and Ducci, 2005; Kendler et al., 2010; Schuckit, Goodwin and Winokur, 1972). Efforts and resources devoted to the identification of genes that contribute to the vulnerability of alcohol-related problems have resulted in a number of positive associations allowing some insight into the pathophysiology of alcohol dependence and related phenotypes (for review see (Kohnke, 2008)). In addition to numerous candidate gene studies, genome-wide association studies (GWAS) utilizing an “unbiased” non-hypothesis driven strategy of genotyping a large number (currently ~ 1, 000,000) of single nucleotide polymorphisms (SNPs) across the human genome have been applied to study genetic risk factors for addiction. However, in the recently published GWAS for alcohol dependence there were no genome-wide significant association findings (Bierut et al., 2010; Edenberg et al., 2010; Treutlein et al., 2009). It is likely a reflection of the fact that with currently available samples, the analysis strategy implemented in these GWAS offers limited power to identify SNPs associated with the complex phenotype of alcohol dependence. This power limitation indicates the need for even larger sample sizes than were used in these studies, as well as new statistical methods and innovative approaches to identify the genes involved in the disease process (Buckland, 2008).
Genome wide studies typically apply analysis approaches that assess the effect of each SNP individually. If a genetic variation causes a major alteration of the protein function its effect size may be large and association with the disease as well as its pathophysiology may be easily identified. It is expected, however, that complex interactions between multiple genetic variations, each with a small effect, contribute to the risk of complex diseases like alcoholism. Thus effect sizes of individual genetic variations are expected to be modest. Assessing the effect of each variation individually is not well suited to the detection of small effects of multiple SNPs. One potential alternative is to focus on physiologically meaningful sets of genes in search for their association with the phenotypes of interest (Holmans, 2010; Wang, Li and Hakonarson, 2010). Unlike more traditional approaches focussed on individual variation, gene-set analyses use an entire gene set as a single entity to test for association with the phenotype of interest. Such approaches have been applied to several neuropsychiatric traits including cognitive ability and bipolar disorder (Holmans et al., 2009; O’Dushlaine et al., 2010; Ruano et al., 2010). By identifying functionally relevant sets of genes corresponding to relevant pathophysiological pathways, results of gene-set analyses can motivate more focused studies of genetic risk factors. This approach therefore has the potential to substantially contribute to the discovery of genetic variants associated with alcohol dependence and related phenotypes.
Model studies have demonstrated that variations in the coding sequence and expression of the mouse Mpdz gene are associated with severity of alcohol withdrawal and seizures. To test the implication of these findings for human alcoholism, we sequenced the human homolog (MPDZ) in search for variation at sites homologous to sites described as being part of a withdrawal seizure-related haplotype in mice (Karpyak et al., 2009). We found no genetic variability in the human MPDZ gene sites homologous to these variability sites in mice. However, the use of a global test of haplotype association revealed a significant difference in haplotype frequencies between alcohol-dependent subjects and controls (p = 0.015), suggesting a potential role of MPDZ gene in alcoholism and or related phenotypes (Karpyak et al., 2009).
The MPDZ (multi-PDZ (PSD95/discs large/ZO-1) domain-containing) protein is known to interact with the protein subunits of glutamate (GRIN2B subunit of NMDA receptor), as well as serotonin (5-HT2A and 5-HT2C), GABA (GABBR2), and dopamine (DRD2, DRD3, and DRD4) receptors (Balasubramanian, Fam and Hall, 2007; Becamel et al., 2001; Griffon et al., 2003; Kim and Sheng, 2004; Krapivinsky et al., 2004; Rama et al., 2008; Ullmer et al., 1998). Of particular interest is the role of MPDZ as a key element of the NMDA-dependent AMPA trafficking cascade (Krapivinsky et al., 2004). As illustrated in Fig. 1, in the synapses of hippocampal neurons, synaptic GTPase-activating protein (SynGAP) and Ca2+/calmodulin-dependent kinase (CaMKII) are brought together by direct physical interaction with the MPDZ domains. Calcium entering via the NMDA receptors triggers changes resulting in SynGAP dissociation from the MPDZ-CaMKII complex, potentiation of synaptic AMPA responses, and an increase in the number of AMPA receptor-containing clusters in hippocampal neuron synapses (Krapivinsky et al., 2004).
Figure 1. A schematic representation of the interactions within NMDA-dependent AMPA trafficking cascade.
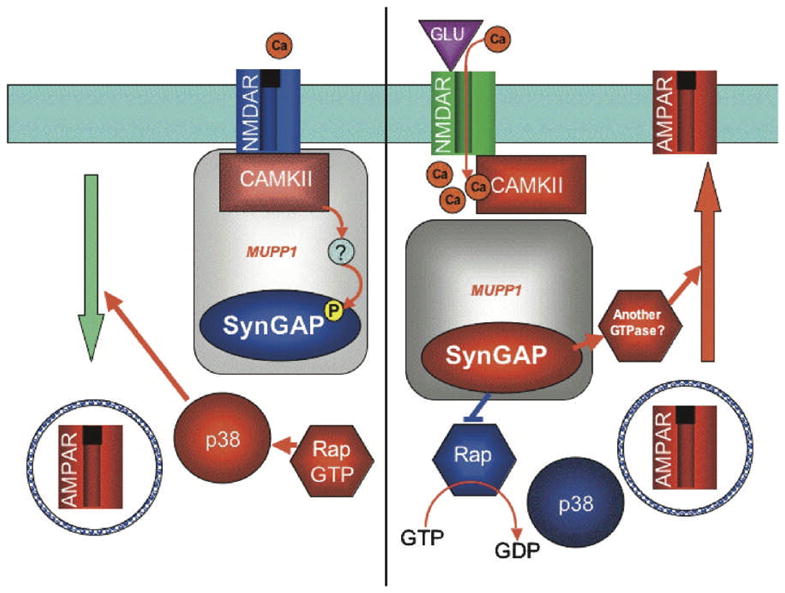
Reprinted with permission from Elsevier (Krapivinsky et al., 2004)
Abbreviations: AMPAR, Alpha-amino-3-hydroxy-5-Methyl-4-isoxazole Propionic Acid-sensitive glutamate receptor; CaMKII, calcium/calmodulin-dependent protein kinase II; GTP, Guanosine-5′-triphosphate; GDP - Guanosine-5′-diphosphate; MUPP1, (MPDZ), multiple PDZ domain protein; NMDAR, N-methyl-D-aspartate receptor; p38, mitogen-activated protein kinase 14; Rap, small Ras family GTPase; SynGAP, Synaptic Ras GTPase activating protein; GLU –glutamate; Ca – calcium.
Evidence from human and animal research implicates glutamate neurotransmission in the pathophysiology of alcohol dependence related craving, withdrawal, neuronal excitotoxicity as well as treatment response (Dahchour and De Witte, 2003; Dodd et al., 2000; Johnson, 2004; Krupitsky et al., 2007; Tsai and Coyle, 1998). Human research supports association of alcohol dependence-related phenotypes with variation in some of the genes (e.g., GRIN1) involved in this cascade (Rujescu et al., 2005; Wernicke et al., 2003). In addition to the MPDZ gene, a recent review of experimental findings emphasizes the importance of NMDA-AMPA receptor-induced synaptic strengthening in different brain areas on alcohol-related behavioral phenotypes (Spanagel, 2009).
Thus, compelling evidence implicates the potential association between alcohol dependence and variability in MPDZ and other genes coding for proteins in the NMDA-dependent AMPA trafficking cascade described by Krapivinsky et al. (2004). To build upon these findings we investigated the association of alcohol dependence with SNP variation in a gene set corresponding to this pathway. This paper presents results of this investigation.
MATERIALS AND METHODS
Selection of the gene set
The NMDA-dependent AMPA trafficking cascade proposed by Krapivinski et al. (2004) includes seven main functional elements presented in Fig. 1 (AMPAR, Alpha-amino-3-hydroxy-5-Methyl-4-isoxazole Propionic Acid-sensitive glutamate receptor; CaMKII, calcium/calmodulin-dependent protein kinase II; MPDZ (also called MUPP1), multiple PDZ domain protein; NMDAR, N-methyl-D-aspartate receptor; p38, mitogen-activated protein kinase 14; Rap, small Ras family GTPase; SynGAP, Synaptic Ras GTPase activating protein). According to contemporary knowledge, some of these functional elements (MPDZ, SynGAP, p38, Rap) are comprised of a single protein while others (NMDA, AMPA, CaMKII) are more complex and include several proteins. Thus, for this study, we defined a gene set that included genes coding for proteins directly shown to be involved in the pathway of interest, as well as genes coding for proteins known to form functional units with proteins that directly interact with the MPDZ protein. Specifically, it has been demonstrated that MPDZ protein directly interacts with the NR2B subunit of the NMDA receptor, and co-immunoprecipitates with CaMKII-alpha and CaMKII-beta proteins (Krapivinsky et al., 2004; Rama et al., 2008). Thus, in addition to the GRIN2B gene that encodes NR2B, we included GRIN1 and GRIN2A genes coding for the NR1 and NR2A subunits, which form heteromeric protein complexes with the NR2B subunit to form an NMDA receptor. Similarly, it has been demonstrated that the GluR1 subunit of AMPA receptor interacts with SynGAP-alpha protein (Krapivinsky et al., 2004; Rama et al., 2008). However, it is known that GluR2-4 subunits assemble homo- or heteromers of functional AMPA receptors together with the GluR1 subunit (Nakagawa et al., 2005; Rosenmund, Stern-Bach and Stevens, 1998). Thus, in addition to the GRIA1 gene encoding the GluR1 subunit, we included GRIA2, GRIA3 and GRIA4 genes encoding for the GluR2-4 subunits.
Accordingly, the final list of candidate genes included in this study is comprised of all the genes that code for the protein elements of this cascade: GRIN1, GRIN2A, GRIN2B, SYNGAP1, CAMK2A, CAMK2B, GRIA1, GRIA2, GRIA3, GRIA4, MPDZ, RAP1GAP and MAPK14, bringing the total number of genes to13.
Data source and SNP selection
This study utilized data from the Study of Addiction: Genetic and Environment (SAGE), obtained through dbGaP (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1). Detailed description of the SAGE data set and its use in a genome-wide association study of alcohol dependence is provided elsewhere (Bierut et al., 2010). The analyses presented here used data from 2544 European American subjects, including 1165 alcohol dependent cases (SSAGA, DSM-IV) and 1379 unrelated, alcohol-exposed, nondependent controls. All subjects were genotyped using the Illumina 1M beadchip, resulting in 839,409 SNPs that passed quality control filters.
SNPs in genes included in the pathway described above were selected from the SAGE data set. All genotyped SNPs in any of these genes, as well as SNPs within 5kb of the first or last exon of one of the genes, were included in the gene set. This selection resulted in 988 SNPs in the 13 genes.
Statistical analysis
Two gene-set analysis approaches were utilized for this study, which we refer to as the onestep and two-step approaches (Biernacka et al., 2010). In the one-step approach, all SNPs in the gene set were used in the analysis without consideration of gene-level association. In the two-step approach, all SNPs in each gene were first used to evaluate association with the gene, followed by aggregation of the gene-level tests, to test for association of the phenotype with the entire gene set.
In the one-step approach, an association test was performed for each SNP using logistic regression assuming log-additive allele effects; the SNP-specific p-values were then combined using Fisher’s method (Chai et al., 2009; Fisher, 1932) to calculate a gene set association statistic. Significance of the gene-set statistic was assessed by permutation to avoid the assumption of independent p-values (Fridley, Jenkins and Biernacka, 2010). Thus, an empirical null distribution of the gene set test statistic was obtained by permuting the phenotype 1000 times, in a way that preserves the linkage disequilibrium between SNPs.
In the two-step approach, the association of the phenotype with variation in each gene was evaluated using principal component (PC) analysis as described by Gauderman et al. (Gauderman et al., 2007); the 13 gene-level association p-values were then combined using Fisher’s method (Fisher, 1932) to calculate the gene set association statistic. The gene-level test consisted of determining the PCs using all SNPs in a gene, followed by a logistic regression analysis with the PCs that explain 80% of the variation within the set of SNPs as predictors in the model. Gene-level association was then evaluated for each gene using a likelihood ratio test of the full PC model compared to the reduced model without the gene-specific PCs. Significance of the gene set association test was again evaluated by permutation. Both approaches used principal components (PCs) to adjust for possible population stratification within the European-American subjects by including the top three genome-wide PCs as covariates in the logistic regression analysis (Price et al., 2006).
RESULTS
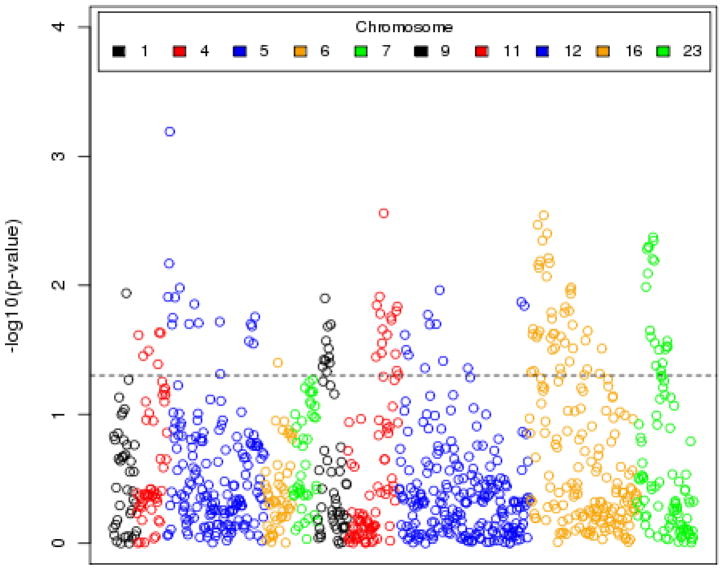
Figure 2 shows a Manhattan plot of individual SNP p-values for association of alcohol dependence with each genotyped SNP in the 13 genes belonging to the NMDA-dependent AMPA receptor trafficking cascade. Individual SNP results showing the lowest p-values are presented in Table 1. Among the 20 SNPs with smallest p-values are nine SNPs in GRIN2A (p<0.01), eight SNPs in GRIA3 (p<0.01), two SNPs in CAMK2A (p=0.00064 for rs980272; p=0.0068 for rs887346), and one SNP in GRIA4 (rs2508467; p=0.0028). None of the SNPs are significantly associated with alcohol dependence after a Bonferroni correction for multiple testing.
Figure 2.
Manhattan plot of individual SNP p-values, for tests of association between alcohol dependence and SNPs in the gene set representing the NMDA-dependent AMPA trafficking cascade pathway.
Table 1.
Results of tests of association of alcohol dependence with individual SNPs: 20 SNPs with smallest p-values.
| SNP | Gene | p-value* |
|---|---|---|
| rs980272 | CAMK2A | 0.00064 |
| rs2508467 | GRIA4 | 0.00276 |
| rs8050843 | GRIN2A | 0.00287 |
| rs2077923 | GRIN2A | 0.00340 |
| rs9933832 | GRIN2A | 0.00397 |
| rs7058062 | GRIA3 | 0.00424 |
| rs6497523 | GRIN2A | 0.00449 |
| rs7058099 | GRIA3 | 0.00451 |
| rs983007 | GRIA3 | 0.00503 |
| rs12557782 | GRIA3 | 0.00504 |
| rs6608062 | GRIA3 | 0.00526 |
| rs7201930 | GRIN2A | 0.00615 |
| rs2157271 | GRIA3 | 0.00626 |
| rs4825849 | GRIA3 | 0.00645 |
| rs16966381 | GRIN2A | 0.00650 |
| rs7204266 | GRIN2A | 0.00672 |
| rs887346 | CAMK2A | 0.00678 |
| rs1833161 | GRIN2A | 0.00692 |
| rs1544604 | GRIN2A | 0.00732 |
| rs989638 | GRIA3 | 0.00810 |
p-values are based on a logistic regression analysis with log-additive allele effects (i.e. 0, 1, 2 genotype coding representing the number of copies of the minor allele).
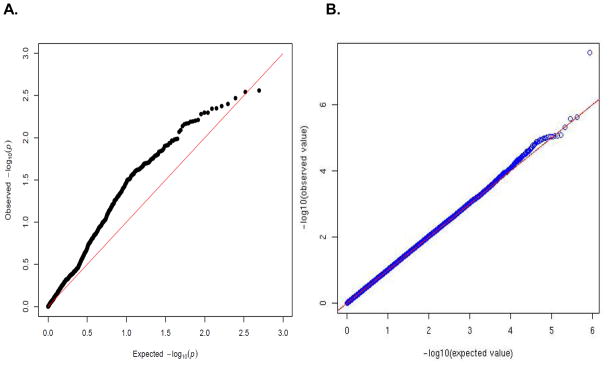
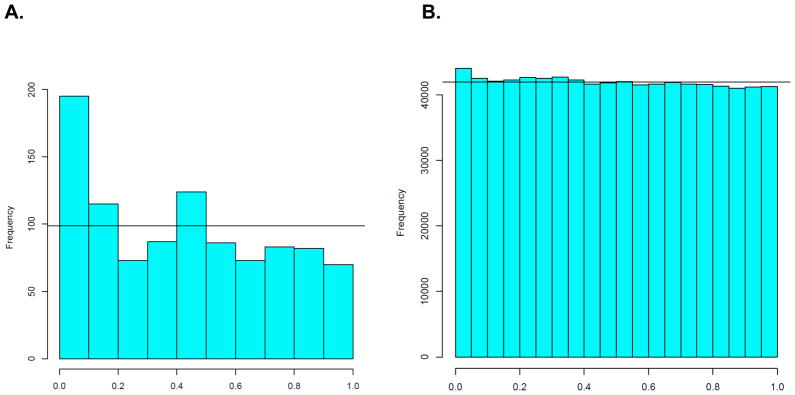
The one-step gene set analysis provided significant evidence of association between alcohol dependence and genetic variation in the NMDA-dependent AMPA receptor trafficking cascade (p=0.003 for the global effect of variation in this pathway). A Q-Q plot of the log-transformed p-values (Figure 3A) for the individual SNPs in the gene set demonstrates a significant deviation form the diagonal line representing the expected distribution of p-values under the null hypothesis. Conversely, a Q-Q plot of all SNPs across the genome presented in Figure 3B shows no such deviation, demonstrating that overall, when considering all genotyped SNPs across the genome, there is no evidence of SNP association with alcohol dependence. Histograms of SNP-specific p-values from the pathway of interest and across the genome are shown in Figures 4A and 4B, respectively. Figure 4A again shows a deviation from the expected distribution under the null hypothesis, demonstrated by a strong shift of the p-values toward the left side of the graph (more small p-values than expected by chance); while Figure 4B demonstrates a distribution of p-values consistent with the null hypothesis (p-values are approximately uniformly distributed between 0 and 1). Thus, both Figures 3 and 4 demonstrate that although no single p-value is smaller than the smallest expected p-value under the null hypothesis, overall, there are more small p-values (e.g. p<0.05) than expected by chance. Of the 988 SNPs in this gene set, 127 were associated with alcohol dependence at a p<0.05 level.
Figure 3.
Q-Q plots of –log10 (p-values) from tests of association between alcohol dependence and SNPs in the NMDA-dependent AMPA trafficking cascade (A) and SNPs across the genome (B).
Figure 4.
Histograms of p-values from tests of association between alcohol dependence and SNPs in the NMDA-dependent AMPA trafficking cascade (A), or across the genome (B).
For the two-step analysis, the overall gene set association test was not statistically significant (p=0.15). Gene-level results from the two-step analysis are displayed in Table 2. The strongest evidence for gene-level association is seen for GRIA2 (p=0.034) and GRIA4 (p=0.055); however, none of the gene-level tests are significant after Bonferroni correction for the 13 genes tested.
Table 2.
Gene-level and pathway results based on two-step analysis, with genes ordered by p-value.
| Gene | Numbers of SNPs | p-value* |
|---|---|---|
| GRIA2 | 52 | 0.034 |
| GRIA4 | 87 | 0.055 |
| GRIN2A | 183 | 0.095 |
| GRIA3 | 97 | 0.240 |
| CAMK2A | 39 | 0.290 |
| GRIA1 | 127 | 0.300 |
| CAMK2B | 42 | 0.330 |
| MAPK14 | 32 | 0.450 |
| GRIN2B | 222 | 0.470 |
| MPDZ | 40 | 0.540 |
| RAP1GAP | 40 | 0.62 |
| SYNGAP1 | 16 | 0.66 |
| GRIN1 | 11 | 0.97 |
| Gene-set | 988 | 0.149 |
Gene-level p-values in the two-step analysis are based on a logistic regression analysis with the top principal components, explaining 80% of all SNP variation in each gene, as the predictors. The gene-set p-value is based on combining the gene-level p-values using Fisher’s method (Fisher, 1932), with significance assessed by permutation (see text for details).
DISCUSSION
We found significant evidence of association between alcohol dependence and variation in the NMDA-dependent AMPA receptor trafficking cascade. While none of the individual SNPs were found to be significantly associated with alcohol dependence after Bonferroni correction for multiple testing, overall the individual p-values were smaller than expected by chance, providing evidence that this gene set is associated with alcohol dependence. This finding supports the original hypothesis that this glutamatergic pathway, which is involved in regulation of excitatory postsynaptic currents in hippocampal neurons (Krapivinsky et al., 2004; Rama et al., 2008), may play an important role in alcohol dependence. Further research is necessary to investigate the specific role this pathway plays in the physiology of alcoholism and to determine which phenotypic and/or endophenotypic traits are directly associated with functional variability in this pathway.
A key motivation for gene-set analysis is the expected gain of power compared to analysis of individual SNPs when many weak or moderate SNP effects within a set of functionally related genes contribute to a phenotypic trait (Holmans, 2010; Wang et al., 2010). The association observed here supports this idea. In fact, out of 839,409 SNPs that passed quality control procedures in the SAGE data, none were significantly associated with alcohol dependence at a genome-wide level of significance. Only fifteen SNPs yielded P < 10−5, and in two independent replication series, no SNP passed a replication threshold of P < 0.05 (Bierut et al., 2010). The significant evidence of gene-set association observed in this study provides further evidence that analyses informed by prior biological knowledge of gene function can be more powerful and provide important insights into disease etiology. It is important to recognize that while a gene set analysis provides a global test for the association of a phenotype with all measured variation in a set of genes, it is not intended to identify specific variants that contribute to the phenotype. However, by identifying pathways that contribute to the phenotype, results of gene set analysis can be used to guide further, more focused, studies of genetic risk factors for complex traits.
It is interesting that the one-step analysis provided higher evidence of association in this study than did the two-step approach. In simulation studies (Biernacka et al., 2010) the two-step approach tended to be more powerful than the one-step approach. However, there were situations when the one-step approach had greater power, including scenarios with moderate SNP-association effects in several genes, or multiple weak effects in each of several genes. The gene-level association results obtained as part of the two-step analysis indicated that there was little evidence of association of alcohol dependence with particular genes in this gene set. One possible interpretation of this finding is that potentially meaningful gene associations may not be detectable by principal component analysis when functionally important SNPs are analyzed in the presence of numerous non-functional SNPs in a gene. On the other hand, the combined effect of SNPs in several functionally-related genes that influence the same phenotype may be easier to detect when a functionally important gene set is considered.
Nine out of 20 SNPs with lowest p-values in our study (Table 1) are in the GRIN2A gene, coding for the NR2A subunit of the NMDA receptor. Previously, a systematic analysis of 10 glutamatergic neurotransmission genes indicated that GRIN2A has the greatest relevance for human alcohol dependence among the investigated genes (Schumann et al., 2008). This association of alcohol dependence with genetic variation in GRIN2A was also replicated in an independent sample (Schumann et al., 2008). Of the remaining 11 of the 20 SNPs with the lowest p-values in our study, eight are in the GRIA3 gene that codes for the GluR3 subunit of the AMPA receptor. Variation in this gene has been shown to be associated with a number of neuro-psychiatric diseases, including schizophrenia (Magri et al., 2008), depression (Utge et al., 2010), sexual disfunction during citalopram treatment of depression (Perlis et al., 2009), and treatment emergent suicidal ideation (Laje et al., 2007). In our analysis of 13 genes in the NMDA-dependent AMPA receptor trafficking cascade, the SNP with the smallest p-value is in CAMK2A, while the one with the second smallest p-value is in GRIA4. SNPs in both CAMK2A and GRIA4 have previously been reported to be associated with clinical phenotypes in studies of schizophrenia (Makino et al., 2003; Need et al., 2009).
In our two-step analysis, the only gene that was associated with alcohol dependence at a nominally significant level was GRIA2 (p=0.034, Table 2). Variability in this gene has been shown to be associated with lithium response in bipolar patients (Perlis et al., 2009). To the best of our knowledge, there have been no reports of association of variability in the GRIA2 gene with alcohol dependence; however, significantly increased expression of the corresponding GluR2 protein was observed in human postmortem brain tissue (hippocampi) from subjects with alcohol abuse histories when compared to a normal control group (Breese, Freedman and Leonard, 1995).
Limitations of the gene set analysis approach include the reliance on a priori defined sets of genes. Thus, its effectiveness is determined by contemporary knowledge of relevant biological pathways. Current gaps in knowledge therefore limit the applicability of this approach in some situations. Moreover, specification of pathways and related gene sets can be over-inclusive or may miss important components. Selection of genes for this study was based on contemporary knowledge about the NMDA-dependent AMPA receptor trafficking cascade and it is possible that the analyzed gene set was over-inclusive or omitted relevant genes. Other limitations include the fact that the analyses made use of available SNP genotypes from the Illumina 1M platform, and did not include other types of variations such as variable number of tandem repeat (VNTR) polymorphisms or copy number variations (CNVs). As the coverage of genes by SNPs on GWAS arrays is not uniform, power to detect association with some genes in the gene set may be lower than for other genes. Finally, although gene set analysis attempts to investigate the overall evidence of association with variation in a set of related genes, currently used statistical methods still fail to account for joint SNP effects beyond simple log-additive effects of individual SNPs. Methods that take into account gene-gene interaction effects may provide greater power to detect gene set association. Nevertheless, the analysis presented here demonstrated a significant gene set association with alcohol dependence.
In conclusion, the statistically significant association of alcohol dependence with genetic variation in the NMDA-dependent AMPA receptor trafficking cascade reported here indicates a potential role of this pathway in alcohol dependence. As in all genetic studies of complex human traits, subsequent replication and functional validation are necessary to fully understand the implications of these findings.
Acknowledgments
The research presented in this paper was supported by the NIAAA (R03 AA019570) and SC Johnson Genomics of Addictions Program at Mayo Clinic. The dataset used for the analyses described in this manuscript was obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgibin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p1. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
Footnotes
Authors Contribution
VK and JB were responsible for the study concept and design. JB designed the analysis plan and oversaw the statistical analysis of the data, while JG and CC managed the data, performed the analyses and prepared results summaries and plots. VK and JB drafted the manuscript. VK, JB, JG, CC, DM critically reviewed and approved the final version of the manuscript.
References
- Balasubramanian S, Fam SR, Hall RA. GABAB receptor association with the PDZ scaffold Mupp1 alters receptor stability and function. J Biol Chem. 2007;282:4162–4171. doi: 10.1074/jbc.M607695200. [DOI] [PubMed] [Google Scholar]
- Becamel C, Figge A, Poliak S, Dumuis A, Peles E, Bockaert J, Lubbert H, Ullmer C. Interaction of serotonin 5-hydroxytryptamine type 2C receptors with PDZ10 of the multi-PDZ domain protein MUPP1. J Biol Chem. 2001;276:12974–12982. doi: 10.1074/jbc.M008089200. [DOI] [PubMed] [Google Scholar]
- Biernacka J, Jenkins G, Wang L, Moyer A, Fridley B. Gene Set Analysis of SNP Data: An Assessment of Methods. Currently. 2010 under review. [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese CR, Freedman R, Leonard SS. Glutamate receptor subtype expression in human postmortem brain tissue from schizophrenics and alcohol abusers. Brain Res. 1995;674:82–90. doi: 10.1016/0006-8993(94)01384-t. [DOI] [PubMed] [Google Scholar]
- Buckland PR. Will we ever find the genes for addiction? Addiction. 2008;103:1768–1776. doi: 10.1111/j.1360-0443.2008.02285.x. [DOI] [PubMed] [Google Scholar]
- Chai HS, Sicotte H, Bailey KR, Turner ST, Asmann YW, Kocher JP. GLOSSI: a method to assess the association of genetic loci-sets with complex diseases. BMC Bioinformatics. 2009;10:102. doi: 10.1186/1471-2105-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahchour A, De Witte P. Excitatory and inhibitory amino acid changes during repeated episodes of ethanol withdrawal: an in vivo microdialysis study. Eur J Pharmacol. 2003;459:171–178. doi: 10.1016/s0014-2999(02)02851-0. [DOI] [PubMed] [Google Scholar]
- Dodd PR, Beckmann AM, Davidson MS, Wilce PA. Glutamate-mediated transmission, alcohol, and alcoholism. Neurochem Int. 2000;37:509–533. doi: 10.1016/s0197-0186(00)00061-9. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr, Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. Statistical methods for research workers. 5. Oliver and Boyd; Edinburgh: 1932. [Google Scholar]
- Fridley BL, Jenkins GD, Biernacka JM. Self-contained gene-set analysis of expression data: an evaluation of existing and novel methods. PLoS ONE. 2010:5. doi: 10.1371/journal.pone.0012693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Murcray C, Gilliland F, Conti DV. Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol. 2007;31:383–395. doi: 10.1002/gepi.20219. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: Uncovering the genes. Nature Reviews Genetics. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Griffon N, Jeanneteau F, Prieur F, Diaz J, Sokoloff P. CLIC6, a member of the intracellular chloride channel family, interacts with dopamine D(2)-like receptors. Brain Res Mol Brain Res. 2003;117:47–57. doi: 10.1016/s0169-328x(03)00283-3. [DOI] [PubMed] [Google Scholar]
- Holmans P. Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Adv Genet. 2010;72:141–179. doi: 10.1016/B978-0-12-380862-2.00007-2. [DOI] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O’Donovan MC, Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA. An overview of the development of medications including novel anticonvulsants for the treatment of alcohol dependence. Expert Opin Pharmacother. 2004;5:1943–1955. doi: 10.1517/14656566.5.9.1943. [DOI] [PubMed] [Google Scholar]
- Karpyak VM, Kim JH, Biernacka JM, Wieben ED, Mrazek DA, Black JL, Choi DS. Sequence Variations of the Human MPDZ Gene and Association With Alcoholism in Subjects With European Ancestry. Alcohol Clin Exp Res. 2009;33:712–721. doi: 10.1111/j.1530-0277.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Dick D, Prescott CA. The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcohol Clin Exp Res. 2010;34:1058–1065. doi: 10.1111/j.1530-0277.2010.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kohnke MD. Approach to the genetics of alcoholism: a review based on pathophysiology. Biochem Pharmacol. 2008;75:160–177. doi: 10.1016/j.bcp.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43:563–574. doi: 10.1016/j.neuron.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Krupitsky EM, Rudenko AA, Burakov AM, Slavina TY, Grinenko AA, Pittman B, Gueorguieva R, Petrakis IL, Zvartau EE, Krystal JH. Antiglutamatergic strategies for ethanol detoxification: comparison with placebo and diazepam. Alcohol Clin Exp Res. 2007;31:604–611. doi: 10.1111/j.1530-0277.2007.00344.x. [DOI] [PubMed] [Google Scholar]
- Laje G, Paddock S, Manji H, Rush AJ, Wilson AF, Charney D, McMahon FJ. Genetic markers of suicidal ideation emerging during citalopram treatment of major depression. Am J Psychiatry. 2007;164:1530–1538. doi: 10.1176/appi.ajp.2007.06122018. [DOI] [PubMed] [Google Scholar]
- Magri C, Gardella R, Valsecchi P, Barlati SD, Guizzetti L, Imperadori L, Bonvicini C, Tura GB, Gennarelli M, Sacchetti E, Barlati S. Study on GRIA2, GRIA3 and GRIA4 genes highlights a positive association between schizophrenia and GRIA3 in female patients. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:745–753. doi: 10.1002/ajmg.b.30674. [DOI] [PubMed] [Google Scholar]
- Makino C, Fujii Y, Kikuta R, Hirata N, Tani A, Shibata A, Ninomiya H, Tashiro N, Shibata H, Fukumaki Y. Positive association of the AMPA receptor subunit GluR4 gene (GRIA4) haplotype with schizophrenia: linkage disequilibrium mapping using SNPs evenly distributed across the gene region. Am J Med Genet B Neuropsychiatr Genet. 2003;116B:17–22. doi: 10.1002/ajmg.b.10041. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Need AC, Keefe RS, Ge D, Grossman I, Dickson S, McEvoy JP, Goldstein DB. Pharmacogenetics of antipsychotic response in the CATIE trial: a candidate gene analysis. Eur J Hum Genet. 2009;17:946–957. doi: 10.1038/ejhg.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dushlaine C, Kenny E, Heron E, Donohoe G, Gill M, Morris D, Corvin A. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- Perlis RH, Laje G, Smoller JW, Fava M, Rush AJ, McMahon FJ. Genetic and clinical predictors of sexual dysfunction in citalopram-treated depressed patients. Neuropsychopharmacology. 2009;34:1819–1828. doi: 10.1038/npp.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rama S, Krapivinsky G, Clapham DE, Medina I. The MUPP1-SynGAPalpha protein complex does not mediate activity-induced LTP. Mol Cell Neurosci. 2008;38:183–188. doi: 10.1016/j.mcn.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Ruano D, Abecasis GR, Glaser B, Lips ES, Cornelisse LN, de Jong AP, Evans DM, Davey Smith G, Timpson NJ, Smit AB, Heutink P, Verhage M, Posthuma D. Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. Am J Hum Genet. 2010;86:113–125. doi: 10.1016/j.ajhg.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujescu D, Soyka M, Dahmen N, Preuss U, Hartmann AM, Giegling I, Koller G, Bondy B, Moller HJ, Szegedi A. GRIN1 locus may modify the susceptibility to seizures during alcohol withdrawal. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:85–87. doi: 10.1002/ajmg.b.30112. [DOI] [PubMed] [Google Scholar]
- Schuckit M, Goodwin D, Winokur G. A study of alcoholism in half siblings. Am J Psychiatry. 1972;128:1132–1136. doi: 10.1176/ajp.128.9.1132. [DOI] [PubMed] [Google Scholar]
- Schumann G, Johann M, Frank J, Preuss U, Dahmen N, Laucht M, Rietschel M, Rujescu D, Lourdusamy A, Clarke TK, Krause K, Dyer A, Depner M, Wellek S, Treutlein J, Szegedi A, Giegling I, Cichon S, Blomeyer D, Heinz A, Heath S, Lathrop M, Wodarz N, Soyka M, Spanagel R, Mann K. Systematic analysis of glutamatergic neurotransmission genes in alcohol dependence and adolescent risky drinking behavior. Arch Gen Psychiatry. 2008;65:826–838. doi: 10.1001/archpsyc.65.7.826. [DOI] [PubMed] [Google Scholar]
- Spanagel R. Alcoholism: a systems approach from molecular physiology to addictive behavior. Physiol Rev. 2009;89:649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide Association Study of Alcohol Dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai G, Coyle JT. The role of glutamatergic neurotransmission in the pathophysiology of alcoholism. Annu Rev Med. 1998;49:173–184. doi: 10.1146/annurev.med.49.1.173. [DOI] [PubMed] [Google Scholar]
- Ullmer C, Schmuck K, Figge A, Lubbert H. Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Lett. 1998;424:63–68. doi: 10.1016/s0014-5793(98)00141-0. [DOI] [PubMed] [Google Scholar]
- Utge S, Soronen P, Partonen T, Loukola A, Kronholm E, Pirkola S, Nyman E, Porkka-Heiskanen T, Paunio T. A population-based association study of candidate genes for depression and sleep disturbance. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:468–476. doi: 10.1002/ajmg.b.31002. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- Wernicke C, Samochowiec J, Schmidt LG, Winterer G, Smolka M, Kucharska-Mazur J, Horodnicki J, Gallinat J, Rommelspacher H. Polymorphisms in the N-methyl-D-aspartate receptor 1 and 2B subunits are associated with alcoholism-related traits. Biol Psychiatry. 2003;54:922–928. doi: 10.1016/s0006-3223(03)00072-6. [DOI] [PubMed] [Google Scholar]