Abstract
Background
Accurate knowledge of a patient's medical problems is critical for clinical decision making, quality measurement, research, billing and clinical decision support. Common structured sources of problem information include the patient problem list and billing data; however, these sources are often inaccurate or incomplete.
Objective
To develop and validate methods of automatically inferring patient problems from clinical and billing data, and to provide a knowledge base for inferring problems.
Study design and methods
We identified 17 target conditions and designed and validated a set of rules for identifying patient problems based on medications, laboratory results, billing codes, and vital signs. A panel of physicians provided input on a preliminary set of rules. Based on this input, we tested candidate rules on a sample of 100 000 patient records to assess their performance compared to gold standard manual chart review. The physician panel selected a final rule for each condition, which was validated on an independent sample of 100 000 records to assess its accuracy.
Results
Seventeen rules were developed for inferring patient problems. Analysis using a validation set of 100 000 randomly selected patients showed high sensitivity (range: 62.8–100.0%) and positive predictive value (range: 79.8–99.6%) for most rules. Overall, the inference rules performed better than using either the problem list or billing data alone.
Conclusion
We developed and validated a set of rules for inferring patient problems. These rules have a variety of applications, including clinical decision support, care improvement, augmentation of the problem list, and identification of patients for research cohorts.
Keywords: Problem list, clinical decision support, data mining, automated inference, methodology
Introduction and background
Having a clear picture of a patient's problems and diagnoses is critical for a variety of reasons. First and foremost, knowledge of a patient's problems facilitates optimal clinical decision making—without understanding the full scope of patients' clinical issues, it is very difficult to take good care of them. However, knowledge of problems is also critical for a variety of other activities, such as clinical decision support,1 quality improvement and measurement, and research.
The most obvious source of information about a patient's problems is the clinical problem list. The concept of a problem list, the central component of the problem-oriented medical record, was first described by Lawrence Weed, MD, in 1968.2 Weed proposed a new method of organizing medical records with problems at the center and data organized around the problems. Clinical problem lists serve a variety of purposes in facilitating care including: promoting continuity of care, describing active diseases, recording patient risk factor assessments, facilitating diagnostic workups and treatment, and helping providers generate care plans and manage preventive care, among others.3–5
Computerized problem lists offer additional advantages over a paper-based list, allowing other patient data such as laboratory results, imaging studies, medications, and allergies to be linked electronically to central problem concepts.2 6 Electronic patient problem lists can also be coded using standard terminologies.7–9 Today, many institutions with electronic health record (EHR) systems utilize either ICD-9, SNOMED, or subsets thereof, as their structured problem vocabulary; and such mappings facilitate automated interpretation of problem data, interoperability, and billing.10–13 Problem lists in modern EHRs are generally maintained manually; however some methods of augmenting the electronic problem list with clinical knowledge and improving its structure, accuracy, and utility have also been proposed,9 14–17 particularly in the area of problem-oriented record visualization and automated knowledge-based linking of problems and data.
An accurate and up-to-date electronic problem list represents the ideal cornerstone of the modern EHR. It provides a succinct clinical picture of the patient, facilitates communication, and enables the electronic record to deliver the appropriate clinical decision support. Clinicians may use the problem list to familiarize themselves with the needs of a patient they are treating for the first time or are covering, as an inventory of conditions that might require management on a particular visit, or as a marker of contraindications for particular therapies. However, despite their importance, patient problem lists are often inaccurate, incomplete, and poorly maintained.18–20 In addition, inaccurate problem lists have been shown to be associated with lower quality of patient care.21 22
The problem list is perhaps even more important for clinical decision support and quality measurement. For example, at Partners Healthcare, a large integrated academic clinical care network, 22% of clinical decision support rules depend on coded problems in the patient problem list.1 In many cases, accurately documented patient problems trigger reminders that help clinicians manage chronic diseases, which account for a large proportion of all costs. Consider, for example, a patient with diabetes. If his diabetes is properly documented, his clinician will receive appropriate alerts and reminders to guide care, the patient will be flagged as eligible for special care management programs, and the quality of care provided to him will be measured and tracked. Without diabetes on the problem list, he might receive none of these benefits.
Given that problem lists are often incomplete, researchers and implementers of clinical information systems have turned to a variety of alternative sources for problem information. Several systems have been reported using natural language processing to infer clinical problems.23–25 Researchers have also used data mining techniques to identify clinical data which can be used as a proxy for problems.26–28 These proxy methods have been especially fruitful in the case of medications: Carpenter and Gorman used medication information to identify possible problem mismatches22 and Poissant et al employ a combination of billing codes, single-indication drugs, and prescription indications to infer problems in an electronic prescribing system.29 30 In addition, the eMerge group has developed natural language processing, proxy and mixed problem inference methods for the purpose of identifying patient phenotypes and selecting cases and controls for genome-wide association studies.31–34
These techniques for inferring patient problems are promising and several have demonstrated positive early results; however, each of the reported systems has one or more limitations. Most use only a single type of data (medications, billing codes, or narrative text) to make their inference, focus on only one clinical problem, or focus on identifying cases (patients who certainly have the disease) and controls (patients who certainly do not have the disease) but leave many patients unclassified. Further, many rely on time consuming manual techniques for generation of their knowledge bases, and none, to our knowledge, have provided their full knowledge base for use or validation by others.
The goal of our project is to describe, in detail, a replicable method for developing problem inference rules, and also to provide a reference knowledge base of these rules for use or validation by other sites.
Methods
The methods we used in this project were designed to be easily replicable by other sites interested in developing their own problem inference rules. We describe a six-step process for rule development designed to yield high quality rules with known performance characteristics. The six steps are:
Automated identification of problem associations with other structured data
Selection of problems of interest
Development of preliminary rules
Characterization of preliminary rules and alternatives
Selection of a final rule
Validation of the final rule.
In the following sections, we present the six steps of this process in detail.
Step 1: Automated identification of problem associations with other structured data
To build inference rules, it is critical to determine what clinical data elements might be useful for predicting problems. Our current project builds on previous work we conducted to identify medication-problem associations and laboratory-problem associations using data mining and co-occurrence statistics.28 The goal of the Automated Patient Problem List Enhancement (APPLE) project was to develop a database of associations using automated data mining tools. In the APPLE study, we performed association rule mining on coded EHR data for a sample of 100 000 patients who received care at the Brigham and Women's Hospital (BWH), Boston, Massachusetts, USA. This dataset included 272 749 coded problems, 442 658 medications, and 11801068 laboratory results for the sample of 100000 patients.
In the previous study, candidate associations were evaluated using five co-occurrence statistics (support, confidence, χ2, interest, and conviction). High scoring medication-problem and laboratory-problem associations (the top 500) were then compared to a gold standard clinical reference (Mosby's Diagnostic and Laboratory Test Reference for laboratory results and Lexi-Comp drug reference database for medications). For medication-problem associations, χ2 was found to be the best performing statistic and for laboratory-problem associations, the highest performing statistic was interest. For medication-problem associations, 89.2% were found to be clinically accurate when compared with the gold standard, as were 55.6% of laboratory-problem associations.
The design, implementation, and results of the APPLE project are discussed in detail in a previous publication.28 The end result of the project was a database of several thousand medication-problem and laboratory-problem associations characterized by multiple co-occurrence statistics. This database was used in the preliminary stages of the current project in the design of inference rules, as described below.
Step 2: Selection of problems of interest
Given our methods and available resources, we wanted to constrain our knowledge base to no more than 20 problems. In order to identify a final set of conditions for inclusion in this project, we assessed a set of 78 potential ‘candidate’ problems. This preliminary list of problems was chosen on the basis of several criteria including: (a) recent related pay-for-performance initiatives at BWH, (b) the existence of relevant problem-dependent clinical decision support rules in the hospital's electronic medical records system (LMR), and (c) the strength of related medication-problem and laboratory-problem associations identified during the APPLE project. To guide the selection process, we developed a simple ranking metric which assigned points for each of the three criteria listed. A final list of 17 study problems (box 1) was chosen based on both the results of this analysis and clinician input. Each of the 17 problems was relevant to at least two of the three criteria described above.
Box 1. Target conditions for creation of problem inference rules.
Conditions (n=17)
ADHD
Asthma/COPD*
Breast cancer
CAD
CHF
Diabetes
Glaucoma
Hemophilia/congenital factor XI/von Willebrand disorder*
Hypertension
Hyperthyroidism
Hypothyroidism
Myasthenia gravis
Osteoporosis/osteopenia*
Renal insufficiency/renal failure*
Rheumatoid arthritis
Sickle cell disease
Stroke
*Multi-condition rules.
ADHD, attention deficit hyperactivity disorder; CAD, coronary artery disease; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease.
Step 3: Development of preliminary rules
Once the list of problems was finalized, we built a preliminary set of inference rules for initial testing. In order to accomplish this task, we first conducted research on each of the selected problems. We began by reviewing the APPLE database to locate all related medication-problem and laboratory-problem associations. Using these automatically-generated inferences as a starting point, we then conducted a thorough review of medical textbooks and online clinical resources, including Harrison's Principles of Internal Medicine, eMedicine, and UpToDate, identifying a list of all laboratory tests and medications relevant to each problem. We also identified all related ICD-9 billing codes for each of the conditions and coded problem list concepts relevant to the problem. Finally, because our EHR system allows for free-text problem entries in addition to coded entries, we also carried out a search for common related free-text entries. We identified all free-text (uncoded) problem phrases appearing five or more times in our sample (2441 in all) and manually reviewed each to determine if it matched a coded term for one of our 17 conditions. In the case of diabetes, for example, related free-text entries we found included ‘diabetes’ (users can enter free-text entries even if they match a coded concept exactly), ‘NIDDM,’ ‘AODM,’ ‘diabetic nephropathy,’ ‘diabetic retinopathy,’ ‘insulin resistance,’ ‘diabetes type II,’ ‘diabetic neuropathy,’ ‘type 2 diabetes,’ ‘type II diabetes,’ ‘diabetes mellitus,’ ‘diet controlled DM,’ ‘IDDM,’ ‘diabetic gastroparesis,’ ‘adult onset diabetes,’ and ‘diabetic complications.’ Many of the benefits of an accurate problem list can only be achieved through the use of coded problem entries. Thus, it was important in the design of our problem inference rules that they be able to identify patients with related free-text problem entries so that a coded entry could be added.
For each of the conditions, we developed a draft condition ‘abstract’ detailing the relevant information identified from the data sources above (a combination of laboratory-problem and medication-problem associations, literature review, ICD-9 codes, and free-text problem entries), and also developed an initial straw man rule. Each rule is comprised of a series of logic statements such as ‘coded or uncoded ADHD entry on problem list OR 1 or more ADHD billing codes OR 1 or more ADHD billing codes AND at least one ADHD medication.’
We presented this set of initial recommendations to an expert panel consisting of three internal medicine physicians (DWB, LIS, HZR). After reviewing the preliminary rules, the committee then recommended changes and proposed alternate rules (eg, additional classes of medications, additional combinations, or modification of thresholds).
In certain cases, the committee had difficulty developing rules that were highly specific for a single condition in our set of interest, particularly when our set of interest contained clinically similar or related diseases. For example, it was feasible to develop a rule that identified patients with either asthma or chronic obstructive pulmonary disease (COPD), but it was difficult to discriminate accurately between the two because of numerous medication overlaps, so the conditions were merged into a rule that predicts asthma or COPD. This strategy was applied in the following final rules: asthma/COPD, osteoporosis/osteopenia, renal insufficiency/renal failure, and hemophilia/congenital factor XI deficiency/von Willebrand disease.
Step 4: Characterization of preliminary rules and alternatives
Up to this point in the design process, the expert panel was working based on APPLE findings, reference information, and their own expertise, but without the benefit of specific characterization of the candidate rules they developed. In order to further inform their deliberations, we then tested each of the preliminary rules they proposed in step 3 as well as several alternate versions of each rule by applying them to a training set of patient records. The training set consisted of a random sample of 100 000 patient records drawn from a population of 839 300 patients with a progress note recorded in the last 2 years in the electronic medical record system at BWH. For each patient record, rules were automatically checked against coded data present in the electronic medical record system and submitted claims from the BWH inpatient and outpatient billing systems.
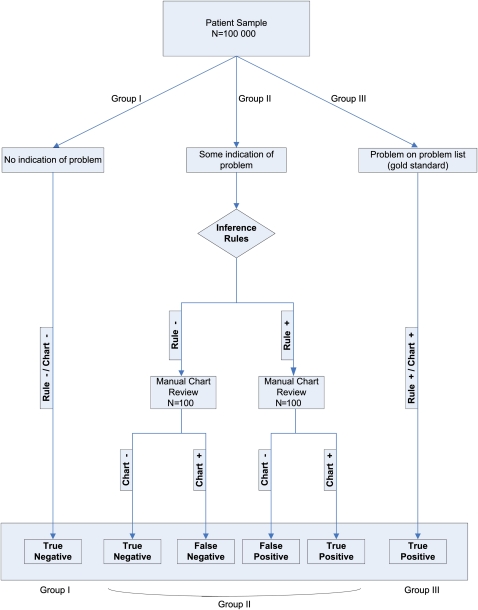
Our method of evaluating each rule is summarized in figure 1. Our gold standard, in each case, was the patient having the problem documented by a clinician in their record—we did not attempt to formulate new diagnoses for patients, or to verify the accuracy of existing diagnoses.
Figure 1.
Patient flow.
Because we had limited resources for chart review, we also made two assumptions to focus our review. First, we considered the presence of a particular condition on the problem list as a gold standard indicator that the patient had a given problem, and thus these patients were counted among the true positives (group III), since they met the criteria for our rules (by having the problem on their problem list) and since we assumed that the problem list assertion was correct. Our second assumption pertained to a subset of patients who had no relevant data in their record for a given condition. This was assessed by checking for the existence of any data that would inform a rule for a particular condition (but not, of course, checked by using the actual rule). For example, for diabetes, if we encountered a patient where no HbA1c test was ever performed, no diabetes-related billing codes were ever submitted, no related problems were on the problem list, and no related medications were on medication list, we assumed that patient did not have diabetes, and these patients were classified as true negatives (group I). Due to the absence of any relevant data, we determined that there would be extremely low yield in reviewing their charts, and instead focused resources on other groups of patients where there was a more reasonable likelihood of a given patient having the problem.
It is almost certainly the case that some patients with a problem on their problem list do not actually have that problem (despite it having been manually added by a clinician) and that some patients with no documented clinical evidence of a particular problem actually do have the problem. However, we believe that these eventualities are rare and that our assumptions are, thus, reasonable. Indeed, to verify or refute them, we would likely need to bring patients in for additional testing and workup, which would be expensive and likely low yield.
The remaining patients (group II) had at least some indication of the condition in their record (eg, any related laboratory investigation ever performed, any related medication prescribed, any related billing code recorded), but did not have the condition documented on their problem list. To be included in this group, a patient needed to have only a single coded laboratory test, medication, billing code or vital sign entry in their record related to the problem in question. For each of these patients, we applied the candidate rule from step 3 of our process, classifying each patient as either having the problem (rule-positive) or not having the problem (rule-negative).
For each condition, we randomly selected 100 rule-positive patients and 100 rule-negative patients from group II for manual chart review. This review was conducted by a team of research assistants under the supervision of the principal investigator, and included complete review of all data in the record, including problems, with a particular focus on free-text components such as progress notes, admission notes, discharge summaries, and consult notes and letters. If a clinician indicated anywhere in the record that the patient had the relevant condition, they were coded as positive for the condition (chart-positive). If there was no mention of the condition, or if the clinician had affirmatively ruled it out, the patient was counted as negative for the condition (chart-negative).
For the sample of 200 patients, this process left us with two data points: rule inference (rule-positive or rule-negative) and gold standard chart interpretation (chart-positive or chart-negative). Patients who were rule- and chart-positive were counted as true positives. Patients who were rule-positive and chart-negative were counted as false positives. Patients who were rule-negative and chart-negative were counted as true negatives. Patients who were rule-negative and chart-positive were counted as false-negatives (figure 1).
After completion of the manual chart review, each of the 100 000 patients was classified as a true-positive, false-positive, true-negative, or false-negative for each of the 17 conditions and associated candidate rules. We used these classifications to compute the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for each rule according to the standard formulas. Because not all charts were manually reviewed, we used inverse probability weights to adjust for our sampling strategy (patients in groups I and III were assigned weights of 1, while patients in group II were divided into rule-positive and rule-negative strata and assigned weights corresponding to the inverse of the stratum sampling fraction).
After computing these quantities for each of the 17 candidate rules, we then evaluated several alternative rules (by varying, for example, laboratory result thresholds, drugs, counts, etc) to determine the performance of each alternative version. For each rule, all versions were tested against the same 200-case set described above.
Step 5: Selection of a final rule
After multiple iterations of each rule were analyzed in this manner against the training set, several versions of each rule (ranging from 3 to 8) were then presented to the expert physician panel for a second time along with the sensitivity, specificity, and positive and negative predictive values for each version. The rule versions presented to the panel represented permutations of the original rule. For example, we might modify various thresholds (looking at HbA1c levels of 7% or 9%, or at 1 vs 2 related medications). The rules were selected to cover a variety of performance characteristics, ranging from rules with high sensitivity and lower PPV to rules with high PPV and lower sensitivity. The panel then chose a final rule from the presented options. In designing these rules, we attempted to maximize sensitivity and PPV overall and the options presented to our expert panel reflected this goal. Due to low overall prevalence of each disease, specificities were consistently very high, making it difficult to discriminate between different versions of each rule using this statistic. As a result, we chose to emphasize PPV over specificity in our analysis. In addition, in order to minimize erroneous inferences, we prioritized PPV while accepting some resultant trade-offs in sensitivity.
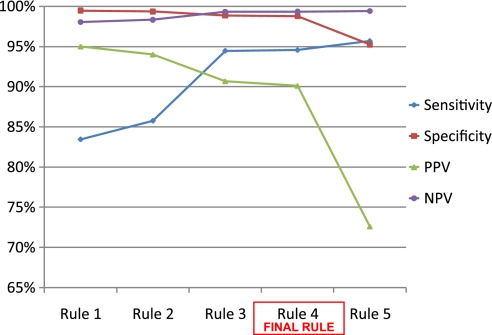
For example, five separate versions of the diabetes rule were presented to the expert panel with results from training set analysis (figure 2). The rules presented were as follows:
-
Rule 1
max A1c ≥9 OR
at least 3 A1c's recorded ≥7 OR
billing codes ≥7 OR
metformin and billing codes ≥2 OR
any insulin OR
any oral anti-diabetic drug OR
diabetes on problem list
-
Rule 2
max A1c ≥9 OR
at least 3 A1c's recorded ≥7 OR
billing codes ≥7 OR
any insulin OR
any oral anti-diabetic drug OR
diabetes on problem list
-
Rule 3
max A1c ≥9 OR
at least 3 A1c's recorded ≥7 OR
billing codes ≥2 OR
any insulin OR
any oral anti-diabetic drug OR
diabetes on problem list
-
Rule 4 (final rule)
max A1c ≥7 OR
billing codes ≥2 OR
any insulin OR
any oral anti-diabetic drug OR
diabetes on problem list
-
Rule 5
max A1c ≥5 OR
billing codes ≥2 OR
any insulin OR
any oral anti-diabetic drug OR
diabetes on problem list.
Figure 2.
Performance statistics for multiple versions of diabetes rule.NPV, negative predictive value; PPV, positive predictive value.
In the case of diabetes, option 1 (the most conservative and complex rule) achieved a high PPV (>95%) but had lower sensitivity (<85%). At the opposite end of the spectrum, option 5 (the most inclusive and simplest rule) achieved a sensitivity of >95% but a PPV of <80%. On the basis of these trade-offs and after confirmation of the clinical accuracy of each version, the panel recommended option 4 as the final rule in the case of diabetes based on a sensitivity of 94.6% and a PPV of 90.1%. This procedure was repeated for each of the 17 rules.
Step 6: Validation of the final rule
To validate the final version of each rule and to guard against over-fitting of the rules against the training set, we repeated the analysis of step 4 on an independent validation set. For this analysis, we drew a second random sample of 100 000 patients from the same population as the initial sample, but excluding patients in the initial sample. For each of the final rules, the same classification and chart review process was carried out, and sensitivity, specificity, and positive and negative predictive values were calculated using the same procedure described in the above section.
Additional analysis
After completing the six-step method described here for each problem, we had 17 fully characterized rules. In order to place these rules in context, we also carried out two additional analyses. First, we computed the sensitivity, specificity, PPV, and NPV of using coded problem list entries only using the method and data of step 6 (we did not conduct an additional chart review, but instead retained the results of the step 6 validation chart review). Because we assumed that all patients with a problem on their problem list had the related condition, the PPV of all such rules was necessarily 100%, but the sensitivity varied. We also computed the sensitivity, specificity, PPV, and NPV of using billing data alone to identify patients with the 17 conditions of interest, again using the methods and data of step 6 (and again retaining the step 6 validation chart review results).
In order to more fully characterize the performance of the rules, we also computed F measures according to the method of van Rijsbergen.35 The F measure is a generalized harmonic mean of the sensitivity and PPV whose parameters can be adjusted to prioritize either variable. We chose to use F0.5 because our goal in developing the rules was to prioritize PPV over sensitivity. The F0.5 measure weights PPV twice as heavily as sensitivity (ie, favoring false negatives over false positives).
Results
A total of 17 problem inference rules were developed (13 single-problem rules and 4 combined-problem rules). The complete list and description of these rules is provided in online appendix A (available as an online data supplement at www.jamia.org). The number of logic statements comprising each rule ranged from two (hemophilia) to five (hypertension, osteopenia/osteoporosis). Final rules used coded and free-text problem recognition and one or more of the following to infer patient problems: (a) related billing codes, (b) related medications, and/or (c) related laboratory data or vital signs.
The results of analysis on the training set for each rule, including sensitivity, specificity, and positive and negative predictive values, are presented on the left side of table 1. When applied to the training set, the average sensitivity and PPV for all 17 rules were 86.4% and 91.1%, respectively. Twelve of the 17 rules had a PPV of over 90% and all were over 65%. For sensitivity, 14 of the 17 rules were over 75% and all were over 65%.
Table 1.
Performance analysis of problem inference rules (training and validation)
| Training set | Validation set | |||||||
| Sens | Spec | PPV | NPV | Sens | Spec | PPV | NPV | |
| ADHD | 67.8 | 100.0* | 99.1 | 99.7 | 62.8 | 100.0* | 96.6 | 99.6 |
| Asthma/COPD | 78.1 | 99.2 | 92.7 | 97.0 | 79.5 | 99.6 | 96.7 | 97.3 |
| Breast cancer | 95.1 | 99.9 | 99.0 | 99.7 | 95.8 | 100.0* | 99.6 | 99.7 |
| CAD | 83.0 | 99.6 | 95.7 | 98.3 | 86.4 | 99.9 | 98.5 | 98.6 |
| CHF | 71.7 | 99.4 | 79.1 | 99.1 | 70.8 | 99.4 | 79.8 | 99.0 |
| Diabetes | 94.6 | 98.8 | 90.1 | 99.4 | 91.3 | 99.3 | 94.9 | 98.8 |
| Glaucoma | 93.8 | 99.9 | 95.0 | 99.9 | 94.4 | 99.9 | 96.2 | 99.9 |
| Hemophilia | 89.7 | 100.0* | 97.7 | 100.0* | 86.5 | 100.0* | 97.8 | 100.0* |
| Hypertension | 80.6 | 96.9 | 92.9 | 90.8 | 81.0 | 96.2 | 89.0 | 93.1 |
| Hyperthyroidism | 83.6 | 99.9 | 87.7 | 99.9 | 86.3 | 99.9 | 88.1 | 99.9 |
| Hypothyroidism | 91.9 | 99.8 | 97.5 | 99.3 | 91.0 | 99.5 | 93.5 | 99.3 |
| Myasthenia gravis | 87.4 | 100.0* | 89.4 | 100.0* | 82.4 | 100.0* | 85.9 | 100.0* |
| Osteoporosis/osteopenia | 73.9 | 99.4 | 94.0 | 96.8 | 70.8 | 99.2 | 90.7 | 97.0 |
| Renal insuf/renal fail | 100.0 | 98.3 | 69.2 | 100.0 | 100.0 | 99.1 | 77.5 | 100.0 |
| Rheumatoid arthritis | 94.8 | 99.8 | 88.2 | 99.9 | 66.5 | 99.9 | 91.7 | 99.3 |
| Sickle cell disease | 95.6 | 100.0* | 90.3 | 100.0* | 96.8 | 100.0* | 91.0 | 100.0* |
| Stroke | 85.8 | 100.0* | 97.4 | 99.7 | 87.3 | 99.9 | 97.9 | 99.7 |
Actual value slightly less than 100%.
Note: CIs were small for each parameter and are thus omitted from the reported results. For example, the CIs for the diabetes rule parameters described above were 86.8% to 94.5% (sensitivity), 98.8% to 99.6% (specificity), 91.0% to 97.3% (PPV), and 98.1% to 99.2% (NPV).
ADHD, attention deficit hyperactivity disorder; CAD, coronary artery disease; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease; fail, failure; insuf, insufficiency; NPV, negative predictive value; PPV, positive predictive value; Sens, sensitivity; Spec, specificity.
The results of validation for each rule, including sensitivity, specificity, and positive and negative predictive values, are presented on the right side of table 1. When applied to the validation set, the average sensitivity and PPV for all 17 rules were 83.9% and 91.7%, respectively. Overall, 12 of the 17 rules had a PPV of over 90% and all were over 75%. Of the 17 rules, 11 had a sensitivity of over 80% and all were greater than 60%.
For each problem, we also assessed the accuracy of two simpler classes of rules including (a) problem list-only rules and (b) related billing code-only rules. The results of this analysis (table 2) showed that our inference rules were more sensitive than the problem list alone, and had better PPV than billing codes alone. Notably, all 17 of our rules had better sensitivity than the problem list (ranging from 12.8% better for hemophilia and related disorders to 95.6% better for renal insufficiency/failure). Fifteen of the 17 rules had better PPV than billing codes alone (glaucoma and renal insufficiency/failure had slightly lower PPV), with the greatest improvements being for relatively rare conditions including hyperthyroidism, myasthenia gravis, and sickle cell disease. For those billing-code only rules with a higher PPV, a significant trade-off was observed with disproportionately lower sensitivities.
Table 2.
Performance of coded problem-only and billing-only rules
| Problem-only | Billing-only | |||||||
| Sens | Spec | PPV | NPV | Sens | Spec | PPV | NPV | |
| ADHD | 45.0 | 100.0 | 100.0 | 99.4 | 73.7 | 99.9 | 91.3 | 99.7 |
| Asthma/COPD | 44.8 | 100.0 | 100.0 | 92.4 | 91.5 | 98.6 | 89.8 | 98.8 |
| Breast cancer | 78.5 | 100.0 | 100.0 | 98.6 | 97.9 | 99.9 | 97.7 | 99.9 |
| CAD | 58.9 | 100.0 | 100.0 | 95.7 | 99.2 | 97.9 | 83.5 | 99.9 |
| CHF | 9.9 | 100.0 | 100.0 | 91.9 | 83.3 | 98.3 | 70.2 | 99.2 |
| Diabetes | 61.9 | 100.0 | 100.0 | 94.5 | 89.4 | 98.5 | 89.8 | 98.4 |
| Glaucoma | 73.4 | 100.0 | 100.0 | 99.4 | 90.0 | 99.9 | 96.7 | 99.8 |
| Hemophilia | 73.7 | 100.0 | 100.0 | 99.9 | 100 | 100.0* | 87.2 | 100 |
| Hypertension | 50.7 | 100.0 | 100.0 | 83.2 | 86.7 | 95.0 | 87.5 | 94.7 |
| Hyperthyroidism | 59.3 | 100.0 | 100.0 | 99.5 | 95.7 | 99.4 | 64.4 | 100.0* |
| Hypothyroidism | 51.8 | 100.0 | 100.0 | 96.7 | 81.6 | 98.2 | 76.4 | 98.7 |
| Myasthenia gravis | 48.6 | 100.0 | 100.0 | 100.0* | 97.3 | 99.9 | 53.3 | 100.0* |
| Osteoporosis/osteopenia | 45.1 | 100.0 | 100.0 | 94.2 | 80.5 | 98.7 | 87.4 | 97.9 |
| Renal insuf/renal fail | 4.7 | 100.0 | 100.0 | 83.5 | 43.3 | 99.6 | 86.7 | 96.4 |
| Rheumatoid arthritis | 23.8 | 100.0 | 100.0 | 97.3 | 90.5 | 99.6 | 84.1 | 99.8 |
| Sickle cell disease | 76.2 | 100.0 | 100.0 | 100.0* | 98.4 | 100.0* | 67.4 | 100.0* |
| Stroke | 72.4 | 100.0 | 100.0 | 99.2 | 100.0 | 99.6 | 86.8 | 100.0 |
Actual value slightly less than 100%.
ADHD, attention deficit hyperactivity disorder; CAD, coronary artery disease; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease; fail, failure; insuf, insufficiency; NPV, negative predictive value; PPV, positive predictive value; Sens, sensitivity; Spec, specificity.
For each rule, we also computed the F0.5 measure as described in the Methods section. Using F0.5, our inference rules outperformed the problem list alone for all 17 conditions and billing codes alone for 12 of the 17 conditions (table 3).
Table 3.
Comparison of problem-only, billing-only, and problem inference rule performance
| F0.5 | |||||
| Billing (b) | Rules (r) | Problems (p) | r>b | r>p | |
| ADHD | 84.6 | 81.9 | 71.1 | N | Y |
| Asthma/COPD | 90.4 | 90.2 | 70.9 | N | Y |
| Breast cancer | 97.8 | 98.3 | 91.6 | Y | Y |
| CAD | 88.2 | 94.1 | 81.1 | Y | Y |
| CHF | 74.1 | 76.6 | 24.8 | Y | Y |
| Diabetes | 89.7 | 93.7 | 83.0 | Y | Y |
| Glaucoma | 94.4 | 95.6 | 89.2 | Y | Y |
| Hemophilia | 91.1 | 93.7 | 89.4 | Y | Y |
| Hypertension | 87.2 | 86.2 | 75.5 | N | Y |
| Hyperthyroidism | 72.3 | 87.5 | 81.4 | Y | Y |
| Hypothyroidism | 78.1 | 92.7 | 76.3 | Y | Y |
| Myasthenia gravis | 62.8 | 84.7 | 73.9 | Y | Y |
| Osteoporosis/osteopenia | 85.0 | 82.9 | 71.1 | N | Y |
| Renal insuf/renal fail | 65.0 | 83.8 | 12.9 | Y | Y |
| Rheumatoid arthritis | 86.1 | 81.4 | 48.4 | N | Y |
| Sickle cell disease | 75.3 | 92.9 | 90.6 | Y | Y |
| Stroke | 90.8 | 94.1 | 88.7 | Y | Y |
ADHD, attention deficit hyperactivity disorder; CAD, coronary artery disease; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease; fail, failure; insuf, insufficiency.
Discussion
In this study, we successfully developed and validated a set of rules that identifies patients who are likely to have a particular problem. These rules were shown to have a high sensitivity and specificity and, in our population, a high positive and negative predictive value.
We found that we were able to generate and validate a large number of rules for important conditions with a relatively small team over a period of several months. The rules generally performed quite well, with high sensitivity and PPV. They also withstood validation on an independent sample of records, suggesting that our rules are not over-fitted to our training data. Thus, these methods appear to be both scalable to more conditions and replicable at other sites. Assessing their generalizability, however, will require additional independent validation at other sites.
The performance of the rules varied depending on the condition, and the performance of the renal insufficiency/failure rules deserves special comment. The National Kidney Foundation defines chronic kidney disease as kidney damage or a glomerular filtration rate of less than 60 for at least 3 months.36 During the chart review, we found many patients meeting these criteria who had no mention of kidney disease anywhere in their record. These patients may have unappreciated renal insufficiency or failure, but were necessarily marked as condition-negative in our analysis (leading to potentially artificially low PPV and specificity). This has two important implications: first, were a different gold standard chosen (eg, evaluation by a nephrologist rather than chart review), the performance of the rules might have been different, although this is more of an issue for some rules like this one than others. Second, although these rules were initially designed to identify problems which are known to providers, they may, in certain instances, also have diagnostic utility in the case of an unappreciated condition. Similar results were reported in another study focusing on renal failure using different methods.25
Comparison to other methods
The problem list and billing data are often used to infer a patient's problems. However, our analysis indicated that each method had shortcomings. The problem list was extremely accurate (ie, one could have a high degree of confidence that a patient has a problem if it appears on the problem list), but it had very low, and variable, sensitivity. In fact, for most problems, the sensitivity was around 50%, meaning that only about half of patients with the problem had it documented on their problem list. This was higher for some chronic conditions, but was extremely low for renal failure (possibly due to the reasons discussed above) and was also quite low for congestive heart failure (CHF). The CHF finding was surprising; however, it seemed that, in many cases, the patient's CHF was so central to their clinical picture that all providers were aware of it but had simply omitted it from the problem list.
Conversely, billing codes were found to have a considerably lower PPV but a high degree of sensitivity, indicating that in some cases patients are billed for problems that they do not have. A review of these data suggests that, in many cases, patients are billed for screening tests under the related problem code (eg, billing with ICD-9 code 250.00 for a diabetes screening) rather than a screening code, which is not an ideal practice,37 although the reasons for it are understandable.
Our research shows that the integration of laboratory, medication, billing, problem, and vital sign data can result in robust rules for inferring patient problems, and that such rules, which take advantage of multiple classes of coded data available in the electronic medical record, have superior performance to single-faceted rules.
Applications
Our methods and validated knowledge base have a variety of applications. First and foremost, they could be used to alert clinicians to potential gaps in the problem list, and could provide clinicians the opportunity to correct these gaps. Additionally, the rules could be used for any application where it is important to know a patient's diagnoses, such as identification of research cohorts, calculation of quality measures, selection of patients for care management programs, and clinical decision support. To apply the rules, developers of such systems would simply modify their inclusion criteria, replacing their current mechanism of problem identification (problem list or billing data) with inference rules such as these. The use of inference rules for identifying patient problems is also of potential value for meeting meaningful use requirements. The Stage 1 goal requires that providers ‘maintain an up-to-date problem list of current and active diagnoses,’ with an ambitious 80% of patients having at least one problem recorded or an indication of no known problems.38 A reliable automated method of increasing problem list use could dramatically improve providers' ability to reach this goal.
One important question for potential users of such rules at other institutions is how to proceed. Ideally, outside sites would utilize our methods to develop and validate their own rules using their own clinical data (and then report the results). However, sites without sufficient clinical data or resources might, instead, choose to use these inference rules (potentially with local modifications). We have provided the complete set of rules in online appendix A. We encourage any site choosing to apply the inference rules to report on their own experience so additional knowledge of the rules' generalizability can be developed.
Limitations
This investigation has several potential limitations. First, it is possible that our sampling assumptions may have introduced a small amount of bias into our results. As discussed in the Methods section, it is possible that a small proportion of patients in group I or group III, whose charts were not reviewed, may have been incorrectly classified as true negatives and true positives, respectively (because they had a problem on their problem list that they do not actually have, or because they have an undiagnosed problem, or a diagnosed problem without any correlated clinical data). It is important to note that provider awareness of a problem (or lack thereof) was our gold standard, rather than the patient's actual pathophysiologic state—in other words, we did not seek to make new diagnoses. We believe that these assumptions are reasonable given that these rules are designed to infer patient problems based on documented clinical data rather than to yield new diagnoses. Given this gold standard, we suspect that the misclassification rate into groups I and III was low; however, to test this assumption it would be necessary to bring patients in for workups to confirm their diagnoses (or lack thereof)—these workups would likely be expensive and low yield. That said, any misclassification in these groups would introduce a small bias in calculated sensitivity and PPV values (systematically increasing them); however, we believe this potential effect to be very small. Additionally, this bias would have the same effect on the statistics for our comparison groups (the problem-only and billing-only measures) in addition to our rules. As such, our comparison between these rule classes is likely unaffected by any bias introduced (and the magnitude of this bias is still likely to be very small).
Second, problems were selected in part based on the strength of laboratory-problem and medication-problem associations. This potentially limits the generalizability of our results with respect to other conditions that have weaker connections to medications and laboratory results. However, in many cases other data (eg, billing codes) may be available to help with prediction, and there are also a number of data types which we have not yet considered (particularly unstructured data such as images and text, as well as patient-reported data).
Third, as described in the Methods section, for each disease we conducted a chart review of a random sample of 100 rule-positive and 100 rule-negative patients to determine the rule's performance. We then tested a number of alternate versions of each inference rule against the same 200-patient sample for each condition. This may have introduced some bias into the performance characterization of the alternative rules, since their sample was influenced by the initial rule. To mitigate this bias, we attempted to select a ‘centrist’ initial rule, and then varied the parameters of the initial rule to create the alternatives, hopefully minimizing bias. Further, and more definitively, once the final rule was chosen for each condition (which could have been the initial rule or one of the alternative rules), the final rule was independently validated on a new randomly selected sample of 200 patient charts (100 rule-positive and 100 rule-negative). As such, the performance measures from the validation set were free from this potential bias.
Fourth, we developed and validated the rules at only a single site—as we mentioned above, we believe that the rules are likely generalizable to other sites, but we encourage other researchers to validate them before use, and also to extend them and report on their results.
Finally, because we included all patients with at least a single note in a 2-year period, a small number of patients in our sample had very little data recorded because they had only a single visit or a low number of visits. We chose to include these patients in order to form a more representative sample; however, our rules fired less frequently for these patients because they were less likely to have sufficient data to meet the rule thresholds (eg, multiple visits with a single billing code). Methods for adjusting inference thresholds based on the volume of data available for a patient merit further study.
Conclusions
We developed and validated a set of problem inference rules. Our findings show that by using laboratory, medication, problem, billing, and vital sign data, patient problems can be accurately inferred, and that the performance of such multi-source rules exceeds the performance of standard sources, such as the problem list or billing codes, alone. Building an improved problem list has a number of downstream potential benefits for delivering good clinical care, improving quality, and conducting research.
Supplementary Material
Acknowledgments
We would like to acknowledge the Partners HealthCare Research Patient Data Registry and Quality Data Management teams for supplying data used in this study and Karen Sax McLoughlin, of the Brigham and Women's Physician Organization, for her work in managing the grant. We are also grateful to Elizabeth S Chen, PhD of the University of Vermont for her participation in preparation of the grant application for this project and design of the methods.
Footnotes
Funding: This work was supported by a grant from the Partners Community HealthCare Incorporated (PCHI) System Improvement Grant Program. PCHI was not involved in the design, execution or analysis of the study or in the preparation of this manuscript.
Competing interests: None.
Ethics approval: This study was approved by the Partners HealthCare Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Wright A, Goldberg H, Hongsermeier T, et al. A description and functional taxonomy of rule-based decision support content at a large integrated delivery network. J Am Med Inform Assoc 2007;14:489–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weed LL. Medical records that guide and teach. New Engl J Med 1968;278:652–7 [DOI] [PubMed] [Google Scholar]
- 3.Lincoln MJ. Developing and implementing the problem list. In: Kolodner R, ed. Cmputerizing Large Integrated Health Networks: The VA Success. New York: Springer, 1997:349–81 [Google Scholar]
- 4.Hurst JW. Ten reasons why Lawrence Weed is right. New Engl J M 1971;284:51–2 [DOI] [PubMed] [Google Scholar]
- 5.Weed LL. The problem-oriented record. In: Hurst JW, Walker HK, eds. The Problem Oriented System. New York: Medcom Press, 1972:23–4 [Google Scholar]
- 6.Safran C, Rury C, Rind DM, et al. A computer-based outpatient medical record for a teaching hospital. MD Comput 1991;8:291–9 [PubMed] [Google Scholar]
- 7.Feinstein AR. The problems of the “problem-oriented medical record”. Ann Intern Med 1973;78:751–62 [DOI] [PubMed] [Google Scholar]
- 8.Brown SH, Miller RA, Camp HN, et al. Empirical derivation of an electronic clinically useful problem statement system. Ann Intern Med 1999;131:117–26 [DOI] [PubMed] [Google Scholar]
- 9.Wang SJ, Bates DW, Chueh HC, et al. Automated coded ambulatory problem lists: evaluation of a vocabulary and a data entry tool. Intern J Med Inform 2003;72:17–28 [DOI] [PubMed] [Google Scholar]
- 10.Fung KW, McDonald C, Srinivasan S. The UMLS-CORE project: a study of the problem list terminologies used in large healthcare institutions. J Am Med Inform Assoc 2010;17:675–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mantena S, Schadow G. Evaluation of the VA/KP problem list subset of SNOMED as a clinical terminology for electronic prescription clinical decision support. AMIA Annu Symp Proc 2007:498–502 [PMC free article] [PubMed] [Google Scholar]
- 12.Nadkarni PM, Darer JA. Migrating existing clinical content from ICD-9 to SNOMED. J Am Med Inform Assoc 2010;17:602–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell JR, Payne TH. A comparison of four schemes for codification of problem lists. Proc Annu Symp Comput Appl Med Care 1994:201–5 [PMC free article] [PubMed] [Google Scholar]
- 14.Weed LL, Zimny NJ. The problem-oriented system, problem-knowledge coupling, and clinical decision making. Phys Ther 1989;69:565–8 [DOI] [PubMed] [Google Scholar]
- 15.Weed LL. Knowledge coupling, medical education and patient care. Crit Rev Med Inform 1986;1:55–79 [PubMed] [Google Scholar]
- 16.Bashyam V, Hsu W, Watt E, et al. Problem-centric organization and visualization of patient imaging and clinical data. Radiographics 2009;29:331–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Vleck TT, Wilcox A, Stetson PD, et al. Content and structure of clinical problem lists: a corpus analysis. AMIA Annu Symp proc 2008:753–7 [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan DM. Clear writing, clear thinking and the disappearing art of the problem list. J Hosp Med 2007;2:199–202 [DOI] [PubMed] [Google Scholar]
- 19.Szeto HC, Coleman RK, Gholami P, et al. Accuracy of computerized outpatient diagnoses in a Veterans Affairs general medicine clinic. Am J Manag care 2002;8:37–43 [PubMed] [Google Scholar]
- 20.Tang PC, LaRosa MP, Gorden SM. Use of computer-based records, completeness of documentation, and appropriateness of documented clinical decisions. J Am Med Inform Assoc 1999;6:245–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartung DM, Hunt J, Siemienczuk J, et al. Clinical implications of an accurate problem list on heart failure treatment. J Gen Intern Med 2005;20:143–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpenter JD, Gorman PN. Using medication list–problem list mismatches as markers of potential error. Proc AMIA Symp 2002:106–10 [PMC free article] [PubMed] [Google Scholar]
- 23.Meystre S, Haug PJ. Natural language processing to extract medical problems from electronic clinical documents: performance evaluation. J Biomed Inform 2006;39:589–99 [DOI] [PubMed] [Google Scholar]
- 24.Meystre SM, Haug PJ. Randomized controlled trial of an automated problem list with improved sensitivity. Int J Medical Inform 2008;77:602–12 [DOI] [PubMed] [Google Scholar]
- 25.Chase HS, Radhakrishnan J, Shirazian S, et al. Under-documentation of chronic kidney disease in the electronic health record in outpatients. J Am Med Inform Assoc 2010;17:588–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burton MM, Simonaitis L, Schadow G. Medication and indication linkage: A practical therapy for the problem list? AMIA Annu Symp Proc 2008:86–90 [PMC free article] [PubMed] [Google Scholar]
- 27.Cao H, Markatou M, Melton GB, et al. Mining a clinical data warehouse to discover disease-finding associations using co-occurrence statistics. AMIA Annu Symp Proc 2005:106–10 [PMC free article] [PubMed] [Google Scholar]
- 28.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform 2010;43:891–901 [DOI] [PubMed] [Google Scholar]
- 29.Poissant L, Tamblyn R, Huang A. Preliminary validation of an automated health problem list. AMIA Annu Symp Proc 2005:1084. [PMC free article] [PubMed] [Google Scholar]
- 30.Poissant L, Taylor L, Huang A, et al. Assessing the accuracy of an inter-institutional automated patient-specific health problem list. BMC Med Inform Decis Mak 2010;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010;26:1205–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pacheco JA, Avila PC, Thompson JA, et al. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. AMIA Annu Symp Proc 2009;2009:497–501 [PMC free article] [PubMed] [Google Scholar]
- 33.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation 2010;122:2016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc 2010;17:568–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Rijsbergen CJ. Information Retrieval. 2nd edn London: Butterworths, 1979 [Google Scholar]
- 36.KDOQI Clinical Practice Guidelines for Chronic Kidney Disease: Evaluation, Classification, and Stratification, 2002. http://www.kidney.org/professionals/KDOQI/guidelines_ckd/p4_class_g1.htm (accessed 30 Sep 2010). [PubMed]
- 37.ICD-9-CM Coding for Diagnostic Tests, 2001. http://www.cms.gov/transmittals/downloads/AB01144.pdf (accessed 30 Sep 2010).
- 38.Comparison of Meaningful Use Objectives Between the Proposed Rule to the Final Rule, 2010. https://www.cms.gov/EHRIncentivePrograms/Downloads/NPRM_vs_FR_Table_Comparison_Final.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.