Abstract
OBJECTIVE
To test the hypothesis that polymorphic variation in the paternally transmitted fetal IGF2 gene is associated with maternal glucose concentrations in the third trimester of pregnancy.
RESEARCH DESIGN AND METHODS
A total of 17 haplotype tag single nucleotide polymorphisms in the IGF2 gene region were genotyped in 1,160 mother/partner/offspring trios from the prospective Cambridge Baby Growth Study (n = 845 trios) and the retrospective Cambridge Wellbeing Study (n = 315 trios) (3,480 samples in total). Associations were tested between inferred parent-of-origin fetal alleles, z scores of maternal glucose concentrations 60 min. after an oral glucose load performed at week 28 of pregnancy, and offspring birth weights.
RESULTS
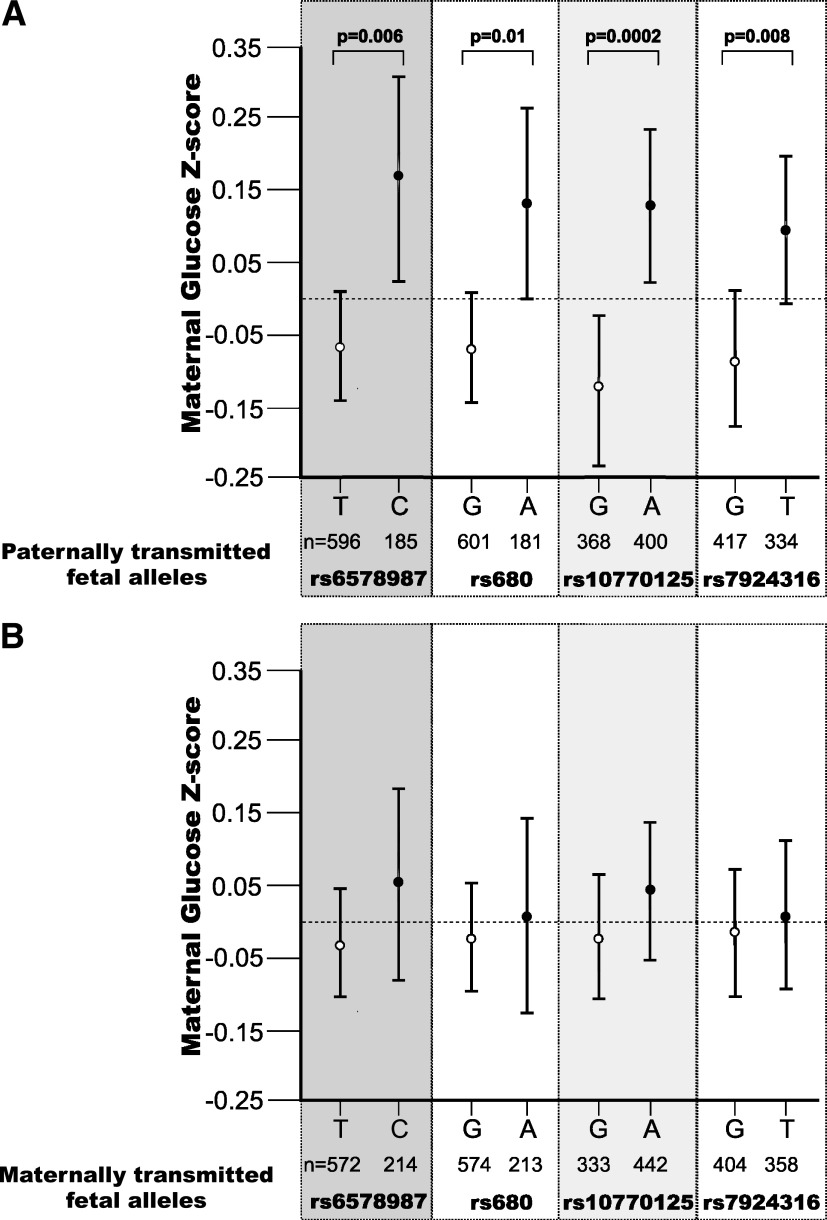
Using the minimum P value test, paternally transmitted fetal IGF2 polymorphisms were associated with maternal glucose concentrations; specifically, paternally transmitted fetal rs6578987 (P = 0.006), rs680 (P = 0.01), rs10770125 (P = 0.0002), and rs7924316 (P = 0.01) alleles were associated with increased maternal glucose concentrations in the third trimester of pregnancy and placental IGF-II contents at birth (P = 0.03). In contrast, there were no associations between maternal glucose concentrations and maternal or maternally transmitted fetal IGF2 genotypes.
CONCLUSIONS
Polymorphic variation in paternally transmitted fetal IGF2 is associated with increased maternal glucose concentrations in pregnancy and could potentially alter the risk of gestational diabetes in the mother. The association may be at least partially mediated by changes in placental IGF2 expression.
Almost 20 years ago it was hypothesized that polymorphic variation in the fetal genome, in particular in fetal growth genes, could lead to alterations in maternal metabolism in pregnancy (1). More recently, we suggested that in addition to the risk associated with polymorphic variation in the mother’s genes, which could affect her glucose concentrations (2), fetal genomic variation could also alter a mother’s risk of developing gestational diabetes mellitus (GDM) (3). Many well-characterized fetal growth genes are imprinted (4), meaning that they are expressed from only either the paternally or the maternally transmitted copy, depending on the gene in question and the stage of development. Haig's kinship, or conflict hypothesis (5,6), suggests that paternally expressed fetal imprinted genes will tend to increase fetal growth, whereas maternally expressed genes will tend to restrain it. This is thought to be achieved through modifying fetal and placental nutritional demand and supply (7), potentially including altering maternal glucose concentrations (1,3).
We recently tested our hypothesis (3) in phenotypically wild type pregnant mice that were carrying pups that had a targeted 13 kb genetic region disrupted, which included the imprinted H19 gene and Igf2 control element (8). Affected pups in this model are born ∼30% heavier than unaffected litter mates (9), principally as a result of biallelic Igf2 expression resulting from the disruption of its control element (10). Consistent with our hypothesis, we found that intraperitoneal glucose tolerance tests indicated that mice carrying knockout offspring had increased circulating glucose concentrations in late pregnancy in comparison with those of genetically matched controls (8).
In humans, the IGF2 gene is located in a region rich in imprinted genes (11p15.5). The IGF2 gene comprises nine exons (codons 7–9 being coding) and four promoters, spanning a region of ∼30 kb (11). Several different RNA molecules are formed upon transcription of the gene within the coding region plus one of the various 5′-untranslated regions arising from exons 1–6. The different transcripts are expressed according to their tissue and the stage of development (11). In the human fetus and placenta, IGF2 is imprinted such that only the paternally transmitted copy of the gene is expressed. Cord blood concentrations of its protein product, IGF-II, are associated with birth weight (12). In contemporary birth cohorts, we found that in first pregnancies, cord blood IGF-II concentrations were also associated with a polymorphic variant in H19, another imprinted gene in the 11p15.5 region in humans and one that may regulate IGF2 gene expression (6) when transmitted from the mother to the fetus (13). This H19 single nucleotide polymorphism (SNP) was also associated with maternal glucose concentrations, suggesting that our findings in mice (8) may be relevant to humans.
The current study was designed to see if the results from our mouse model (8), indicating that functional variation in the fetal IGF2 gene alters maternal glucose concentrations in pregnancy, could also be observed in an analogous manner in humans. Specifically, we tested the hypothesis that polymorphic variation in paternally transmitted fetal IGF2 is associated with alterations in maternal glucose concentrations at week 28 of pregnancy and offspring birth weight.
RESEARCH DESIGN AND METHODS
Cohorts and assessment of glucose concentrations.
The Cambridge Baby Growth Study (prospective and longitudinal) recruited mothers (and their partners and offspring) attending ultrasound clinics during early pregnancy at the Rosie Maternity Hospital, Cambridge, U.K., between 2001 and 2009 (14,15). At 28 weeks of gestation, all the mothers underwent a 75-g oral glucose tolerance test after an overnight fast. Venous blood was collected after fasting and at 60 min. after the glucose load for the measurement of plasma glucose concentrations (using a routine glucose oxidase–based method). Capillary blood glucose measurements were made at 0, 30, 60, 90, and 120 min using an Abbott Freestyle Mini (Abbott Diagnostics, Maidenhead, U.K.). Blood and/or mouth swab samples for DNA extraction were collected from the father and the offspring after its birth. In total, 845 DNA trios were collected from the families of 1,074 mothers recruited to the study for whom oral glucose tolerance test data were available. In this cohort, 96.9% of the offspring were of Caucasian ethnicity, 0.8% were of mixed race, 0.6% were black (African or Caribbean), 0.8% were Oriental, and 0.9% were Indo-Asian.
The Cambridge Wellbeing Study (retrospective) recruited mothers, fathers, and children where the pregnant mother had delivered a full-term, singleton baby at the Rosie Maternity Hospital, Cambridge, U.K., between the years 1999 to 2000 (13,14). Routinely collected clinical data were available on offspring birth weight and mother's whole blood glucose levels measured 60 min after the oral consumption of 50-g glucose at 27 to 29 weeks of gestation. Exclusion criteria were maternal hypertension and treatment for diabetes during pregnancy. In this cohort, all the offspring were of Caucasian ethnicity. We sought permission from the mother’s general practitioner to approach the mother, partner, and child to collect their DNA sample by postal mouth swab kits. In total, 315 DNA trios were collected out of a total of 563 women who consented; glucose concentrations were available from >3,000 women, many of whom were not approached to provide consent for genetic analysis. Tables 1 and 2 show characteristics of the study participants from the two cohorts.
TABLE 1.
Study participant demographics for the 845 family trios in the Cambridge Baby Growth Study
| Demographic | |
|---|---|
| Maternal age at delivery (years) | 33.5 (4.3) |
| Maternal height (cm) | 165.9 (7.2) |
| Maternal prepregnancy weight (kg) | 66.3 (13.5) |
| Maternal prepregnancy BMI (kg/m2) | 24.1 (4.6) |
| 28-week maternal fasting plasma glucose concentration (mmol/L) | 4.3 (0.6) |
| 28-week maternal glucose plasma glucose concentration (60 min after the consumption of a 75-g oral glucose load) (mmol/L) | 6.8 (1.7) |
| Prepregnancy gravidity | 1.8 (0.9) |
| Prepregnancy parity | 0.4 (0.8) |
| Twin pregnancies (%) | 2.0 |
| Maternal smoking in pregnancy (%) | 5.4 |
| Gestational age at delivery (weeks) | 39.8 (1.6) |
| Offspring ethnicity (% Caucasian) | 96.9 |
| Offspring sex (% male) | 51.6 |
| Offspring birth weight (g) | 3,479 (533) |
| Paternal age at delivery (years) | 35.4 (5.3) |
Data are mean (SD) or percentages.
TABLE 2.
Study participant demographics for the 315 family trios in the Cambridge Wellbeing Study
| Demographic | |
|---|---|
| Maternal age at delivery (years) | 33.1 (4.1) |
| 28-week maternal glucose plasma glucose concentration (60 min after the consumption of a 50-g oral glucose load) (mmol/L) | 5.6 (1.4) |
| Prepregnancy parity | 0.8 (0.8) |
| Twin pregnancies (%) | 0* |
| Gestational age at delivery (weeks) | 39.6 (1.3) |
| Offspring ethnicity (% Caucasian) | 100* |
| Offspring sex (% male) | 50.8 |
| Offspring birth weight (g) | 3,562 (495) |
Data are mean (SD) or percentages. *Inclusion criteria.
Ethical approval.
Both the Cambridge Baby Growth Study and the Cambridge Wellbeing Study were approved by the local ethics committee, Addenbrooke’s Hospital, Cambridge, U.K. Written informed consent was obtained from the parents, including consent for inclusion of their infants in the study.
Measurement of birth weight.
For both cohorts, we used the routine measurement of birth weight made at delivery by midwives and recorded in the hospital notes. Approximately 420 of the mothers gave birth during the daytime, and their term placentas were collected and processed within an hour of delivery, snap frozen, and then stored at −20°C until aliquots were removed for protein and/or RNA extraction.
SNP selection.
A total of 17 IGF2 region (i.e., IGF2-INS-TH cluster) SNPs were selected based on the 11 haplotype tag SNPs identified by Rodríguez et al. (16), plus 6 SNPs from the Centre d'Etude du Polymorphisme Humain (CEU) population (Utah residents with ancestry from Northern and Western Europe) of the HapMap Project Build 36 using Haploview (17) (Hapmap accessed 19 January 2010). We force included the 11 Rodríguez et al. SNPs, using pairwise tagging r2 >0.8 and a minor allele frequency of at least 0.2 as the acceptance criteria (Fig. 1), and identified 5 haplotype blocks tagged by 14 SNPs (Table 3 and Supplementary Fig. 1). Three of the SNPs used by Rodríguez et al. were not required to define these haplotype blocks but were included for genotyping.
FIG. 1.
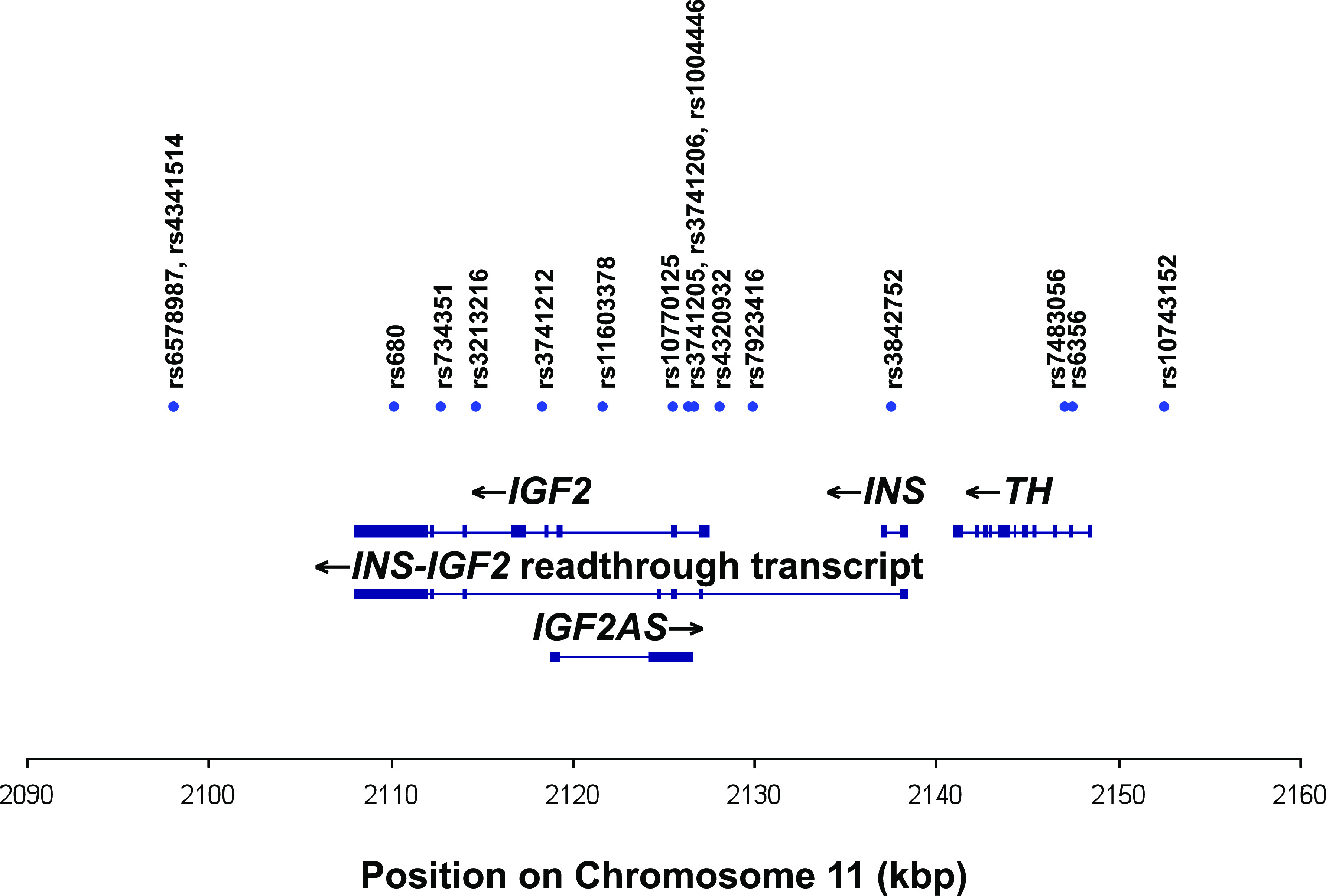
Schematic diagram of the region of chromosome 11 genotyped in this study, showing the associated genes (IGF2, INS, and TH) and transcripts (IGF2, INS, TH, antisense IGF2, and INS-IGF2 readthrough transcript). Exons are shown as blocks. Chromosomal position based on National Center for Biotechnology Information Genome Reference Consortium Human genome build 36. (A high-quality color representation of this figure is available in the online issue.)
TABLE 3.
IGF2 region SNPs genotyped in the Cambridge Baby Growth and Wellbeing studies
| dbSNP Number | Position on chromosome 11* (bp) | Relative position (bp) | Haplotype block |
|---|---|---|---|
| rs6578987 | 2098162 | 0 | 1 |
| rs4341514 | 2098179 | 17 | 1 |
| rs680 | 2110210 | 12048 | N/A |
| rs734351 | 2112789 | 14627 | N/A |
| rs3213216 | 2114755 | 16593 | 2 |
| rs3741212 | 2118434 | 20272 | 2 |
| rs11603378 | 2121749 | 23587 | 2 |
| rs10770125 | 2125590 | 27428 | 2 |
| rs3741206 | 2126440 | 28278 | 3 |
| rs3741205 | 2126460 | 28298 | 3 |
| rs1004446 | 2126719 | 28557 | 3 |
| rs4320932 | 2128177 | 30015 | 3 |
| rs7924316 | 2130023 | 31861 | N/A |
| rs3842752 | 2137649 | 39487 | 4 |
| rs7483056 | 2147095 | 48933 | 4 |
| rs6356 | 2147527 | 49365 | 5 |
| rs10743152 | 2152557 | 54395 | 5 |
N/A SNPs were analyzed in isolation as the only genetic variants added to the linear regression statistical models.
*Chromosomal position based on National Center for Biotechnology Information Genome Reference Consortium Human genome build 36 (reference assembly).
Genotyping.
Genomic DNA was extracted from blood samples or mouth swabs using an Autopure LS Machine (Qiagen Ltd., Crawley, U.K.). Of the 17 IGF2 SNPs, 15 were genotyped using KASPar assays, which are competitive allele-specific PCR SNP genotyping assays using FRET quencher cassette oligonucleotides (designed and performed by KBioscience, Hoddesdon, U.K.). The remaining two SNPs were genotyped by PCR restriction fragment–length polymorphism analysis as described below.
rs11603378 was genotyped using 10 ng genomic DNA, 1 × NH4 buffer (Bioline, London, U.K.), 200 mmol/L each deoxyribonucleotide triphosphate, 2.5 mmol/L magnesium, 0.6 pmol/L each oligonucleotide primer (forward 5′-CCAGGCCACATAGTGTAAGTCTGGAA-3′ and reverse 5′-CCAGGGCGCTCCGCGCTA-3′), 10% v/v glycerol, and 0.5 units Taq DNA polymerase (Bioline) in a total volume of 10 μL. This PCR mix was incubated at 94°C for 5 min followed by 20 cycles of 94°C for 45 s, 64°C for 45 s (dropping 0.5°C per cycle), and 72°C for 45 s. This was followed by 15 cycles of 94°C for 45 s, 54°C for 45 s, and 72°C for 45 s, followed by a 10-min incubation at 72°C. This was digested for 16 h at 37°C with 1 unit of StyI (New England Biolabs, Hitchin, U.K.). Separating the samples on a 2% w/v agarose gel by electrophoresis gave a 373 bp band for the G allele and 287 and 86 bp bands for the T allele.
rs3741206 was genotyped using a similar method but with different primers (forward 5′-GAGCCGGGGGCCCGAGATTC-3′ and reverse 5′-GTGCCAGGGAGGCTGGGAGA-3′), only 1 mmol/L magnesium and no glycerol in the PCR mix, and annealing temperatures that were 2°C higher throughout. The PCR products were digested by incubation for 16 h at 37°C with 1 unit of HpyCH4 V (New England Biolabs). Separating the samples on a 2% w/v agarose gel by electrophoresis gave a 245 bp band for the A allele and 195 and 50 bp bands for the G allele.
All SNP genotypes (for the offspring and parents separately), with the exception of rs3741205, were shown to be consistent with Hardy Weinberg equilibrium (P > 0.05 using the χ2 test) and had a repeat genotyping discordancy rate of <1.0%.
Placental RNA extraction and gene expression studies.
Of the 420 available placentas, 29 were from pregnancies where the fetus had all four paternally transmitted IGF2 alleles that were associated with higher maternal glucose concentrations; 32 were from pregnancies where the fetus had all four alleles that were associated with lower glucose concentrations. Of these, the 21 (higher) and 29 (lower) most recently collected placentas were used for IGF-II protein measurements. For the transcription studies, equivalent numbers were 16 and 19.
While keeping the tissue frozen, three samples were cut from the basal plate of each placenta, and the outer 3 mm of each was discarded. Samples were then pooled before homogenization and extraction using Qiagen Tissueruptor and RNeasy fibrous mini kits as per the manufacturer’s instructions. RNA was quantified using a Nanodrop (Labtech Ltd., Ringmer, U.K.) and the integrity was assessed using an Agilent Bioanalyzer (Agilent Technologies, Stockport, U.K.). Only samples with a 260/280 nm absorbance ratio >2.0 and an integrity score >6.4 were used. Synthesis of cDNA was performed using the Bioline cDNA Synthesis Kits according to the manufacturer’s instructions (using 2 μg RNA per reaction, reverse transcriptase and random hexamers, and incubating at 42°C for 30 min.). Controls containing no reverse transcriptase were included for each sample. TaqMan Gene Expression Assays (Applied Biosystems, Warrington, U.K.) were purchased for various IGF2 transcripts and the validated placental reference genes YWHAZ, TOP1, and UBC (18) (Supplementary Table 1). The quantitative PCR reactions were performed using TaqMan Universal Master Mix (Applied Biosystems) on a Stratagene Mx3005P Real-Time PCR system (Agilent Technologies). Data were analyzed using geNorm software (19) to check the stability of the three reference genes. M values (geNorm software) for TOP1, UBC, and YWHAZ were 0.453, 0.542, and 0.429, respectively, with a pairwise variation value of 0.172. The resulting normalization factors were used to convert the raw expression values.
IGF-II protein was extracted overnight from 400 mg mechanically pulverized placental samples in 4 mL 0.18 mol/L HCl/75% ethanol solution at 4°C. The following morning, the samples were spun for 15 min at 12,000g at 4°C. Then 100 μL of the upper layer was carefully removed and neutralized with 100 μL 1 mol/L Tris-buffered saline pH 7.4. IGF-II contents were measured in 20 μL of these neutralized extracts using ELISA kits according to the manufacturer’s instructions (Stratech Scientific, Newmarket, U.K.). Extract total protein contents were estimated by measuring their absorbance at 280 nm.
Statistical analysis.
In light of the different glucose loads used in each cohort, postload maternal glucose responses were standardized by calculating z scores separately in each cohort (using the mean and SD for glucose concentrations from all women in each cohort) and then analyzed as a single group of >1,100 family DNA trios with maternal glucose z scores. IGF2 SNP genotypes from both parents and their child were used to infer parental transmission according to Supplementary Table 2. Initial association analyses were performed separately for each haplotype block (or SNP that was not part of a block) using ANOVA models with maternal glucose z score or birth weight as the dependent variable and all the fetal SNP alleles from that block as independent covariates. For haplotype blocks (or SNPs) reaching P < 0.05 in these ANOVA models, the relevant fetal alleles were taken forward to a general linear model containing the relevant dependent variable adjusted for parity and offspring sex (as potential confounders) (2) for maternal glucose z scores. Birth weight models were adjusted for gestational age, parity, and offspring sex, which account for 25% of the variance in birth weight in our populations. Areas under the capillary blood glucose curves were calculated using the trapezoid rule. The critical value for statistical significance corrected for testing all 17 genotyped IGF2 SNPs was 0.003, calculated using the minimum P value test (20), which has been shown to be robust in this type of analysis (21). A P value of <0.017 was used to define statistical significance for association with specific SNPs by applying a Bonferroni correction to the use of paternally and maternally transmitted fetal alleles and maternal genotypes. Placental data were analyzed using the Mann-Whitney U test, with a P value of <0.05 considered statistically significant.
RESULTS
Haplotype block 1.
Haplotype block 1 fetal alleles were associated with maternal glucose z score (P = 0.048) but not with offspring birth weight (P = 0.7). The paternally transmitted fetal rs6578987 C allele was associated with increased maternal glucose z score (P = 0.006; without significant interaction with offspring sex, P = 0.5) (Fig. 2). The maternally transmitted fetal rs6578987 allele and the rs4341514 alleles (irrespective of their parent of origin) were not associated with maternal glucose z scores.
FIG. 2.
Error bar graph showing mean (95% CI) maternal glucose concentration z scores 60 min postglucose load according to 1) paternally transmitted and 2) maternally transmitted fetal IGF2 alleles at rs6578987, rs680, rs10770125, and rs7924316.
Haplotype block 2.
Haplotype block 2 fetal alleles were associated with maternal glucose z score (P = 0.018) but not with offspring birth weight (P = 1.0). The paternally transmitted fetal rs10770125 A allele was associated with increased maternal glucose z score (P = 0.0002, with an effect size of 0.268 [95% CI 0.164–0.372] z scores, ∼0.4 mmol/L glucose; there was no significant interaction with offspring sex, P = 0.7) (Fig. 2). In contrast, the maternally transmitted fetal rs10770125 allele and all the alleles for SNPs rs3213216, rs3741212, or rs11603378 were not associated with maternal glucose z scores (data not shown).
Haplotype blocks 3–5.
Maternal glucose z score was not associated with fetal alleles from haplotype blocks 3 (P = 0.5), 4 (P = 0.06), or 5 (P = 0.2). In a similar manner, offspring birth weight was not associated with fetal alleles from haplotype blocks 3 (P = 0.5), 4 (P = 0.8), or 5 (P = 0.2).
Other SNPs not defining haplotype blocks.
The paternally transmitted but not the maternally transmitted fetal rs680 A allele was associated with increased maternal glucose z score (P = 0.01; without significant interaction with offspring sex, P = 0.7) (Fig. 2). Neither fetal rs680 allele was associated with offspring birth weight. None of the fetal rs734351 alleles were associated with either maternal glucose z score or with offspring birth weight. The paternally transmitted fetal rs7924316 T allele was associated with increased maternal glucose z score (P = 0.01; without significant interaction with offspring sex, P = 0.4) (Fig. 2), unlike the allele transmitted from the mother. Neither fetal rs7924316 allele was associated with offspring birth weight.
Associations in the two separate cohorts.
Paternally transmitted fetal rs10770125 was significantly associated with maternal glucose concentrations in both the Cambridge Baby Growth Study (P = 0.01) and the Cambridge Wellbeing Study (P = 0.006) (Supplementary Table 3). The other SNPs that had significant associations in the combined cohort did not have significant associations in the modestly sized Cambridge Wellbeing Study (although the trends were in the same direction). The one exception to this was rs7924316, which had a stronger association in the Cambridge Wellbeing Study (P = 0.04) than in the larger Cambridge Baby Growth Study (P = 0.07).
Fasting plasma venous glucose and areas under the capillary glucose curves.
Paternally transmitted fetal rs6578987, rs680, rs10770125, and rs7924316 also showed associations with fasting plasma glucose concentrations (P = 0.002, 0.008, 0.046, and 0.007, respectively) and areas under the capillary glucose curves (P = 0.005, 0.026, 0.020, and 0.059, respectively) in the Cambridge Baby Growth Study (Supplementary Table 4).
Maternal IGF2 genotypes.
None of the maternal (as opposed to the maternally transmitted fetal) IGF2 genotypes were associated with either their own glucose z scores or offspring birth weights (data not shown).
Linkage disequilibrium between fetal IGF2 alleles that were associated with maternal glucose concentrations.
There was variable but significant linkage disequilibrium between the four fetal SNPs whose paternally transmitted alleles were associated with maternal glucose concentrations (D′ = 0.81–0.96, r2 = 0.30–0.90) (Supplementary Fig. 2).
Placental IGF2 expression and IGF-II protein content at term.
Placental IGF-II (protein) contents at birth were raised by 39% in those placentas whose corresponding fetal genotypes were associated with higher glucose concentrations (P = 0.03) (Table 4). There was no significant difference in the relative expression of the IGF2 transcripts 1 or 2 or antisense IGF2 transcripts; the INS-IGF2 readthrough transcript was undetectable in all placentas. Relative expression of IGF2 transcript 3 was reduced by 46% in those placentas having genotypes associated with higher maternal glucose concentrations (P = 0.02).
TABLE 4.
IGF-II protein contents and IGF2 transcript expression (expressed relative to a composite expression score of TOP1, UBC, and YWHAZ) in term placentas from the Cambridge Baby Growth Study having genotypes associated with either higher or lower maternal glucose concentrations at week 28 of the pregnancy
| Placentas with IGF2 genotypes associated with higher maternal glucose concentrations | Placentas with IGF2 genotypes associated with lower maternal glucose concentrations | P value | |
|---|---|---|---|
| IGF-II protein (% of total protein) | 0.93 (0.70–1.44) (n = 21) | 0.67 (0.58–0.99) (n = 29) | 0.028 |
| IGF2 transcript variant 1 expression | 0.143 (0.098–0.190) (n = 16) | 0.091 (0.074–0.174) (n = 19) | 0.2 |
| IGF2 transcript variant 2 expression | 0.101 (0.072–0.149) (n = 16) | 0.074 (0.053–0.131) (n = 19) | 0.3 |
| IGF2 transcript variant 3 expression | 0.040 (0.023–0.069) (n = 16) | 0.074 (0.043–0.117) (n = 19) | 0.018 |
| Antisense IGF2 expression | 0.065 (0.040–0.103) (n = 16) | 0.065 (0.053–0.092) (n = 19) | 0.7 |
The INS-IGF2 readthrough transcripts were undetectable throughout. Data are median (interquartile range) analyzed using Mann-Whitney U tests.
DISCUSSION
Our results show that polymorphic variation in the fetal IGF2 gene, specifically in the copy of the gene transmitted from the father, is associated with increased maternal glucose concentrations in the third trimester of pregnancy. Consistent with their expected imprinted status, we found no associations with the maternally transmitted fetal alleles. Neither was the maternal IGF2 genotype associated with glucose concentrations, unlike the findings in one previous study assessing maternal IGF2 restriction fragment–length polymorphisms in 96 women with GDM (22). The significant associations between paternally transmitted fetal alleles and maternal glucose z scores are unique findings, in terms of both IGF2 and finding associations between genetic variation in paternally transmitted genes and maternal glucose tolerance in pregnancy. In one study, mothers carrying offspring with Beckwith Wiedemann syndrome, where probands have abnormally increased IGF2 expression (23), showed a trend toward an increased risk of developing GDM (24). Also, in our mouse model, we showed that pregnant mice carrying pups with a doubling in Igf2 expression levels have worse glucose tolerance than controls in the final week of their 3-week pregnancies (8). Unlike recent findings for a maternal progesterone receptor gene polymorphism (25) and peroxisome proliferator–activated receptor-γ (26), in our study, there was no significant interactive effect of fetal sex on the associations with maternal glucose concentrations, suggesting that these associations are mediated by different mechanisms. They are all examples of fetuses having associations with changes in maternal glucose concentrations in pregnancy, however (2,3).
The kinship hypothesis suggests that paternally expressed imprinted genes will tend to boost fetal growth and that in contrast, maternally expressed imprinted genes will tend to limit it (5,6). The significant associations between certain paternally transmitted fetal IGF2 alleles and increased maternal glucose concentrations in our study are partially consistent with the kinship hypothesis (5,6), therefore, given that only the paternally transmitted IGF2 allele is active in fetal life and maternal glucose concentrations in pregnancy are linked to fetal growth (27). The results are not fully consistent, however, because there were no associations between any of the IGF2 fetal or maternal variants and offspring birth weight, probably because the effect sizes on maternal glucose concentrations were too small to make a detectable difference in birth weight in our sample. Indeed, using data from the Hyperglycemia and Adverse Pregnancy Outcome Study (27), our largest effect size would have given an odds ratio of only 1.11 for a birth weight above the 90th percentile. The lack of association with birth weight is in general agreement with other studies that have sought but not found association between fetal IGF2 SNP variants and birth weight (28–30), although associations have been found in some populations (31–33). Cord blood IGF-II protein concentrations are associated with birth weight (12,13), but its genetic regulation is not solely due to polymorphic IGF2 variation because other genes in 11p15.5, including the insulin gene variable number of tandem repeats (34), and H19 (13) can influence it.
We found four paternally transmitted IGF2 fetal alleles that are associated with maternal glucose concentrations in week 28 of pregnancy. Using standard interpretations of D′ >0.7 and/or r2 >0.3 (35,36), these four markers appear to be in strong linkage disequilibrium with each other and could therefore be markers of the same causal signal. The strongest candidate that we have for this marker is paternally transmitted fetal rs10770125 (and the SNPs that it tags, rs1003483 and rs4244808), because it gave the largest differential in maternal glucose z scores between the two alleles, the lowest P value, and also was the only marker that was independently statistically significant in both the Cambridge Baby Growth Study and the more modestly sized Cambridge Wellbeing Study (Supplementary Table 3). Human splicing finder (37) predicts that rs10770125 is an exonic splicing or enhancer site plus a micro RNA binding site. In a similar manner, for rs1003483 and rs4244808, it predicts alterations in exonic splicing or enhancer sites, albeit with fewer potential sites than for rs10770125. As well as all three SNPs being present in the IGF2 and antisense IGF2 (IGF2AS) transcripts, they are also present in the INS-IGF2 readthrough transcript. Indeed, rs10770125 codes for a missense nonsynonymous leucine-to-proline mutation in this readthrough transcript. However, this transcript was undetectable in our placentas and so is unlikely to explain the associations with maternal glucose concentrations despite its supposed placental expression (38). In contrast, those placentas whose genotypes were associated with higher maternal glucose concentrations had significantly increased IGF-II protein contents and nonsignificant increases in median mRNA expression of IGF2 transcript variants 1 and 2 of a similar magnitude, which may more easily explain the associations given that in mouse models, placental Igf2 expression relates to that of maternal hormones thought to increase maternal glucose concentrations (38). They also had lower expression of the IGF2 low copy number transcript variant 3, although the significance of this is unclear because the IGF-II isoform 2 it codes for is present at approximately eightfold lower levels in human placenta than isoform 1 coded for by transcript variants 1 and 2 (39,40).
The limitations of our study include the relatively modest size of the cohorts used. One explanation for this is the practical difficulties in collecting samples from three family members rather than just one. It should be noted that inferring the parental origins of fetal alleles for >1,100 trios requires genotyping of >3,300 individuals. For our primary analyses, we combined data collected from two separate cohorts to increase statistical power because they were recruited from patients treated at the same hospital and so were drawn from the same geographic and ethnic (largely Caucasian) pool. A further limitation of our study is the lack of paternity testing, although the confounding effects of this are likely to be minimal as 1) paternally transmitted fetal alleles can be positively inferred even without paternal DNA samples unless maternal and offspring SNP genotypes are heterozygous and 2) in our cohorts using data from 31 SNPs, DNA trio genotypes were not consistent in only 1.5% of the cases (data not shown). We only had maternal BMI available in a subset of family trios (which was not different between groups inheriting our key SNPs) and, thus, these data were not included in the main analyses. However, we cannot totally exclude a confounding effect of maternal BMI in those samples for which this information was not available. In a similar manner, we did not have details of paternal age and BMI or maternal smoking and hypertension for all the trios, so we cannot entirely exclude a confounding effect of these either. The glucose load used was also different in the two cohorts (50 g for the Cambridge Wellbeing Study and 75 g for the Cambridge Baby Growth Study), but we overcame this by standardizing glucose concentration z scores separately in each cohort. Furthermore, not all DNA trios were informative because parental transmission could not be inferred if all members of a trio were heterozygous for a particular SNP. The statistical analyses could therefore have been biased by informative omission from heterozygous trios. This is unlikely to have had a major effect, though, because not all heterozygous parents or children were considered uninformative (e.g., even if both parents were heterozygous for a particular SNP, they were considered uninformative only if their offspring were also heterozygous). Ultimately, however, for our association of paternally transmitted fetal rs10770125 with maternal glucose concentrations or z scores, statistical significance was reached in the number of samples genotyped whether or not data from our two birth cohorts were combined. Replication in additional populations is needed to strengthen this finding.
In conclusion, while associations with maternal genes have been described (2), we report for the first time association between polymorphic variation in a paternally transmitted fetal gene, namely, IGF2, and maternal glucose concentrations in pregnancy. These findings are consistent with recent findings from animal models (7,8) and support the hypothesis that variations in fetal imprinted genes regulate the maternal availability of nutrients (3).
ACKNOWLEDGMENTS
The genotyping and placental RNA work in this study was funded by a grant from the Evelyn Trust and Diabetes UK (11/0004241). Funding for other aspects of the Cambridge Baby Growth Study and Cambridge Wellbeing Study has come from the Medical Research Council, the European Union Framework 5, the World Cancer Research Fund, Newlife Birth Defects, the Mothercare Group Foundation, and Wellbeing of Women.
No potential conflicts of interest relevant to this article were reported.
C.J.P. designed the study, performed the genotyping and statistical analysis, wrote and edited the manuscript, and guarantees the contents of this article. R.V.S. performed the genotyping and placental analyses and commented on the manuscript. D.L.W. and L.M. performed the genotyping and commented on the manuscript. C.L.A., K.K.O., I.A.H., and D.B.D. designed the study, including the establishment of the cohorts, and edited the manuscript.
The authors thank all the families in the Cambridge Baby Growth and Cambridge Wellbeing studies, University of Cambridge research nurses (Suzanne Smith, Amie Wardell, and Karen Forbes), staff at the Addenbrooke’s Wellcome Trust Clinical Research Facility, the Cambridge National Institute for Health Research Biomedical Research Centre, and midwives at the Rosie Maternity Hospital. The authors also thank Karen Whitehead of the University of Cambridge for excellent technical assistance.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0689/-/DC1.
REFERENCES
- 1.Haig D. Placental hormones, genomic imprinting, and maternal–fetal communication. J Evol Biol 1996;9:357–380 [Google Scholar]
- 2.Petry CJ. Gestational diabetes: risk factors and recent advances in its genetics and treatment. Br J Nutr 2010;104:775–787 [DOI] [PubMed] [Google Scholar]
- 3.Petry CJ, Ong KK, Dunger DB. Does the fetal genotype affect maternal physiology during pregnancy? Trends Mol Med 2007;13:414–421 [DOI] [PubMed] [Google Scholar]
- 4.Dunger DB, Petry CJ, Ong KK. Genetics of size at birth. Diabetes Care 2007;30(Suppl. 2):S150–S155 [DOI] [PubMed] [Google Scholar]
- 5.Haig D, Westoby M. Parent-specific gene expression and the triploid endosperm. Am Nat 1989;134:147–155 [Google Scholar]
- 6.Haig D. Genetic conflicts in human pregnancy. Q Rev Biol 1993;68:495–532 [DOI] [PubMed] [Google Scholar]
- 7.Reik W, Constância M, Fowden A, et al. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J Physiol 2003;547:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petry CJ, Evans ML, Wingate DL, et al. Raised late pregnancy glucose concentrations in mice carrying pups with targeted disruption of H19delta13. Diabetes 2010;59:282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 1995;375:34–39 [DOI] [PubMed] [Google Scholar]
- 10.Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res 2006;113:188–193 [DOI] [PubMed] [Google Scholar]
- 11.O’Dell SD, Day IN. Insulin-like growth factor II (IGF-II). Int J Biochem Cell Biol 1998;30:767–771 [DOI] [PubMed] [Google Scholar]
- 12.Ong K, Kratzsch J, Kiess W, Costello M, Scott C, Dunger D; The ALSPAC Study Team. Avon Longitudinal Study of Pregnancy and Childhood. Size at birth and cord blood levels of insulin, insulin-like growth factor I (IGF-I), IGF-II, IGF-binding protein-1 (IGFBP-1), IGFBP-3, and the soluble IGF-II/mannose-6-phosphate receptor in term human infants. J Clin Endocrinol Metab 2000;85:4266–4269 [DOI] [PubMed] [Google Scholar]
- 13.Petry CJ, Ong KK, Barratt BJ, et al. ; ALSPAC Study Team. Common polymorphism in H19 associated with birthweight and cord blood IGF-II levels in humans. BMC Genet 2005;6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ong KK, Diderholm B, Salzano G, et al. Pregnancy insulin, glucose, and BMI contribute to birth outcomes in nondiabetic mothers. Diabetes Care 2008;31:2193–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong KK, Langkamp M, Ranke MB, et al. Insulin-like growth factor I concentrations in infancy predict differential gains in body length and adiposity: the Cambridge Baby Growth Study. Am J Clin Nutr 2009;90:156–161 [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez S, Gaunt TR, O’Dell SD, et al. Haplotypic analyses of the IGF2-INS-TH gene cluster in relation to cardiovascular risk traits. Hum Mol Genet 2004;13:715–725 [DOI] [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265 [DOI] [PubMed] [Google Scholar]
- 18.Cleal JK, Day P, Hanson MA, Lewis RM. Measurement of housekeeping genes in human placenta. Placenta 2009;30:1002–1003 [DOI] [PubMed] [Google Scholar]
- 19.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tippett LHC. The Methods of Statistics. 1st ed. London, Williams and Norgate, 1931 [Google Scholar]
- 21.Chapman J, Whittaker J. Analysis of multiple SNPs in a candidate gene or region. Genet Epidemiol 2008;32:560–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ober C, Xiang KS, Thisted RA, Indovina KA, Wason CJ, Dooley S. Increased risk for gestational diabetes mellitus associated with insulin receptor and insulin-like growth factor II restriction fragment length polymorphisms. Genet Epidemiol 1989;6:559–569 [DOI] [PubMed] [Google Scholar]
- 23.Choufani S, Shuman C, Weksberg R. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet 2010;154C:343–354 [DOI] [PubMed] [Google Scholar]
- 24.Wangler MF, Chang AS, Moley KH, Feinberg AP, Debaun MR. Factors associated with preterm delivery in mothers of children with Beckwith-Wiedemann syndrome: a case cohort study from the BWS registry. Am J Med Genet A 2005;134A:187–191 [DOI] [PubMed] [Google Scholar]
- 25.Hocher B, Chen YP, Schlemm L, et al. Fetal sex determines the impact of maternal PROGINS progesterone receptor polymorphism on maternal physiology during pregnancy. Pharmacogenet Genomics 2009;19:710–718 [DOI] [PubMed] [Google Scholar]
- 26.Hocher B, Schlemm L, Haumann H, et al. Interaction of maternal peroxisome proliferator-activated receptor gamma2 Pro12Ala polymorphism with fetal sex affects maternal glycemic control during pregnancy. Pharmacogenet Genomics 2010;20:139–142 [DOI] [PubMed] [Google Scholar]
- 27.Metzger BE, Lowe LP, Dyer AR, et al. ; HAPO Study Cooperative Research Group. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med 2008;358:1991–2002 [DOI] [PubMed] [Google Scholar]
- 28.Gomes MV, Soares MR, Pasqualim-Neto A, Marcondes CR, Lôbo RB, Ramos ES. Association between birth weight, body mass index and IGF2/ApaI polymorphism. Growth Horm IGF Res 2005;15:360–362 [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez S, Gaunt TR, Dennison E, et al. Replication of IGF2-INS-TH*5 haplotype effect on obesity in older men and study of related phenotypes. Eur J Hum Genet 2006;14:109–116 [DOI] [PubMed] [Google Scholar]
- 30.Souren NY, Paulussen AD, Steyls A, et al. Parent-of-origin specific linkage and association of the IGF2 gene region with birth weight and adult metabolic risk factors. Int J Obes (Lond) 2009;33:962–970 [DOI] [PubMed] [Google Scholar]
- 31.Kaku K, Osada H, Seki K, Sekiya S. Insulin-like growth factor 2 (IGF2) and IGF2 receptor gene variants are associated with fetal growth. Acta Paediatr 2007;96:363–367 [DOI] [PubMed] [Google Scholar]
- 32.Nagaya K, Makita Y, Taketazu G, et al. Paternal allele of IGF2 gene haplotype CTG is associated with fetal and placental growth in Japanese. Pediatr Res 2009;66:135–139 [DOI] [PubMed] [Google Scholar]
- 33.Adkins RM, Somes G, Morrison JC, et al. Association of birth weight with polymorphisms in the IGF2, H19, and IGF2R genes. Pediatr Res 2010;68:429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson LA, Docherty HM, MacKenzie AE, Docherty K. An engineered zinc finger protein reveals a role for the insulin VNTR in the regulation of the insulin and adjacent IGF2 genes. FEBS Lett 2009;583:3181–3186 [DOI] [PubMed] [Google Scholar]
- 35.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet 2002;3:299–309 [DOI] [PubMed] [Google Scholar]
- 36.Weiss KM, Clark AG. Linkage disequilibrium and the mapping of complex human traits. Trends Genet 2002;18:19–24 [DOI] [PubMed] [Google Scholar]
- 37.Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009;37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishida M, Ohashi S, Kizaki Y, Naito J, Horiguchi K, Harigaya T. Expression profiling of mouse placental lactogen II and its correlative genes using a cDNA microarray analysis in the developmental mouse placenta. J Reprod Dev 2007;53:69–76 [DOI] [PubMed] [Google Scholar]
- 39.Monk D, Sanches R, Arnaud P, et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet 2006;15:1259–1269 [DOI] [PubMed] [Google Scholar]
- 40.De Ceuninck F, Willeput J, Corvol M. Purification and characterization of insulin-like growth factor II (IGF II) and an IGF II variant from human placenta. J Chromatogr B Biomed Appl 1995;666:203–214 [DOI] [PubMed] [Google Scholar]