Abstract
OBJECTIVE
Hyperglycemia plays a pivotal role in the development and progression of vascular complications, which are the major sources of morbidity and mortality in diabetes. Furthermore, these vascular complications often persist and progress despite improved glucose control, possibly as a result of prior episodes of hyperglycemia. Epigenetic modifications mediated by histone methyltransferases are associated with gene-activating events that promote enhanced expression of key proinflammatory molecules implicated in vascular injury. In this study, we investigated genetic polymorphisms of the SETD7, SUV39H1, and SUV39H2 methyltransferases as predictors of risk for micro- and macrovascular complications in type 1 diabetes.
RESEARCH DESIGN AND METHODS
In the Finnish Diabetic Nephropathy Study (FinnDiane) cohort, 37 tagging single nucleotide polymorphisms (SNPs) were genotyped in 2,991 individuals with type 1 diabetes and diabetic retinopathy, diabetic nephropathy, and cardiovascular disease. Seven SNPs were genotyped in the replication cohorts from the Steno Diabetes Center and All Ireland/Warren 3/Genetics of Kidneys in Diabetes (GoKinD) U.K. study.
RESULTS
In a meta-analysis, the minor T allele of the exonic SNP rs17353856 in the SUV39H2 was associated with diabetic retinopathy (genotypic odds ratio 0.75, P = 1.2 × 10−4). The same SNP showed a trend toward an association with diabetic nephropathy as well as cardiovascular disease in the FinnDiane cohort.
CONCLUSIONS
Our findings propose that a genetic variation in a gene coding for a histone methyltransferase is protective for a diabetic microvascular complication. The pathophysiological implications of this polymorphism or other genetic variation nearby for the vascular complications of type 1 diabetes remain to be investigated.
Micro- and macrovascular complications develop in a subset of individuals with type 1 diabetes. The severity of comorbid complications is especially striking in patients with established diabetic nephropathy, who have an 18.3-fold increased all-cause mortality rate when compared with the general population (1). Diabetic nephropathy clusters in families (2), which emphasizes the importance of an inherited genetic component. Diabetic retinopathy is another microvascular complication that has an inherited susceptibility (heritability of h2 = 0.52 in the Finnish population) (3).
Along with a genetic background, the lifetime glucose exposure increases the risk of diabetes complications. In the follow-up of the Diabetes Control and Complications Trial, known as the Epidemiology of Diabetes Interventions and Complications (EDIC) study, patients with conventional blood glucose control were more prone to diabetes than patients with intensive glucose control (4). Data from the EDIC and other clinical trials, including the UK Prospective Diabetes Study (5), indicate that diabetes complications continue to develop and progress even in individuals who have managed to improve their glycemic control, a phenomenon referred to as hyperglycemic memory or the legacy effect. Epigenetic modifications on the DNA and histone proteins represent potential mechanisms to explain the hyperglycemic memory (6).
In a series of recent experimental studies, it was demonstrated that hyperglycemia can induce specific and long-lasting gene-activating epigenetic changes. In particular, transient hyperglycemia mediates specific changes to histone 3 lysine 4 (H3K4) and lysine 9 (H3K9) (7,8). In human vascular cells previously exposed to hyperglycemia, the nuclear factor-κB (NF-κB) subunit RELA (alias p65) gene expression is increased partly as a result of monomethylation of H3K4 (H3K4me1) and decreased di- and trimethylation of H3K9 (H3K9me2 and H3K9me3) in the gene promoter. This upregulation of RELA gene expression with concomitant NF-κB activation results in increased expression of inflammatory genes such as VCAM1 and MCP1 downstream in the same pathway. Activated NF-κB has also been detected in a variety of renal cell types in patients with diabetic nephropathy as well as in retinal pericytes in diabetic conditions (9,10).
The gene coding for a histone methyltransferase for H3K4 is the SET domain containing lysine methyltransferase 7 (SETD7). The SETD7 protein is also a regulator of DNA (cytosine-5-)-methyltransferase 1 (DNMT1), which is responsible for maintaining DNA methylation patterning in cell divisions (11). Suppressor of variegation 3–9 homolog 1 (SUV39H1) is a histone methyltransferase that catalyzes the methylation of H3K9, which is associated with the suppression of transcription. SUV39H2 is another H3K9-selective methyltransferase that shares strong sequence homology with SUV39H1. On the basis of previous experimental studies, it appears that these enzymes (SETD7 and SUV39H1/H2) are playing a pivotal role in the glucose-mediated inflammatory response and are therefore candidate genes for diabetic vascular complications, conditions where chronic inflammation is often observed.
In this study, genetic variation of SETD7 (chr4q31.1), SUV39H1 (chrXp11.23), and SUV39H2 (chr10p13) was studied in relation to micro- and macrovascular complications in a large well-characterized population of Finnish patients with type 1 diabetes. Selected single nucleotide polymorphisms (SNPs) were genotyped in replication cohorts from the Steno Diabetes Center and All Ireland/Warren 3/Genetics of Kidneys in Diabetes (GoKinD) U.K. study.
RESEARCH DESIGN AND METHODS
Discovery population.
Patients were from the nationwide Finnish Diabetic Nephropathy Study (FinnDiane). At the baseline visit, patients underwent a thorough clinical investigation in conjunction with a regular visit to the attending physician. A total of 2,991 patients with type 1 diabetes were included. Type 1 diabetes was defined as age at onset ≤35 years and permanent insulin treatment initiated within a year from diagnosis. Patients were divided into three groups according to urinary albumin excretion rate (AER) in two out of three consecutive overnight or 24-h urine collections. Normal AER was defined as an AER <20 μg/min or <30 mg/24 h, microalbuminuria as an AER ≥20 and <200 μg/min or ≥30 and <300 mg/24 h, and macroalbuminuria as an AER ≥200 μg/min or ≥300 mg/24 h. End-stage renal disease (ESRD) was defined as patient either having received a kidney transplant or undergoing dialysis. Patients with inadequate AER data were excluded from all analyses (n = 22, <1% of total cohort). There were altogether 810 case subjects with diabetic nephropathy (macroalbuminuria or ESRD) and 1,070 control subjects with AER within the normal range despite diabetes duration ≥15 years (Table 1). Diabetic retinopathy was defined as patient having undergone laser treatment (n = 1,180), and control subjects were patients without laser treatment (n = 1,785). Cardiovascular disease (CVD) (n = 298) was defined as any of the following events: myocardial infarction, coronary artery procedure, stroke, limb amputation, or peripheral artery procedure. CVD control subjects (n = 341) had no CVD and normal AER despite an age of ≥45 years.
TABLE 1.
Clinical characteristics of the FinnDiane patients
| Nephropathy |
Retinopathy |
|||
|---|---|---|---|---|
| Case subjects | Control subjects | Case subjects | Control subjects | |
| n | 810 | 1,070 | 1,167 | 1,785 |
| Sex (% male) | 59.4 | 43.8 | 57.3 | 48.3 |
| Age (years) | 42.0 ± 9.4 | 41.4 ± 11.4 | 43.1 ± 9.8 | 35.1 ± 11.5 |
| Age at diagnosis (years) | 11.6 ± 7.0 | 14.1 ± 8.2 | 11.9 ± 7.3 | 15.9 ± 8.6 |
| Duration of diabetes (years) | 30.3 ± 8.1 | 27.2 ± 9.2 | 31.2 ± 8.3 | 19.2 ± 10.6 |
| BMI (kg/m2) | 25.3 ± 3.9 | 25.0 ± 3.2 | 25.3 ± 3.8 | 24.9 ± 3.3 |
| SBP (mmHg) | 147 ± 21 | 132 ± 17 | 143 ± 20 | 130 ± 16 |
| A1C (%) | 8.9 ± 1.6 | 8.2 ± 1.3 | 8.7 ± 1.5 | 8.4 ± 1.5 |
| Current smoker (%) | 22.7 | 17.8 | 21.1 | 24.3 |
| Triglycerides (mmol/L) | 1.5 (0.4−11.3) | 0.9 (0.3−10.2) | 1.2 (0.3−11.3) | 1.0 (0.3−10.2) |
| eGFR (mL ⋅ min−1 ⋅ 1.73 m−2) | 56 ± 33 | 98 ± 17 | 70 ± 34 | 101 ± 21 |
| Nephropathy status | ||||
| Normal AER (%) | 0 | 100 | 23.3 | 81.4 |
| Microalbuminuria (%) | 0 | 0 | 17.7 | 12.1 |
| Macroalbuminuria (%) | 66.9 | 0 | 36.9 | 6.1 |
| ESRD (%) | 33.1 | 0 | 22.0 | 0.4 |
| Retinopathy (%)* | 84.9 | 24.8 | 100 | 0 |
| CVD (%) | 25.8 | 5.9 | 21.2 | 2.6 |
Data are means ± SD or medians (range) unless otherwise indicated.
*Laser-treated diabetic retinopathy.
Clinical examination.
Blood pressure was measured twice, and the average was calculated. Mean arterial pressure (MAP) was estimated from systolic blood pressure (SBP) and diastolic blood pressure (DBP) measurements. Height and weight were measured and BMI was calculated. Current smoking was defined as smoking at least one cigarette per day. The estimated glomerular filtration rate (eGFR) was calculated using the CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) equation (12). A blood sample was obtained to determine A1C and serum lipids and for DNA extraction. A1C was measured by standardized immunoassays. Serum lipids and lipoprotein concentrations were measured by automated enzymatic methods.
Replication populations.
Danish patients with type 1 diabetes were recruited from the Steno Diabetes Center. Diabetic nephropathy was defined as macroalbuminuria or ESRD and no other kidney disease. Altogether, 452 case subjects with diabetic nephropathy and 432 control subjects with normal AER and diabetes duration ≥15 years were included. Diabetic retinopathy was graded as nil, simplex, or proliferative retinopathy, and patients with proliferative retinopathy were compared with patients with no retinopathy. Cardiovascular events studied were stroke and myocardial infarction. Clinical characteristics of the Steno cohort are presented in Table 2.
TABLE 2.
Clinical characteristics of the replication populations
| Steno cohort |
GoKind U.K. cohort |
|||||||
|---|---|---|---|---|---|---|---|---|
| Nephropathy |
Retinopathy |
Nephropathy |
Retinopathy |
|||||
| Case subjects | Control subjects | Case subjects | Control subjects | Case subjects | Control subjects | Case subjects | Control subjects | |
| n | 452 | 432 | 416 | 159 | 718 | 749 | 556 | 382 |
| Sex (% male) | 60.8 | 53.2 | 56.0 | 49.7 | 57.8 | 42.7 | 49.5 | 45.3 |
| Age (years) | 45.3 ± 11.5 | 42.0 ± 10.4 | 44.5 ± 11.1 | 44.9 ± 11.7 | 48.0 ± 10.4 | 43.5 ± 11.0 | 45.2 ± 10.4 | 44.3 ± 11.6 |
| Age at diagnosis (years) | 13.7 ± 8.2 | 17.4 ± 8.8 | 13.5 ± 8.1 | 18.6 ± 9.3 | 14.8 ± 7.7 | 15.5 ± 7.9 | 14.4 ± 7.6 | 15.8 ± 7.9 |
| Duration of diabetes (years) | 28.3 ± 8.8 | 27.9 ± 10.0 | 31.0 ± 8.7 | 26.3 ± 10.8 | 33.3 ± 9.4 | 28.1 ± 9.0 | 30.9 ± 9.1 | 28.5 ± 9.2 |
| BMI (kg/m2) | 24.2 ± 3.3 | 24.2 ± 3.1 | 24.4 ± 3.5 | 23.6 ± 2.5 | 26.3 ± 4.7 | 26.1 ± 4.2 | 26.0 ± 4.0 | 26.0 ± 4.0 |
| SBP (mmHg) | 144 ± 22 | 134 ± 19 | 145 ± 21 | 130 ± 17 | 145 ± 21 | 125 ± 15 | 135 ± 21 | 128 ± 17 |
| A1C (%) | 9.4 ± 1.5 | 8.4 ± 1.1 | 9.2 ± 1.0 | 8.2 ± 1.4 | 8.9 ± 1.8 | 8.6 ± 1.5 | 8.9 ± 1.7 | 8.7 ± 1.6 |
| Smoking (%) | 41.6 | 38.9 | 37.7 | 47.2 | 42.8 | 35.4 | 38.8 | 36.9 |
| Triglycerides (mmol/L) | 1.3 (0.3−9.9) | 0.8 (0.3−5.4) | 1.1 (0.3−9.9) | 0.8 (0.3−3.5) | NA | NA | NA | NA |
| eGFR (mL ⋅ min−1 ⋅ 1.73 m−2) | 78 ± 32 | 102 ± 15 | 80 ± 30 | 104 ± 13 | NA | NA | NA | NA |
| Nephropathy status | ||||||||
| Normal AER (%) | 0 | 100 | 27.6 | 96.2 | 0 | 100 | 41.7 | 90.6 |
| Macroalbuminuria (%) | 94.7 | 0 | 67.3 | 3.8 | 73.1 | 0 | 32.7 | 9.4 |
| ESRD (%) | 5.3 | 0 | 5.0 | 0 | 26.9 | 0 | 25.5 | 0.3 |
| Retinopathy (%)* | 67.9 | 26.6 | 100 | 0 | 71.7 | 52.6 | 100 | 0 |
| CVD (%) | 9.5 | 3.7 | 9.9 | 2.5 | NA | NA | NA | NA |
Data are means ± SD or medians (range) unless otherwise indicated.
*Proliferative retinopathy diagnosis (in Steno) and self-reported proliferative retinopathy (in GoKind U.K.).
The other replication population was recruited as part of the All Ireland/Warren 3/GoKinD U.K. resources, previously described elsewhere (13), and referred to as the GoKinD U.K. cohort in this article. In brief, 718 case subjects with diabetic nephropathy and 749 control subjects with no renal disease were included. Retinopathy data were self-reported; 556 were case subjects with proliferative retinopathy and 382 were control subjects (Table 2).
The study protocol was approved by the ethics committees of all participating FinnDiane centers, a local ethical committee in Denmark, and research ethics committees in the U.K. and Ireland. The protocol follows the Declaration of Helsinki. Each patient participating in the study provided written informed consent.
Power calculations.
Power calculations were made using the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc) for the FinnDiane cohort. For diabetic retinopathy, the prevalence was set to 0.4. With a marker allele frequency of 0.05, we have >90% power (P = 0.001) in an allelic association study with odds ratios (OR) of 1.35 for marker allele heterozygotes and 1.7 for homozygotes. For diabetic nephropathy, the same effect sizes were approximated as for diabetic retinopathy and prevalence was set to 0.3. With a marker allele frequency of 0.05, our study cohort gives >85% power (P = 0.05) to detect a difference in an allelic association study. For genotype-based comparisons between case subjects and control subjects a marker allele frequency of 0.05 is sufficient to reach 80% power (P = 0.05). With the sample set for CVD, we have 80% power (P = 0.05) in an allelic association analysis with a minor allele frequency (MAF) of 0.1 and an OR 1.7 in a dominant model.
SNP selection.
Study regions were selected to include the genes and 10 kb outside both the 3′ and 5′ ends of SETD7, SUV39H1, and SUV39H2. SNP genotype data typed in the HapMap CEU (Utah residents with Northern and Western European ancestry from the CEPH collection) population were downloaded from the HapMap (Phase II, Data Release 24). All SNPs with a MAF ≥0.05 and a Hardy-Weinberg equilibrium (HWE) P value ≥ 0.001 were included in further selection. Tagger program in Haploview 4.1 software (http://www.broadinstitute.org/mpg/haploview) (14) was used to select markers for genotyping. Tagger was run with a r2 threshold of 0.8 and pairwise tagging only settings. Twenty-five tagSNPs for SETD7, one tagSNP for SUV39H1, and 11 tagSNPs for SUV39H2 were selected to capture variability of 43, 1, and 15 SNP markers, respectively. The average r2 between tagged SNP and tagSNPs was 0.906 for SETD7 and 0.885 for SUV39H2. Three last 3′ tagSNPs for SUV39H2 were in the last intron of the adjacent gene DCLRE1C. The selected set of SNPs was genotyped in the FinnDiane cohort. All SNPs showing an association (P ≤ 0.05) with diabetes complications in this cohort were selected for replication in the Steno and GoKinD U.K. cohorts.
Genotyping.
SNP genotyping of the FinnDiane samples was performed with the Sequenom’s MassARRAY iPLEX system (Sequenom, San Diego, CA) except for one SNP. This SNP, as well as all samples from the Steno cohort, were genotyped with TaqMan genotyping technology using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Two control samples in four replicates and at least eight zero samples (H2O) were included in the 384-genotyping plates. After TaqMan PCR, an allelic discrimination was conducted, and genotypes were called with SDS 2.3 software (Applied Biosystems). For the control samples, 100% genotyping accuracy was required. Predesigned TaqMan SNP genotyping assays and one custom assay for rs12572872 were obtained from Applied Biosystems. GoKinD U.K. cohort SNP genotyping was performed with MassARRAY iPLEX Gold (Sequenom).
Statistical analysis.
Genotype and allele frequencies in case and control subjects were compared with χ2 test or with the Fisher exact test where appropriate. X-chromosomal SNP rs3373 was studied separately for men and women. Correction for multiple testing was carried out by calculating the effective number of independent tests using the SNPSpD program (http://gump.qimr.edu.au/general/daleN/SNPSpD) and the equation from Li and Ji (15). The significance limit of the study was assessed first by calculating the effective number of SNPs (21.23) and then multiplying by the number of studied phenotypes (3). Thus, the significance limit of the study was P = 7.8 × 10−4 (P = 0.05/64). Logistic regression was used to give an OR and 95% CI for the SNP when analyzed with other risk factors for diabetic nephropathy, retinopathy, or CVD. In brief, a major allele homozygote was the reference genotype in all models, and minor allele homozygotes and heterozygotes were combined. All covariates added to the models were significant (P < 0.05) in the univariate logistic regression models. To make the logistic regression model more accurate for the majority of data, outliers were removed before including the SNP data in the models (standardized residuals examined, 0.01 level). For diabetic retinopathy, diabetes duration, A1C, BMI, SBP, and ln triglycerides (information on the triglyceride concentrations was not available from the GoKinD U.K. cohort) were the phenotypic variables in the regression models. For diabetic nephropathy, the variables of diabetes duration, A1C, smoking (yes/no, current smoker in FinnDiane and Steno, and an ever-smoker in the GoKinD U.K. cohort), SBP, and ln triglycerides were inserted in the regression models. CVD was studied with the covariates of diabetes duration, A1C, ln triglycerides, MAP, sex, smoking history (yes/no), and eGFR. In the Steno cohort, the variables in the regression model for CVD were limited to eGFR, A1C, and ln triglycerides because of the smaller number of case subjects.
The Hardy-Weinberg equilibrium was tested with Haploview 4.1. Haplotype analyses were conducted with Haploview, and correction for multiple testing was calculated with a permutation for 100,000 tests. Haplotypes were set with CIs. The Fisher method was used for combining P values from χ2 tests from three populations studied. A meta-analysis was performed using a fixed-effects meta-analysis in Plink 1.07 software (http://pngu.mgh.harvard.edu/~purcell/plink) (16). In silico gene expression analysis was performed with GeneSapiens (www.genesapiens.org) (17). Exonic splicing enhancer motifs (ESEs) were evaluated with the ESEfinder 3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi), and the untranslated (UTR) region analysis was performed with UTRScan (itbtools.ba.itb.cnr.it/utrscan). MiRNA binding was studied in silico with TargetScan (www.targetscan.org) and miRbase (www.mirbase.org).
RESULTS
Genotyping.
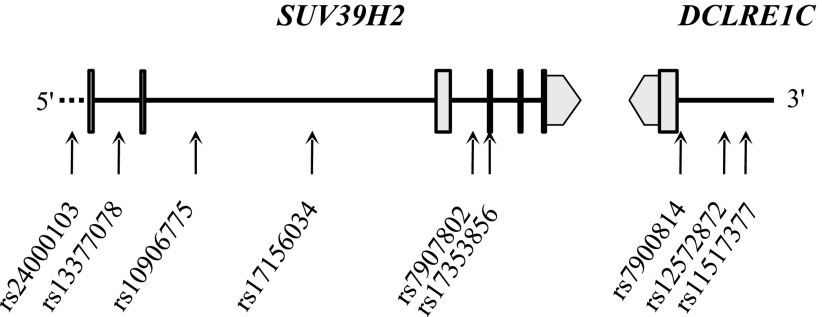
A total of 23 SNPs out of 25 tagSNPs within SETD7 were successfully genotyped in the discovery cohort. The rs7680948 was left out from the iPlex because of primer failures, and rs4863655 located 9.7 kb from the gene failed genotyping and was excluded from the analysis. The rs3373 in SUV39H1 was successfully genotyped, but one male patient was heterozygous for this X-chromosomal SNP and was excluded from the analysis. Nine tagSNPs out of 11 in SUV39H2 were successfully genotyped (Fig. 1). The rs17156024 failed in genotyping and rs7922341 gave unreliable genotyping results that were studied further with PCR and sequencing. The results still showed unreliable genotype calling (data not shown), and rs7922341 genotypes were excluded. Altogether, genotypes were successfully called for 33 SNPs with average success rates of 0.982, 0.982, and 0.984 for SETD7, SUV39H1, and SUV39H2, respectively (genotypes provided in Supplementary Tables 2–4). All samples with a genotyping success rate of ≥0.75 were included in further analyses.
FIG. 1.
Genotyped SNPs in the chr10:14,959,424−14,996,308 location (NCBI36 assembly). All putative exons are marked with boxes, and UTR regions are marked with arrows.
No deviation from the Hardy-Weinberg equilibrium was observed (P > 0.001). All SNPs showing an association (P ≤ 0.05) with any diabetes complication in the FinnDiane were selected for replication. All seven SNPs were successfully genotyped in the Steno and GoKinD U.K. replication cohorts (genotype counts provided in Supplementary Tables 5–7).
Diabetic retinopathy.
Exonic SNP rs17353856 in the SUV39H2 and two SNPs rs7900814 and rs12572872 in the last intron of the adjacent gene DCLRE1C were associated with diabetic retinopathy in FinnDiane (P < 0.003 for genotypic association, Table 3). The minor allele A of rs12572872 was significantly associated with diabetic retinopathy after adjustment for clinical risk factor diabetes duration, A1C, BMI, SBP, and triglycerides (genotypic OR 0.78 [95% CI 0.62–0.97], P = 0.029). These three SNPs were then analyzed in the Steno and GoKinD U.K. cohorts (Table 3). Although the association with rs12572872 could not be replicated, the results followed the same trend in the Steno (OR 0.71 [0.42–1.26], P = 0.245) and GoKinD U.K. cohorts (OR 0.89 [0.64–1.23], P = 0.478). The minor allele T of rs17353856 was significantly associated with a decreased risk of diabetic retinopathy in the GoKinD U.K. cohort (OR 0.69 [0.50–0.95], P = 0.023). In a meta-analysis, this SNP was significantly associated with retinopathy (allelic OR 0.77, P = 2.6 × 10−4, and genotypic OR 0.75, P = 1.2 × 10−4). The haplotype consisting of SNPs rs7907802, rs17353856, and rs7900814 was not significantly associated with diabetic retinopathy (haplotype blocks in Supplementary Fig. 1 and retinopathy association in Supplementary Table 1) when corrected for clinical risk factors (data not shown).
TABLE 3.
Diabetic retinopathy association in discovery and replication cohorts and in a meta-analysis
|
SUV39H2 |
MAF (%) |
Allelic association |
Genotype association |
||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Cohort | Case subjects | Control subjects | P* | OR (95% CI)† | P† | P* | OR (95% CI)† | P† |
| rs17353856 | FinnDiane | 10.5 | 12.7 | 0.015 | 0.82 (0.65–1.04) | 0.096 | 0.0017 | 0.76 (0.61–1.01) | 0.064 |
| Steno | 12.6 | 14.5 | 0.413 | 0.90 (0.53–1.53) | 0.687 | 0.703 | 0.83 (0.45–1.52) | 0.546 | |
| GoKind U.K. | 13.8 | 17.2 | 0.047 | 0.69 (0.50–0.95) | 0.023 | 0.151 | 0.68 (0.47–0.97) | 0.034 | |
| Combined‡ | 0.012 | 0.77 | 2.6 × 10−4 | 0.008 | 0.75 | 1.2 × 10−4 | |||
| rs7900814§ | FinnDiane | 21.0 | 23.8 | 0.013 | 0.90 (0.76–1.08) | 0.252 | 0.0028 | 0.84 (0.68–1.04) | 0.100 |
| Steno | 28.1 | 31.8 | 0.240 | 0.80 (0.53–1.20) | 0.279 | 0.399 | 0.71 (0.42–1.22) | 0.216 | |
| GoKind U.K. | 26.2 | 23.5 | 0.190 | 1.15 (0.93–1.43) | 0.23 | 0.097 | 0.93 (0.71–1.20) | 0.190 | |
| Combined‡ | 0.94 | 0.352 | 0.0054 | 0.85 | 0.015 | ||||
| rs12572872§ | FinnDiane | 15.6 | 18.7 | 0.003 | 0.83 (0.68–1.01) | 0.065 | 6.0 × 10−4 | 0.78 (0.62–0.97) | 0.029 |
| Steno | 19.5 | 23.6 | 0.148 | 0.80 (0.50–1.27) | 0.340 | 0.241 | 0.71 (0.40–1.26) | 0.245 | |
| GoKind U.K. | 26.4 | 27.2 | 0.697 | 0.88 (0.68–1.14) | 0.335 | 0.710 | 0.89 (0.64–1.23) | 0.478 | |
| Combined‡ | 0.013 | 0.84 | 0.007 | 0.005 | 0.79 | 0.001 | |||
*Two-tailed P value from the χ2 test. A combined P value is calculated with the Fisher method if the effect is in the same direction.
†Logistic regression model with diabetes duration, A1C, BMI, SBP, and ln triglycerides (triglycerides not in the GoKind U.K. cohort).
‡For combined data, a fixed-effects meta-analysis P value and OR are given.
§SNP is in the last intron of the adjacent gene DCLRE1C.
Diabetic nephropathy.
In the FinnDiane cohort, rs3373 close to SUV39H1, rs17353856 in the SUV39H2, and three SNPs in SETD7 were associated with diabetic nephropathy either in an allelic association analysis or when genotypes were compared (Table 4). When adjusted for relevant risk factors (diabetes duration, male sex, SBP, A1C, smoking, and triglycerides), rs17353856 in the SUV39H2 and rs11100112 and rs2725790 in the SETD7 remained significantly associated with diabetic nephropathy (OR 0.69 [95% CI 0.50–0.97], P = 0.030; 0.67 [0.51–0.88], P = 0.004; and 0.76 [0.58–0.99], P = 0.043, respectively) (Table 4). Seven SNPs were then studied for association with diabetic nephropathy in the replication populations. The rs2592970 in the SETD7 was found to be associated with diabetic nephropathy in the GoKinD U.K. cohort (P = 0.036). This association, however, did not remain after adjustment for clinical risk factors. In a meta-analysis, rs11100112 was significantly associated with decreased risk of diabetic nephropathy (OR 0.76, P = 2.5 × 10−4), but there was significant heterogeneity between the populations (Cochran Q test, P = 0.036, Supplementary Table 8). Therefore, the fixed-effect meta-analysis results should be interpreted with caution. A random-effects meta-analysis was tested, but no significant associations were observed.
TABLE 4.
Diabetic nephropathy association in discovery and replication cohorts and in a meta-analysis
| Gene |
MAF (%) |
Allelic association |
Genotype association |
||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Cohort | Case subjects | Control subjects | P* | OR (95% CI)† | P† | P* | OR (95% CI)† | P† |
| SETD7 | |||||||||
| rs11100112 | FinnDiane | 18.0 | 20.7 | 0.044 | 0.75 (0.60–0.95) | 0.015 | 0.122 | 0.67 (0.51–0.88) | 0.004 |
| Steno | 19.0 | 21.4 | 0.210 | 0.91 (0.66–1.26) | 0.572 | 0.461 | 0.84 (0.57–1.24) | 0.381 | |
| GoKind U.K. | 21.8 | 22.3 | 0.778 | 0.99 (0.76–1.29) | 0.932 | 0.486 | 1.15 (0.83–1.59) | 0.404 | |
| Combined‡ | 0.130 | 0.83 | 0.0051 | 0.303 | 0.76 | 2.5 × 10−4 | |||
| rs2592970 | FinnDiane | 37.0 | 39.3 | 0.154 | 0.88 (0.73–1.06) | 0.177 | 0.004 | 0.94 (0.72–1.21) | 0.612 |
| Steno | 35.2 | 35.8 | 0.800 | 0.99 (0.75–1.30) | 0.919 | 0.301 | 1.10 (0.76–1.59) | 0.628 | |
| GoKind U.K. | 35.1 | 38.9 | 0.036 | 0.86 (0.68–1.09) | 0.205 | 0.110 | 0.86 (0.65–1.24) | 0.501 | |
| Combined‡ | 0.094 | 0.89 | 0.047 | 0.007 | 0.95 | 0.523 | |||
| rs2725790 | FinnDiane | 21.0 | 22.9 | 0.161 | 0.78 (0.63–0.98) | 0.029 | 0.053 | 0.76 (0.58–0.99) | 0.043 |
| Steno | 18.8 | 19.3 | 0.794 | 1.02 (0.73–1.42) | 0.925 | 0.252 | 1.10 (0.74–1.63) | 0.650 | |
| GoKind U.K. | 18.3 | 20.6 | 0.113 | 0.96 (0.73–1.27) | 0.761 | 0.290 | 0.93 (0.67–1.29) | 0.657 | |
| Combined‡ | 0.205 | 0.86 | 0.022 | 0.085 | 0.84 | 0.030 | |||
| SUV39H1 | |||||||||
| rs3373 (men) | FinnDiane | 34.3 | 28.5 | 0.060 | 1.51 (1.02–2.25) | 0.042 | |||
| Steno | 27.1 | 29.5 | 0.561 | 0.93 (0.55–1.57) | 0.780 | ||||
| GoKind U.K. | 24.6 | 25.3 | 0.820 | 0.96 (0.69–1.35) | 0.760 | ||||
| Combined‡ | 1.02 | 0.840 | |||||||
| SUV39H2 | |||||||||
| rs17353856 | FinnDiane | 9.9 | 12.0 | 0.043 | 0.71 (0.53–0.97) | 0.029 | 0.001 | 0.69 (0.50–0.97) | 0.030 |
| Steno | 12.5 | 13.6 | 0.519 | 0.93 (0.62–1.39) | 0.709 | 0.198 | 0.94 (0.60–1.47) | 0.777 | |
| GoKind U.K. | 14.1 | 14.7 | 0.652 | 0.95 (0.69–1.31) | 0.748 | 0.186 | 1.00 (0.70–1.44) | 0.988 | |
| Combined‡ | 0.206 | 0.81 | 0.0079 | 0.0023 | 0.80 | 0.012 | |||
| rs7900814§ | FinnDiane | 20.4 | 22.2 | 0.189 | 0.92 (0.74–1.15) | 0.456 | 0.005 | 0.87 (0.67–1.13) | 0.289 |
| Steno | 28.5 | 29.8 | 0.561 | 0.98 (0.73–1.32) | 0.883 | 0.480 | 0.96 (0.66–1.40) | 0.817 | |
| GoKind U.K. | 26.7 | 28.0 | 0.410 | 0.93 (0.79–1.10) | 0.450 | 0.400 | 1.03 (0.84–1.27) | 0.430 | |
| Combined‡ | 0.394 | 0.93 | 0.230 | 0.031 | 0.95 | 0.506 | |||
| rs12572872§ | FinnDiane | 15.9 | 16.8 | 0.465 | 1.00 (0.78–1.28) | 0.990 | 0.044 | 1.01 (0.76–1.34) | 0.951 |
| Steno | 20.7 | 20.3 | 0.857 | 1.11 (0.79–1.56) | 0.543 | 0.913 | 1.12 (0.75–1.67) | 0.587 | |
| GoKind U.K. | 26.3 | 25.8 | 0.734 | 1.05 (0.81–1.36) | 0.728 | 0.709 | 1.07 (0.78–1.47) | 0.681 | |
| Combined‡ | 0.934 | 1.04 | 0.667 | 0.377 | 1.08 | 0.555 | |||
*Two-tailed P value from χ2 test. A combined P value is calculated with Fisher method if the effect is in the same direction.
†Logistic regression model with diabetes duration, A1C, sex, SBP, smoking, and ln triglycerides (triglycerides not in the GoKind U.K. cohort).
‡Fisher method P values and fixed-effects meta-analysis P values and ORs are given.
§SNP is in the last intron of the adjacent gene DCLRE1C.
CVD.
In the FinnDiane cohort, rs3373 close to SUV39H1 in men and rs17353856 in SUV39H2 were associated with CVD (P = 0.037 and P = 0.017, respectively), but the associations did not remain when adjusted for eGFR, A1C, diabetes duration, triglycerides, sex, smoking history, and MAP (OR 2.06 [95% CI 0.96–4.53], P = 0.073, and 0.66 [0.40–1.10], P = 0.114, respectively) (Table 5). These results could not be replicated in the Steno cohort.
TABLE 5.
CVD association in the FinnDiane and Steno cohorts
| Gene |
MAF (%) |
Allelic association |
Genotype association |
||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Cohort | Case subjects | Control subjects | P* | OR (95% CI)† | P† | P* | OR (95% CI)† | P† |
| SUV39H1 | |||||||||
| rs3373 (men) | FinnDiane | 35.4 | 24.6 | 0.037 | 2.06 (0.96–4.53) | 0.073 | |||
| Steno | 25.0 | 31.2 | 0.509 | 0.41 (0.08–2.02) | 0.273 | ||||
| SUV39H2 | |||||||||
| rs17353856 | FinnDiane | 8.6 | 12.9 | 0.017 | 0.66 (0.40–1.10) | 0.114 | 0.046 | 0.66 (0.37–1.15) | 0.140 |
| Steno | 14.0 | 14.2 | 0.956 | 1.02 (0.33–3.19) | 0.975 | 0.769 | 1.17 (0.33–4.15) | 0.807 | |
| rs7900814‡ | FinnDiane | 20.1 | 23.6 | 0.132 | 0.96 (0.66–1.39) | 0.821 | 0.129 | 1.00 (0.64–1.57) | 0.996 |
| Steno | 27.7 | 31.9 | 0.404 | 0.52 (0.20–1.31) | 0.165 | 0.522 | 0.47 (0.14–1.53) | 0.209 | |
| rs12572872‡ | FinnDiane | 14.4 | 18.3 | 0.067 | 0.89 (0.58–1.35) | 0.577 | 0.130 | 0.91 (0.56–1.41) | 0.694 |
| Steno | 20.0 | 19.8 | 0.967 | 0.91 (0.32–2.61) | 0.864 | 0.956 | 0.96 (0.28–3.28) | 0.951 | |
*Two-tailed P value from χ2 test.
†ORs and 95% CI are from logistic regression models with eGFR, A1C, diabetes duration, ln triglycerides, sex, smoking history, and MAP in the FinnDiane cohort and eGFR, A1C, and ln triglycerides in the Steno cohort.
‡SNP is in the last intron of the adjacent gene DCLRE1C.
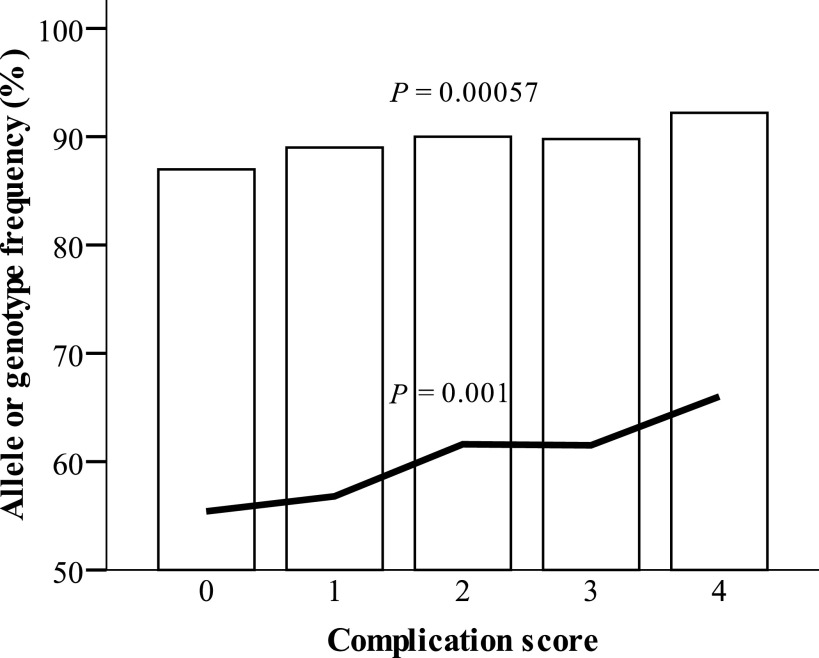
Interestingly, rs17353856 is the same exonic SNP associated with diabetic retinopathy as well as diabetic nephropathy in the FinnDiane cohort. Therefore, we calculated a complication score for each patient. Groups were assessed by the presence of diabetic vascular complications: 0 = no vascular complication; 1 = retinopathy or nephropathy; 2 = retinopathy + nephropathy or CVD; 3 = retinopathy and CVD or nephropathy and CVD; and 4 = retinopathy, nephropathy, and CVD. The rs17353856 major allele C frequencies increased with increasing complication score (P = 5.7 × 10−4) (Fig. 2 and Supplementary Table 9). However, a similar trend could not be replicated in the Steno cohort.
FIG. 2.
The rs17353856 C allele frequencies (white bars) and genotype CGG/CGG frequencies (black line) in the complication score groups. The CGG genotype consists of rs17353856 C allele (major), rs7900814 G allele (major), and rs12572872 G allele (major). The complication score groups are formed as follows: 0 = no vascular complications; 1 = diabetic nephropathy or retinopathy; 2 = diabetic nephropathy and retinopathy or CVD; 3 = CVD + diabetic nephropathy or retinopathy; and 4 = CVD, diabetic nephropathy, and retinopathy. P values are from the χ2 linear by linear association test.
DISCUSSION
In this article, we present suggestive evidence that an exonic SNP rs17353856 in the SUV39H2 is associated with diabetic retinopathy in patients with type 1 diabetes. In addition, a variant in the SETD7 showed a trend toward an association with diabetic nephropathy. The association between the rs17353856 and diabetic nephropathy was borderline significant, but the association between the SNP and diabetic retinopathy was confirmed in a meta-analysis of three studied populations. Because this same SNP showed a trend toward an association with CVD in the FinnDiane cohort, a complication score variable was created. It revealed that the major allele C (risk allele) of this SNP and the adjacent two SNPs in the same haplotype block were more frequent in patients with several vascular complications. This trend was not replicated in the Steno cohort, possibly because of the smaller size of this particular patient cohort.
The synonymous SNP rs17353856 is located in the third or fourth exon of SUV39H2 depending on the splicing variant. In silico analysis of exonic splicing enhancers around rs17353856 (ESEFinder 3.0) suggests that a putative binding site of the SRSF1 protein is lost with the rs17353856 T allele in the DNA sequence. ESE-binding proteins are involved in precursor mRNA splicing, and according to the Ensembl database automatic transcript annotation pipeline (www.ensembl.org), SUV39H2 precursor mRNA could be alternatively spliced by deleting multiple exons and exons carrying rs17353856 among them. In addition, the same sequence in the reverse DNA strand is located in the long last intron of an alternative transcript of the adjacent gene DCLRE1C (ENST00000378289).
The rs17353856 is in low linkage disequilibrium with any SNPs nearby in the HapMap CEU population except rs11594111 (A/G) (r2 = 1.0, HapMap Data Releases 24 and 28 checked). The 1000 Genomes Project Pilot 1 Phase has not come up with new SNPs that would be in high linkage disequilibrium with rs17353856 (data accessed through SNAP SNP Annotation and Proxy Search, www.broadinstitute.org/mpg/snap/ldsearch.php). rs11594111 is in the 3′ UTR of SUV39H2. According to UTRScan, this region does not bind any known UTR-binding proteins. There is a microRNA miR101-1 target sequence in the rs11594111 region, and with the G-allele of rs11594111, the continuous complementary to miR101-1 is extended from 7 nucleotides to 11 nucleotides (TargetScan, miRbase). However, SUV39H2 mRNA is not among the predicted targets for miR101-1 (TargetScan).
Interestingly, rs17353856 in the SUV39H2 is located next to a G-nucleotide in the DNA sequence (ATA[C/T]GGC). It is of note that a CG-dinucleotide is a target for DNA methylation, which is an epigenetic process that can influence gene expression when the DNA sequence near the transcription start site becomes methylated. Less is known about the role of DNA methylation outside the transcription start sites. In a DNA sequence that contains the minor T allele, no site of DNA methylation exists because a CG-dinucleotide is changed to a TC-dinucleotide. If and how this truly affects DNA methylation or gene expression remains an open question.
The rs11100112 is located in the third intron of the SETD7. In a meta-analysis, the SNP showed significant heterogeneity when ORs corrected for clinical risk factors were combined. Therefore, the significant effect observed in the meta-analysis should be interpreted with caution. However, this trend-like result is interesting, because at least the allelic effect of the SNP was in the same direction in patients with diabetic nephropathy in all three studied populations.
Logistic regression is a well-established method to analyze SNP data when adjusting for clinical features. To avoid small group sizes of the minor homozygote group, a dominant genotypic model was chosen in the logistic regression analysis. It can, however, be argued that the dominant model does not represent the true effect of the genetic variation on a polygenegic disease such as diabetes complications. In addition, this approach was chosen to adjust for a sufficient number of clinical risk factors for diabetes complications in the regression models and because heterozygous genotype frequencies rather than minor allele homozygous genotype frequencies (for example, for the SNPs rs11100112 and rs17353856) differed more in case subjects and control subjects regarding diabetic nephropathy in the FinnDiane cohort. This result indicates a possible drift toward a dominant genetic effect.
Diabetic vascular complications occur as a result of complex pathological processes, which are activated by chronic hyperglycemia. It is now evident that metabolic memory, whereby diabetes complications continue to develop and progress in some individuals despite their ability to improve their glycemic control, could have long-lasting effects. Several clinical observations support the concept that an earlier period of hyperglycemia can lead to subsequent sustained and long-lasting effects within the vasculature, ultimately resulting in ongoing organ injury and dysfunction. The emerging picture is one of remarkable complexity and, in this context of various vascular complications of diabetes, the increasingly recognized phenomenon of hyperglycemic memory is perhaps the most clinically important and unsolved problem in diabetes (18).
The recent discoveries that epigenetic modifications are implicated in studies of glucose-mediated injury and specifically that histone modifications are important determinants regulating the longevity of gene-activating events in the context of hyperglycemia have emphasized the potential role of epigenetic events in the pathogenesis of diabetes complications (19). Our current view of epigenetic changes is mainly derived from primary cell culture experiments and rodent models of hyperglycemic variability. The striking parallel is the identification of proinflammatory molecules that are associated with both transient and chronic hyperglycemia with demonstrable macrovascular disease (8). Although the initial studies on histone methylation in response to hyperglycemia were performed in aortic endothelial cells, subsequent studies were performed in microvascular endothelial cells with similar glucose-induced changes in histone methylation described. Thus, it is likely that the epigenetic modifications originally reported in cells of a macrovascular origin are also relevant to endothelial cells from other sites including retina and kidney.
The phenomenon of metabolic memory appears to be at least partly attributed to sustained gene-activating epigenetic events associated with SETD7 methyltransferase (H3K4me) and decreased gene-silencing epigenetic events by SUV39H1 (H3K9me2, H3K9me3) (7). SUV39H2, a close homolog of the SUV39H1, has not been shown to play a proven role in hyperglycemia-induced epigenetic processes in the vasculature. In silico analysis with GeneSapiens shows SUV39H2 expression in a variety of human tissues including the eye and the kidney, but at a lower level when compared with SUV39H1. SUV39H1 and SUV39H2 are essential in all cell types because they regulate telomere length in mammalian cells (20). Importantly, both SUV39H1 and SUV39H2 are upregulated in human microvascular endothelial cells (21).
We suggest that the altered function of histone methyltransferases might be an early event in diabetic vascular complications because hyperglycemia induces histone methylation alterations that activate NF-κB RELA (p65) and further increase the expression of other inflammatory genes. In turn, this inflammatory response can trigger further epigenetic chromatin modifications, thus promoting altered gene regulation of key pathological genes implicated in diabetes complications (22). It is known that another epigenetic modification, histone acetylation by EP300, increases expression of extracellular matrix (ECM) genes in endothelial cells in the kidney and retina in response to hyperglycemia (23,24). Most importantly, such an overexpression of ECM genes in mesangial cells of the kidney has been associated with decreased repressive histone marks (H3K9me2/3) and increased activating histone marks (H3K4me by SETD7) in the gene promoters (25).
Bone morphogenic protein (BMP) signaling is another pathway that is associated with diabetic nephropathy and vascular diseases (26). This pathway also links SUV39H1/H2 to complications of diabetes because SUV39H1 and SUV39H2 are shown to repress target genes with SMAD1/SMAD5 proteins in the BMP pathway (27). Genetic variabilty in SMAD and BMP 2/4/7 genes do not associate with diabetic nephropathy (28,29). Thus, our current study further proves that, at least in diabetic nephropathy, genetic variants in this pathway do not strongly affect the complication. However, the studies in diabetic retinopathy remain sparse.
A well-characterized phenotype is important in genetic case-control studies. Retinopathy was diagnosed slightly differently in the three study populations, which can be seen as a limitation of the study. In the FinnDiane cohort, laser treatment was the diagnostic criterion for diabetic retinopathy, and the diagnosis was verified by the attending physician. Even self-reported laser treatment was shown to be a good indicator of severe retinopathy (30). In the Steno cohort, a precise diagnosis was done of proliferative retinopathy, whereas in the GoKinD U.K. cohort, the retinopathy diagnosis was self-reported by the patient. Despite these differences, consistent SNP association results could be found across these populations. Another difficulty in the retinopathy study was the definition of control subjects. There is no consensus of an established diabetes duration that could be used and required for the control subjects. In diabetic nephropathy, type 1 diabetes duration of 15 years is the standard used, although it can be argued not to be strong enough for the control definition. We chose not to exclude patients either with albuminuria or with short diabetes duration from diabetic retinopathy control subjects. This approach obviously weakened the control phenotype accuracy but gave more power to the association analyses with a larger numbers of subjects examined. After the original analyses, the effect of diabetes duration in the control subjects was carefully studied in the FinnDiane cohort. It became evident that when type 1 diabetes duration filters were added, the number of control subjects and apparent significance decreased, but at the same time, case and control subjects became genetically more different. The same effect was seen in the replication populations (data not shown).
In conclusion, we found a polymorphism in the histone methyltransferase SUV39H2 to be associated with diabetic retinopathy. This gene was not previously linked to complications of diabetes, although it is a close homolog of SUV39H1 that has been shown to play a role in the hyperglycemia-induced inflammatory response. This genetic study of three populations proposes that a variation in this particular histone methyltransferase is protective for a diabetic microvascular complication. It remains to be determined as to the pathophysiological implications of this specific finding and in particular how it relates to the ultimate development of diabetes vascular complications.
ACKNOWLEDGMENTS
The study in Finland was supported by grants from the Folkhälsan Research Foundation, Samfundet Folkhälsan, and the Wilhelm och Else Stockmann Foundation. The Warren 3/GoKinD U.K. Study Group was jointly funded by Diabetes UK and the Juvenile Diabetes Research Foundation and includes the following individuals: Belfast: Professor A.P. Maxwell, Dr. A.J. McKnight, and Dr. D.A. Savage; Edinburgh: Dr. J. Walker; London: Dr. S. Thomas and Professor G.C. Viberti; Manchester: Professor A.J.M. Boulton; Newcastle: Professor S. Marshall; Plymouth: Professor A.G. Demaine and Dr. B.A. Millward; and Swansea: Professor S.C. Bain.
No potential conflicts of interest relevant to this article were reported.
A.S. researched data, wrote the manuscript, and contributed to discussion. A.E.-O. wrote, reviewed, and edited the manuscript and contributed to discussion. C.F. contributed to discussion and reviewed and edited the manuscript. N.S. researched data. M.P. reviewed the manuscript. L.T. researched data and reviewed and edited the manuscript. H.-H.P. researched data. A.J.M. researched data, contributed to discussion, and reviewed and edited the manuscript. A.P.M. and M.E.C. reviewed and edited the manuscript. P.-H.G. contributed to discussion and reviewed and edited the manuscript.
Parts of this study were presented in oral form at the Annual Meeting of the European Diabetic Nephropathy Study Group (EDNSG), Poitiers, France, 20–22 May 2010.
The authors acknowledge all study subjects who participated in the study in Finland, Denmark, the U.K., and Ireland. The authors thank all the physicians and nurses at each center participating in the collection of FinnDiane patients (Supplementary Table 10). The authors also thank the Danish Diabetes Association and the Sehested Hansen Foundations for their continuous support of the study.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0073/-/DC1.
REFERENCES
- 1.Groop PH, Thomas MC, Moran JL, et al. FinnDiane Study Group. The presence and severity of chronic kidney disease predicts all-cause mortality in type 1diabetes. Diabetes 2009;7:1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J. Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 2004;53:2449–2454 [DOI] [PubMed] [Google Scholar]
- 3.Hietala K, Forsblom C, Summanen P, Groop PH. FinnDiane Study Group: heritability of proliferative diabetic retinopathy. Diabetes 2008;8:2176–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Writing Team for the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA 2003;290:2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med 2008;359:1577–1589 [DOI] [PubMed] [Google Scholar]
- 6.Pirola L, Balcerczyk A, Okabe J, El-Osta A. Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol 2010;6:665–675 [DOI] [PubMed] [Google Scholar]
- 7.El-Osta A, Brasacchio D, Yao D, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 2008;205:2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brasacchio D, Okabe J, Tikellis C, et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 2009;58:1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mezzano S, Aros C, Droguett A, et al. NF-κB activation and overexpression of regulated genes in human diabetic nephropathy. Nephrol Dial Transplant 2004;10:2505–2512 [DOI] [PubMed] [Google Scholar]
- 10.Romeo G, Liu W, Asnaghi V, Kern TS, Lorenzi M. Activation of nuclear factor-κB induced by diabetes and high glucose regulates a proapoptotic program in retinal pericytes. Diabetes 2002;7:2241–2248 [DOI] [PubMed] [Google Scholar]
- 11.Estève PO, Chin HG, Benner J, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U S A 2009;106:5076–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levey AS, Stevens LA, Schmid CH, et al. CKD-EPI (Chronic Kidney Disease Epidemilogy Collaboration): a new equation to estimate glomerular filtration rate. Ann Intern Med 2009;9:604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKnight AJ, Patterson CC, Pettigrew KA, et al. Warren 3/U.K. Genetics of Kidneys in Diabetes (GoKinD) Study Group: a GREM1 gene variant associates with diabetic nephropathy. J Am Soc Nephrol 2010;5:773–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265 [DOI] [PubMed] [Google Scholar]
- 15.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 2005;95:221–227 [DOI] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 2007;3:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilpinen S, Autio R, Ojala K, et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol 2008;9:R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalmers J, Cooper ME. UKPDS and the legacy effect. N Engl J Med 2008;359:1618–1620 [DOI] [PubMed] [Google Scholar]
- 19.Tonna S, El-Osta A, Cooper ME, Tikellis C. Metabolic memory and diabetic nephropathy: potential role for epigenetic mechanisms. Nat Rev Nephrol 2010;6:332–341 [DOI] [PubMed] [Google Scholar]
- 20.García-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 2004;36:94–99 [DOI] [PubMed] [Google Scholar]
- 21.Lukk M, Kapushesky M, Nikkilä J, et al. A global map of human gene expression. Nat Biotechnol 2010;28:322–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol 2010;299:F14–F25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu B, Chiu J, Feng B, Chen S, Chakrabarti S. PARP activation and the alteration of vasoactive factors and extracellular matrix protein in retina and kidney in diabetes. Diabetes Metab Res Rev 2008;24:404–412 [DOI] [PubMed] [Google Scholar]
- 24.Chen S, Feng B, George B, Chakrabarti R, Chen M, Chakrabarti S. Transcriptional coactivator p300 regulates glucose-induced gene expression in endothelial cells. Am J Physiol Endocrinol Metab 2010;298:E127–E137 [DOI] [PubMed] [Google Scholar]
- 25.Sun G, Reddy MA, Yuan H, Lanting L, Kato M, Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 2010;21:2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maciel TT, Kempf H, Campos AH. Targeting bone morphogenetic protein signaling on renal and vascular diseases. Curr Opin Nephrol Hypertens 2010;19:26–31 [DOI] [PubMed] [Google Scholar]
- 27.Frontelo P, Leader JE, Yoo N, et al. Suv39h histone methyltransferases interact with Smads and cooperate in BMP-induced repression. Oncogene 2004;23:5242–5251 [DOI] [PubMed] [Google Scholar]
- 28.McKnight AJ, Woodman AM, Parkkonen M, et al. Investigation of DNA polymorphisms in SMAD genes for genetic predisposition to diabetic nephropathy in patients with type 1 diabetes mellitus. Diabetologia 2009;52:844–849 [DOI] [PubMed] [Google Scholar]
- 29.McKnight AJ, Pettigrew KA, Patterson CC, Kilner J, Sadlier DM, Maxwell AP. Investigation of the association of BMP gene variants with nephropathy in type 1 diabetes mellitus. Diabet Med 2010;27:624–630 [DOI] [PubMed]
- 30.Grassi MA, Mazzulla DA, Knudtson MD, et al. Patient self-report of prior laser treatment reliably indicates presence of severe diabetic retinopathy. Am J Ophthalmol 2009;147:501–504 [DOI] [PMC free article] [PubMed] [Google Scholar]