Abstract
Split-hand/foot malformation with long-bone deficiency (SHFLD) is a relatively rare autosomal-dominant skeletal disorder, characterized by variable expressivity and incomplete penetrance. Although several chromosomal loci for SHFLD have been identified, the molecular basis and pathogenesis of most SHFLD cases are unknown. In this study we describe three unrelated kindreds, in which SHFLD segregated with distinct but overlapping duplications in 17p13.3, a region previously linked to SHFLD. In a large three-generation family, the disorder was found to segregate with a 254 kb microduplication; a second microduplication of 527 kb was identified in an affected female and her unaffected mother, and a 430 kb microduplication versus microtriplication was identified in three affected members of a multi-generational family. These findings, along with previously published data, suggest that one locus responsible for this form of SHFLD is located within a 173 kb overlapping critical region, and that the copy gains are incompletely penetrant.
Keywords: split-hand/foot malformation, SHFM, SHFLD, microduplication, microarray, conserved regulatory element
Introduction
The Split-hand/foot malformations (SHFMs) are a heterogeneous group of malformations, in which the hand and/or foot findings may occur in isolation or with other anomalies. Split-hand/foot malformation with long-bone deficiency (SHFLD) (OMIM # 119 100, 610 685, and 612 576) is a rare autosomal-dominant disorder with variable expressivity and incomplete penetrance, the clinical presentation of which may range from isolated hypoplastic halluces to classic SHFM to monodactyly with tibial aplasia.1 Other phenotypes with SHFM and long-bone involvement include SHFM with fibular deficiency (OMIM 113 310) and SHFM with ulnar defects (OMIM 314 360). Given that these entities seem to be distinct, there may be multiple loci involved in these malformations.
Isolated SHFM has been mapped to six loci, and the genetic causes are well understood for only three: SHFM1 maps to 7q21, SHFM2 maps to Xq26, SHFM3 is associated with microduplications in 10q24, SHFM4 is caused by dominant mutations in P63 in 3q27, SHFM5 maps to 2q31 (reviewed in Basel et al2), and SHFM6 is caused by recessive mutations in WNT10B in 12q13.3 The known causes of syndromic forms of SHFM include dominant P63 mutations in the ectrodactyly, ectodermal dysplasia, cleft lip/palate (EEC3) syndrome4 (OMIM # 604 292) and related phenotypes, and recessive CDH3 mutations in the ectrodactyly, ectodermal dysplasia, and macular dystrophy (EEM) syndrome5 (OMIM # 225 280). It is of note that genomic rearrangements are a relatively frequent finding in some forms of SHFM and have been described for SHFM1, SHFM3, and SHFM5. SHFM1 has been associated with deletions, translocations, and inversions involving 7q21,6 SHFM3 involves duplications of BRTC, POLL, and all or part of FBXW,7, 8 and SHFM5 has been reported in association with deletions centromeric to EVX2.9
Studies to date suggest that SHFLD is genetically distinct from the isolated forms of SHFM. Although no specific genetic cause has yet been elucidated, three loci have been proposed. Chromosome 2q14.2 has been implicated in a single individual with SHFLD arising from a de novo translocation without disrupting any known genes, but corresponding to the homologous region in mouse which gives rise to the ‘dominant hemimelia' (Dh) limb malformation.10 Both 1q42.2-q43 and 6q14.1, designated as loci SHFLD1 and SHFLD2, respectively, were shown to be linked to SHFLD in a large consanguineous Arab family.11, 12 SHFLD3 was mapped in a Brazilian family to an 861 kb interval at 17p13.1-17p13.3.13
In this study we report three independent and overlapping microduplications at 17p13.3 in individuals with SHFLD. The first, a 254 kb duplication, segregates with the disorder in a three-generation family and manifests with variable expressivity and incomplete penetrance. The second, a 527 kb duplication, was found in an affected child and inherited from her clinically unaffected mother. The third, a 430 kb duplication versus triplication, segregates with SHFLD in three affected members of a multi-generational family. These duplications share a 173 kb region of overlap and are located within the SHFLD3 region described by Lezirovitz et al.13
Materials and methods
Clinical data
Three kindreds were involved in the study. Family 1 participants were examined by CA and GG. Digital photographs and radiographs of hands and feet were obtained for all affected individuals and obligate carriers. Photographs and radiographs for the individual(s) from families 2 and 3 were taken at the time of clinical assessment, some dating back a number of years.
DNA samples and chromosome analysis
DNA samples were obtained/used following research ethics approval from Children's Hospital of Eastern Ontario for family 1 and research ethics approval from the Institutional Review Board of Self Regional Healthcare (Greenwood, SC, USA) for families 2 and 3. DNA was extracted from whole blood using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA) in family 1, the FlexiGene DNA kit (Qiagen) in family 2 and one member of family 3, and a high-salt precipitation method14 in the other members of family 3. Pediatric samples for family 1 were obtained from saliva using Oragene saliva extraction kits (DNA Genotek, Kanata, Ontario, Canada) according to the manufacturer's instructions. Chromosome analysis from peripheral blood was carried out according to standard protocols.15
Microarray analysis
The proband's sample from family 1 was analysed with the Affymetrix 500K SNP platform (Affymetrix, Santa Clara, CA, USA). An additional family member from family 1, the affected proband and unaffected mother from family 2, and three affected members of family 3 were analysed using the Affymetrix Genome-wide Human SNP 6.0 array (Affymetrix). For each array, DNA (250 ng) was processed using reagents and protocols according to manufacturer's instructions (Affymetrix). After hybridisation of the DNA onto the chip, the arrays were washed and stained in the GeneChip Fluidics Station 450 (Affymetrix) and scanned using the Affymetrix 500K SNP/Genome-wide SNP 6.0 array protocol (Affymetrix) and the GeneChip Scanner 3000 7G. Copy-number analysis was carried out with Affymetrix's Genotyping Console 4.0 using the in silico control of 270 HapMap samples. Affymetrix's Genotyping Console 4.0 (GTC 4.0) was used to standardise array intensity files to a default reference and detect genotype.
Haplotyping
Family 1 was genotyped using the six microsatellite markers D17S1308, UT269, UT137, AFM022XB6, D17S1533, and D17S1528. Each marker was PCR amplified as described previously16 and electrophoresed on a 6% acrylamide gel using the LI-COR DNA sequencer Model 4000 (LI-COR, Lincoln, NE, USA). Genotypes were determined using RFLPscan software (version 3.0; Scanalytics, Rockville, MD, USA) and haplotypes were generated using Cyrillic software (version 2.1; Cyrillic Software, Oxfordshire, UK).
Quantitative PCR analysis (qPCR)
Quantitative PCR (qPCR) was used to test for the 17p13.3 duplication in all three families. Relative quantitation of genomic dosage was determined using an EP RealPlex4 mastercycler (Eppendorf AG, Hamburg, Germany) and calculated by the relative threshold cycle (ΔΔCT) method.17 PCR amplicons were generated in triplicate for each individual and four controls. Genomic dosage for three genes located within the duplicated interval, TIMM22, ABR, and TUSC5, was determined by SYBR green incorporation using Taqman RNAseP reference (ABI, Carlsbad, CA, USA). Six primer pairs were chosen based on sequences that were predicted to be amenable to qPCR and informative for microarray confirmation (Figure 3). Relative genomic dosage was calculated as 2−ΔΔCt where ΔCt=(mean CtTarget)−(mean CtReference) and ΔΔCT=ΔCtpatient−ΔCtcontrol. Analyses were carried out for selected members of family 1, the affected proband and both parents from family 2, and all three available individuals from family 3.
Phylogenetic footprinting
To infer patterns of evolution and identify uncharacterized functional elements, we compared the human SHFLD locus with orthologous and well-annotated sequences of mouse, opossum, chicken, and zebrafish using PipMaker and MultiPipmaker (http://bio.cse.psu.edu/pipmaker/).18, 19 Sequences corresponding to the duplicated region were obtained from Ensembl Genome Data Resources from the following contigs: human, NCBI36/hg18, ch17: 956201–1128916; mouse, NCBIM37, ch11: 75 400 000–76 800 000; opossum, BROADO5, ch2: 515 100 000–517 000 000; chicken WASHUC2, ch19: 5 100 000–7 000 000; zebrafish, Zv8, ch15: 24 000 000:25 500 000.
Results
Family 1
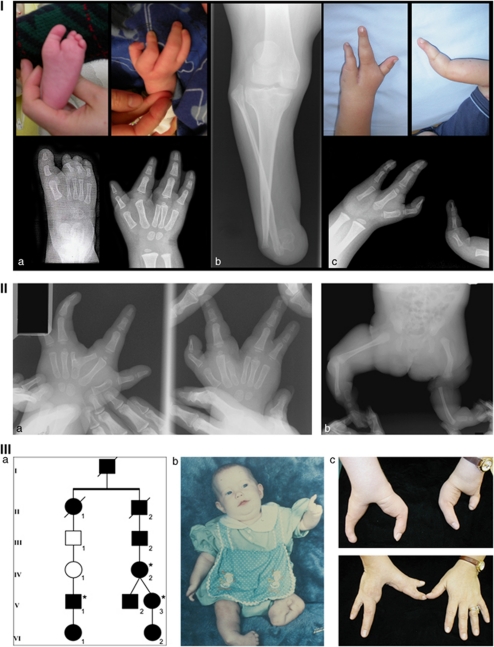
The proband was evaluated for bilateral split hands and a split left foot (Figure 1, panel I). In total, 28 individuals spanning three generations of his extended Mennonite family participated, and five had limb anomalies (Figure 2). In three, the anomalies were limited to the hands; two had tibial hypoplasia or aplasia (Figure 1). Inheritance was consistent with an autosomal-dominant pattern, with incomplete penetrance and variable expressivity. No other significant health issues were reported in the family; cognitive abilities and development were apparently normal in all individuals.
Figure 1.
Panel I: clinical findings in family 1. (a) Hand and foot findings in the proband. (b) Tibial hypoplasia in individual 107. (c) Monodactyly and oligodactyly with split hand in individual S108.2 who also has tibial hypoplasia (not shown). Pedigree of Family 1 can be found in Figure 2. Panel II: clinical findings in the isolated affected individual from family 2 showing hand malformations (a) including six metacarpals, bifid metacarpal with split hand and (b) tibial aplasia. Radiographs courtesy of Shriners Hospital for Children, Greenville, SC. Panel III: partial pedigree and clinical findings in family 3. (a) Relationships between the individuals from whom samples were available (asterisks). (b) Preoperative photograph of individual V-3 showing hand malformations, distal hypoplasia of right tibia, aplasia of left tibia, and clubfeet. (c) Hand findings in individuals V-3 (upper) and IV-2 (lower) as adults. Individual V-3 was born with only a thumb on the left hand (b), and the lateral digit shown here is a transplanted hallux.
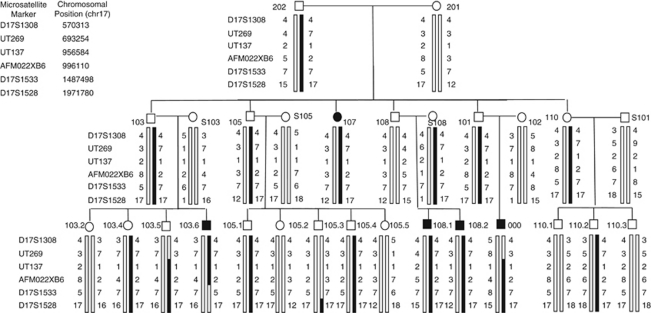
Figure 2.
Pedigree with haplotypes of the region on 17p13.3. Circles (○)=females and squares (□)=males. Solid symbols=affected; unshaded symbols=unaffected. Legend to the left of each generation indicates microsatellite marker used and list (top left) indicates genomic co-ordinates (chr 17) of each. The black portion of each haplotype bar indicates haplotype that segregates with the duplication. Individuals 000 and S108 were found to carry the duplication by array (corresponding to region spanning markers UT137 and AFM022XB6). qPCR confirmed array results and also confirmed that individuals 107, 108.2, 103.5, and 103.6 carry the duplication. Three offspring of individuals 101 and 102 and their children did not participate; two sons (reportedly unaffected) with eight children between them (sexes unknown, also reportedly unaffected), and one daughter (reportedly with a ‘short toe') and her five children (reportedly unaffected).
Mutation analysis of the P63 exons previously implicated in SHFM420, 21 (exons 5–8, 13, and 14) was carried out as part of an initial clinical diagnostic workup and no sequence alterations were detected in the proband. The proband was analysed with the 500K Affymetrix array and individual S108 (Figure 2) was analysed with the 6.0 Affymetrix array. In both individuals, this identified a 254 kb duplication at 17p13.3 bordered by SNP_A-4268793 and SNP A-8640248 (6.0 array), corresponding to genomic co-ordinates 956 201–1 210 473 NCBI36/hg18 (Human Mar. 2006) assembly (Figure 3). This duplication has not been reported as a copy-number variant in the database of genomic variants (http://projects.tcag.ca/variation/), NCBI (http://www.ncbi.nlm.nih.gov) or the Sanger database (http://www.sanger.ac.uk/humgen/cnv/); however, some genomic variants have been reported (both duplications and deletions) in ABR, located at the telomeric end of the duplication. Segregation analysis using six microsatellite markers revealed that the haplotype containing the duplication was present in all affected individuals and obligate carriers (Figure 2). Individual (S108), who married into the family, also possessed the duplication and the disease haplotype. Although we could not determine her ancestral relationship to the family, such a finding is not surprising as this population is somewhat genetically isolated and has a high coefficient of inbreeding.22 Analysis of the proband (individual 000 in Fig 2) and other family members with the segregating haplotype, including individuals 107, S108, 108.2, 103.5, and 103.6 (Figure 2) by qPCR showed that each had the duplication and confirmed the breakpoints found by microarray (Figure 3 and Supplementary Table and Figure).
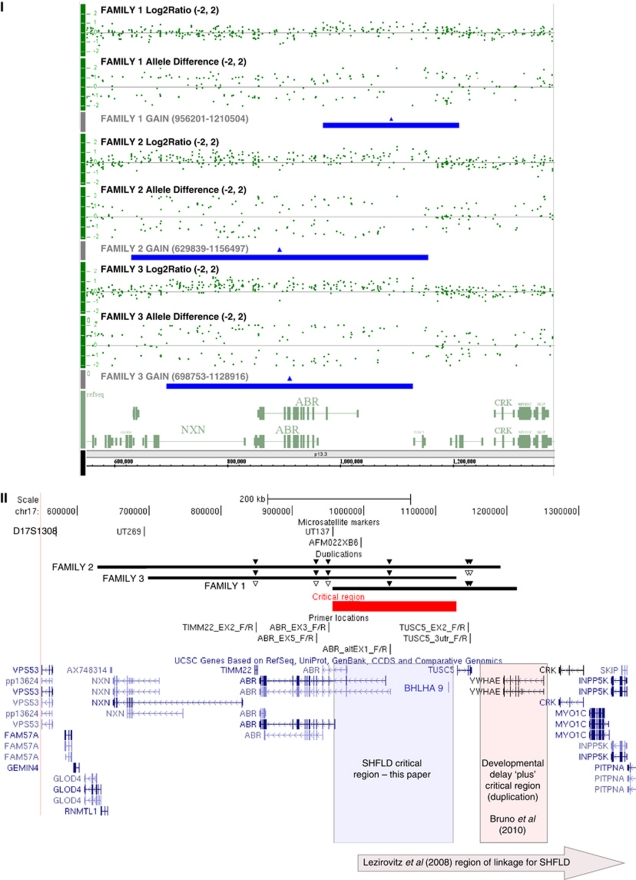
Figure 3.
Panel I: Affymetrix Genome-wide SNP 6.0 microarray results (NCBI36/hg18 build). Chromosome 17p13.3 (550000-1380000) showing the copy-number gain in representative affected family members from families 1 (top), 2 (middle), and 3 (bottom). Data for each patient are represented by log2 ratio and allele calls, and the region of copy gain is denoted as a solid bar below the data for each patient. Panel II: relevant area of chromosome 17 from UCSC (http://genome.ucsc.edu/) showing annotated genes.37, 38 Beneath scale co-ordinates at top are the microsatellites used with their respective positions. Marker D17S1528 is located more centromeric than can be shown in the figure. Duplications as indicated (genomic co-ordinates in panel I). Overlapping area extends over 172 715 nucleotides and is indicated by solid line. Solid and open triangles above each of the families' duplications indicate confirmation of increased and normal dosage, respectively, by qPCR (qPCR data in Supplementary Figure and Table). SHFLD critical region indicated by labelled shaded box. Critical region described by Bruno et al25 involving YWHAE indicated as labelled. Area of linkage reported by Lezirovitz et al13 indicated by labelled shaded arrow at bottom.
Family 2
The proband was evaluated because of congenital limb anomalies (Figure 1 panel II). Radiographs of the upper limbs revealed six metacarpals in the right hand with a thumb, three additional digits, and a deep central cleft between digits 2 and 3. The left hand contained five metacarpals, the third being bifid, but was otherwise similar to the right. Radiographs of the lower limbs revealed aplasia of the tibiae. The right foot had only four metatarsals and four toes with a central cleft, and the left foot had three metatarsals with two toes and a central cleft. Development has been normal. Selected exons of P63 (5–8, 13, and 14) were sequenced on a clinical diagnostic basis but no mutations were found, and high-resolution chromosomes were normal. Affymetrix Genome-wide Human SNP 6.0 array in the proband and subsequently in her mother revealed a 527 kb duplication at 17p13.3 with genomic co-ordinates 629 839–1 156 497, NCBI36/hg18 (Human Mar. 2006) assembly (Figure 3). The mother had no evidence of any limb or digit anomalies. Analysis of the proband and both parents by qPCR revealed maternal inheritance of this duplication and confirmed the breakpoints found on microarray (Figure 3 and Supplementary Table and Figure). Of note, use of the Affymetrix software to determine the copy-number state in both the proband and mother showed a copy number of 3 across part of the region (suggestive of a duplication) and a copy number of 4 across the remainder of the region (suggestive of a triplication); however, the qPCR data (Supplementary Table) were most consistent with a duplication across the entire region.
Family 3
One affected individual (individual V-1, Figure 1, panel IIIa), now 30 years old, was first evaluated at age 6 for congenital limb abnormalities and an extensive family history consistent with SHFLD. As an adult, he had normal high-resolution chromosomes and P63 sequencing (exons 5–8, 13, and 14) performed on a clinical basis. He was recently seen in follow-up with his 3-year-old daughter (individual VI-1, Figure 1, panel IIIa), who also has SHFLD, and had an Affymetrix Genome-wide Human SNP 6.0 array that revealed a 430 kb copy gain at 17p13.3 with genomic co-ordinates 698 753–1 128 916, NCBI36/hg18 (Human March 2006) assembly (Figure 3). A second duplication spanning 596 kb was identified at 20p12.1 with genomic co-ordinates 16 641 447–17 237 736. Other members of his extended family with SHFLD had also been previously evaluated, including two of his third cousins (a pair of dizygotic twins, individuals V-2 and V-3), the mother of the twins (individual IV-2), and the daughter of the female twin (individual VI-2). All had SHFLD, with variable upper limb and foot involvement and tibial a/hypoplasia requiring amputations and prostheses (Figure 1, panel IIIb). DNA samples were available from two of these individuals (IV-2 and V-3). Previous research-based testing in one of these individuals excluded the presence of a P63 mutation via SSCP analysis of all coding exons and an SHFM3 rearrangement by pulsed-field gel electrophoresis. Analysis using the Affymetrix Genome-wide Human SNP 6.0 array showed the same 17p13.3 copy gain in both individuals, and no evidence of the 20p12.1 duplication in either individual. qPCR analysis of all three affected individuals showed the 17p13.3 copy gain in each of them and confirmed the breakpoints found by microarray (Figure 3 and Supplementary Table and Figure). Of note, use of the Affymetrix software to determine the copy-number state in all three individuals showed a copy number of 4 across the region (suggestive of a triplication); however, it was unclear whether this was due to signal noise, and it could not be conclusively determined from the qPCR data (Supplementary Table) whether the copy number was higher than that of a duplication.
Sequence alignments and phylogenetic analysis
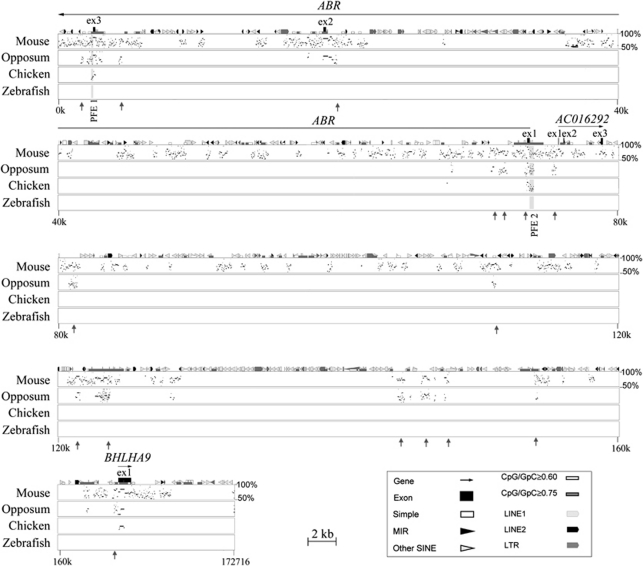
The overlap of the three microduplications is defined by genomic co-ordinates chr 17: 956 201–1 128 916 in the NCBI36/hg18 human genome assembly. We performed cross-species comparison to identify regions of evolutionary conservation that likely harbour DNA elements with important biological functions. This region contains BHLHA9, a highly conserved novel protein-coding gene identified by virtual library screening, AC016292.1, a putative processed transcript, and 3 of the 24 coding exons of ABR (Figure 4). We screened for unannotated regions of conservation between human, mouse, and opossum that extend beyond 150 bp. We identified 18 regions that exhibit more than 50% of sequence identity across all the mammalian species examined and may signify uncharacterized functional elements. Two of the identified putative functional elements (PFE1 and PFE2) exhibit over 75% sequence identity in the mammalian and avian species examined, but given their locations these may represent regulators of ABR. The other 16 regions are conserved in human, mouse, and opossum, but are not present in chicken. Some of these regions may correspond to yet unidentified functional elements that evolved after the separation of the mammalian and avian lineages (Figure 4).
Figure 4.
Cross-species comparative sequence analysis of the SHFLD critical region. The graph compares the percent identity between human genomic sequence of 173 kb with corresponding sequences from mouse, opossum, chicken, and zebrafish.18 Aligned segments that show over 50% percent of identity are plotted as a series of dots (stippling) according to their length and position within the human sequence. The human sequence is shown on the horizontal axis and the percent identities with non-human sequences are shown on the vertical axis. Annotations correspond to the human sequence (top of plot). Gene sequences are represented by solid-black arrows indicating the direction of transcription, and exons are indicated by black boxes. ABR and BHLHA9 are known protein-coding genes; AC016292.1 is a putative processed transcript. Shaded regions depict putative functional elements, PFE1 and PFE2, which are conserved between human, mouse, opossum, and chicken. Grey vertical arrows depict regions of conservation between human, mouse, and chicken. The analysis also detects and annotates various repetitive sequences in the reference sequence (human). These include simple repeats, long terminal repeats (LTRs), mammalian interspersed elements (MIRs), long interspersed nuclear elements (LINEs), and short interspersed nuclear elements (SINEs). Such sequences are commonly present in multiple copies per genome and are excluded from sequence comparisons. For further details on PIP analyses, see http://bio.cse.psu.edu/pipmaker.
Discussion
SNP array copy-number variation (CNV) analysis has become a useful tool for identifying duplications associated with rare disorders (reviewed in Berg et al23 and Vissers et al24). Here we report three overlapping microduplications within the previously defined locus for SHFLD,13 thereby significantly narrowing the SHFLD critical region to 173 kb at 17p13.3. The possibility of a microtriplication in family 3 could not be completely excluded based on the available data. Both microduplications and microdeletions involving 17p13.3 manifest with a specific but variable phenotype including autism, developmental delay, facial dysmorphism, and hand/foot abnormalities (reviewed in Bruno et al25). Although the majority of the reported microduplications25, 26, 27 overlap our 173 kb region, the 72 kb critical region necessary for the non-SHFLD phenotype is just centromeric to the SHFLD duplication. Bruno et al25 suggest that YWHAE (encoding a 14-3-3e –protein) is critical to the neurobehavioural phenotype. Only a portion of YHWAE, five of six total exons, is included in the duplication found in family 1. The lack of complete YHWAE duplication may explain why none of the individuals in this family have any apparent neurobehavioural phenotype. The hand/foot anomalies in the patients reported and reviewed by Bruno et al25 are generally mild and not in the SHFM spectrum; however, one patient is described as having a bilateral groove between the first and second toes and two other patients are described as having ‘sandal gaps'. CRK is duplicated in all of their patients who had limb anomalies and was postulated to be a candidate gene for these findings. None of the microduplications described herein involves CRK.
Chromosomal rearrangements are a well-documented cause of SHFM and other skeletal disorders.6, 7, 9, 28, 29, 30 Similar to the findings reported herein, the primary genetic findings in patients with SHFM3 are genomic duplications. Those duplications involve a minimal 325 kb region that includes BTRC, POLL, and a portion of the FBXW4 (human dactylin) gene. In the SHFM3 ‘like' dactylaplasia mouse model, a genomic rearrangement leads to a reduced level of a normal Fbxw4 transcript.31 However, in human cell lines from individuals with SHFM3, two genes BTRC and SUFU, show increased expression with normal FBXW4 expression.8 Lyle et al8 suggest that the genomic rearrangement in the area disrupts normal expression pattern of genes through alteration of long-range control mechanisms, but the exact mechanism causing SHFM is unclear.
The SHFLD arising from the duplicated 173 kb critical region described herein may occur secondary to a direct effect on a gene contained within the duplicated region. This region contains exons 1–3 of ABR, BHLHA9 and a putative processed transcript AC016292.1. ABR contains a GTPase-activating protein domain,32 and functional studies in mice suggest that the protein has a role in vestibular morphogenesis.33 Public databases report a number of rare CNVs, both deletions and duplications, the majority of which are more distal to exon 1 of ABR. However, ABR may possess an as yet unidentified function relevant to SHFLD and the presence of genomic variants in the reference population does not absolutely rule out ABR as a potential candidate. BHLHA9 encodes a basic helix-loop-helix protein and exhibits >75% of sequence identity in pairwise comparisons between human, opossum, and chicken (Figure 4). However, little is known about its function.
Genetic alterations affecting cis-acting regulatory elements are a known cause of limb malformation syndromes. Duplications of regulatory elements have been shown to be responsible for at least three different limb phenotypes; a tandem duplication of a 5.5 kb region 3′ of BMP2 is associated with brachydactyly type A230 and microduplications of the ZRS, a long-range cis-acting regulatory element of sonic hedgehog (SHH), lead to triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV.28, 29 Within the duplicated region described here, evolutionarily conserved sequences of unknown function (Figure 4) may represent cis-acting regulatory elements altering the activity of a neighbouring gene, or the duplications may disrupt the interaction between a nearby enhancer and its target gene. One possible candidate gene, CRK, seems to be involved in several signalling pathways and is thought to be necessary for normal embryonic development and possibly normal skeletal development.34 A more promising candidate is NXN, which is located telomeric to the critical region and functions as a negative regulator of Wnt via dishevelled (Dvl); in Xenopus altered expression of NXN leads to a bent body phenotype.35 Thus, future experiments may include examining transcript levels of putative downstream target genes, obtaining functional data from in vivo cellular reporter assays, and altering any regulatory elements or genes in animal models.
As the individuals described in Bruno et al25 with hand/foot anomalies had microduplications that extended across our critical region, their limb anomalies may represent a subtle skeletal phenotype that, at the most extreme, can manifest as SHFLD. Given the decreased penetrance seen with the microduplications, there is also the possibility that a second genetic locus is involved. Such a mechanism might explain why other duplications that overlap the SHFLD critical region defined herein do not result in SHFLD. We could not account for the disease severity or incomplete penetrance by segregation of the microsatellites in those who possess the duplication in family 1, suggesting that if a second locus exists, it is not in the region covered by the microsatellites examined. Alternatively, it may be that the duplication must involve a shorter length of critical sequence in order to cause the abnormal limb phenotype. Large duplications that involve the terminal band of chromosome 7q, which contains both the ZRS and SHH, cause neither preaxial polydactyly nor syndactyly, as seen with microduplications of the ZRS.36 If SHFLD only occurs when an enhancer, but not its target gene, is present in a double dose on a single chromosome, then a larger duplication involving both the enhancer and target gene would not produce an abnormal limb. Similarly, SHFLD might only occur when the duplication resides between a limb enhancer and its target gene, such that the two sequences become separated by a critical distance that affects their ability to interact. In this case, the larger duplications described by Bruno et al might preserve the normal physical relationship between the enhancer and its target gene, and thus not lead to SHFLD.
In summary we present three independent overlapping microduplications, which suggest that at least one molecular mechanism underlying SHFLD involves the 173 kb critical region described herein. Based on current understanding of the molecular mechanisms underlying other limb anomalies associated with duplications, we hypothesise that duplication of this critical region either alters the dosage of a regulatory element involved in limb development or disrupts the interaction between a nearby regulatory element and its target gene(s). Of the 18 conserved regions identified, two exhibit over 75% sequence identity in mammalian and avian species (Figure 4, PFE1 and PFE2), whereas the other 16 are conserved in human, mouse, and opossum (Figure 4, arrows). Some of these may represent regulatory elements, the function of which may be altered by the SHFLD duplications. Alternatively, duplication of BHLHA9 or AC016292 may cause SHFLD. Further studies that include functional analysis of the duplicated region, as well as analyses of other SHFLD/SHFM cases for similar duplications, will hopefully clarify the molecular pathogenesis of this disorder.
Acknowledgments
We wish to acknowledge the participation of the families, without whom this work would not have been possible. We also thank Cindy Skinner, RN for assistance with sample coordination. This work was funded by a grant from the Children's Hospital of Eastern Ontario Research Institute, the Clinical Teachers' Association of Queen's University, and support from the South Carolina Birth Defects Foundation, the Genetics Endowment of South Carolina, and the South Carolina Department of Disabilities and Special Needs. OJ holds HSF/Pfizer Postdoctoral fellowship.
Note added in proof
The nonpenetrant mother with the duplication in Family 2 has bilateral hearing loss and has worn hearing aids since 3 years of age.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Majewski F, Kuster W, ter Haar B, Goecke T. Aplasia of tibia with split-hand/split-foot deformity. Report of six families with 35 cases and considerations about variability and penetrance. Hum Genet. 1985;70:136–147. doi: 10.1007/BF00273072. [DOI] [PubMed] [Google Scholar]
- Basel D, Kilpatrick MW, Tsipouras P. The expanding panorama of split hand foot malformation. Am J Med Genet A. 2006;140:1359–1365. doi: 10.1002/ajmg.a.31304. [DOI] [PubMed] [Google Scholar]
- Ugur SA, Tolun A. Homozygous WNT10b mutation and complex inheritance in split-Hand/Foot malformation. Hum Mol Genet. 2008;17:2644–2653. doi: 10.1093/hmg/ddn164. [DOI] [PubMed] [Google Scholar]
- Celli J, Duijf P, Hamel BC, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–153. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- Kjaer KW, Hansen L, Schwabe GC, et al. Distinct CDH3 mutations cause ectodermal dysplasia, ectrodactyly, macular dystrophy (EEM syndrome) J Med Genet. 2005;42:292–298. doi: 10.1136/jmg.2004.027821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, Poorkaj P, Massa H, et al. Physical mapping of the split hand/split foot locus on chromosome 7 and implication in syndromic ectrodactyly. Hum Mol Genet. 1994;3:1345–1354. doi: 10.1093/hmg/3.8.1345. [DOI] [PubMed] [Google Scholar]
- de Mollerat XJ, Gurrieri F, Morgan CT, et al. A genomic rearrangement resulting in a tandem duplication is associated with split hand-split foot malformation 3 (SHFM3) at 10q24. Hum Mol Genet. 2003;12:1959–1971. doi: 10.1093/hmg/ddg212. [DOI] [PubMed] [Google Scholar]
- Lyle R, Radhakrishna U, Blouin JL, et al. Split-hand/split-foot malformation 3 (SHFM3) at 10q24, development of rapid diagnostic methods and gene expression from the region. Am J Med Genet A. 2006;140:1384–1395. doi: 10.1002/ajmg.a.31247. [DOI] [PubMed] [Google Scholar]
- Goodman FR, Majewski F, Collins AL, Scambler PJ. A 117-kb microdeletion removing HOXD9-HOXD13 and EVX2 causes synpolydactyly. Am J Hum Genet. 2002;70:547–555. doi: 10.1086/338921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbs C, Heller R, Everman DB, et al. A new locus for split hand/foot malformation with long bone deficiency (SHFLD) at 2q14.2 identified from a chromosome translocation. Hum Genet. 2007;122:191–199. doi: 10.1007/s00439-007-0390-7. [DOI] [PubMed] [Google Scholar]
- Naveed M, Al-Ali MT, Murthy SK, et al. Ectrodactyly with aplasia of long bones (OMIM; 119 100) in a large inbred arab family with an apparent autosomal dominant inheritance and reduced penetrance: Clinical and genetic analysis. Am J Med Genet A. 2006;140:1440–1446. doi: 10.1002/ajmg.a.31239. [DOI] [PubMed] [Google Scholar]
- Naveed M, Nath SK, Gaines M, et al. Genomewide linkage scan for split-hand/foot malformation with long-bone deficiency in a large arab family identifies two novel susceptibility loci on chromosomes 1q42.2-q43 and 6q14.1. Am J Hum Genet. 2007;80:105–111. doi: 10.1086/510724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezirovitz K, Maestrelli SR, Cotrim NH, Otto PA, Pearson PL, Mingroni-Netto RC. A novel locus for split-hand/foot malformation associated with tibial hemimelia (SHFLD syndrome) maps to chromosome region 17p13.1-17p13.3. Hum Genet. 2008;123:625–631. doi: 10.1007/s00439-008-0515-7. [DOI] [PubMed] [Google Scholar]
- Schwartz CE, Ulmer J, Brown A, Pancoast I, Goodman HO, Stevenson RE. Allan-herndon syndrome. II. linkage to DNA markers in Xq21. Am J Hum Genet. 1990;47:454–458. [PMC free article] [PubMed] [Google Scholar]
- Barch MJ, Knutsen T, Spurbeck JL.(eds).: Cytogenetics Laboratory Manual3rd edn.New York: Lippincott-Raven; 1997127–130. [Google Scholar]
- Armour CM, McCready ME, Baig A, Hunter AG, Bulman DE. A novel locus for brachydactyly type A1 on chromosome 5p13.3-p13.2. J Med Genet. 2002;39:186–188. doi: 10.1136/jmg.39.3.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, et al. PipMaker – a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarinova O, Hatch G, Poitras L, et al. Functional resolution of duplicated hoxb5 genes in teleosts. Development. 2008;135:3543–3553. doi: 10.1242/dev.025817. [DOI] [PubMed] [Google Scholar]
- Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet. 2000;67:59–66. doi: 10.1086/302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne T, Hamel B, van Bokhoven H, Brunner HG. Pattern of p63 mutations and their phenotypes – update. Am J Med Genet A. 2006;140:1396–1406. doi: 10.1002/ajmg.a.31271. [DOI] [PubMed] [Google Scholar]
- Agarwala R, Schaffer AA, Tomlin JF. Towards a complete North American Anabaptist genealogy II: Analysis of inbreeding. Hum Biol. 2001;73:533–545. doi: 10.1353/hub.2001.0045. [DOI] [PubMed] [Google Scholar]
- Berg JS, Potocki L, Bacino CA. Common recurrent microduplication syndromes: Diagnosis and management in clinical practice. Am J Med Genet A. 2010;152A:1066–1078. doi: 10.1002/ajmg.a.33185. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Vries BB, Veltman JA. Genomic microarrays in mental retardation: From copy number variation to gene, from research to diagnosis. J Med Genet. 2010;47:289–297. doi: 10.1136/jmg.2009.072942. [DOI] [PubMed] [Google Scholar]
- Bruno DL, Anderlid BM, Lindstrand A, et al. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J Med Genet. 2010;47:299–311. doi: 10.1136/jmg.2009.069906. [DOI] [PubMed] [Google Scholar]
- Bi W, Sapir T, Shchelochkov OA, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos L, Jonch AE, Kjaergaard S, et al. A new microduplication syndrome encompassing the region of the Miller-Dieker (17p13 deletion) syndrome. J Med Genet. 2009;46:703–710. doi: 10.1136/jmg.2008.065094. [DOI] [PubMed] [Google Scholar]
- Klopocki E, Ott CE, Benatar N, Ullmann R, Mundlos S, Lehmann K. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J Med Genet. 2008;45:370–375. doi: 10.1136/jmg.2007.055699. [DOI] [PubMed] [Google Scholar]
- Sun M, Ma F, Zeng X, et al. Triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV are caused by genomic duplications involving the long range, limb-specific SHH enhancer. J Med Genet. 2008;45:589–595. doi: 10.1136/jmg.2008.057646. [DOI] [PubMed] [Google Scholar]
- Dathe K, Kjaer KW, Brehm A, et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am J Hum Genet. 2009;84:483–492. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidow A, Bulotsky MS, Kerrebrock AW, et al. A novel member of the F-box/WD40 gene family, encoding dactylin, is disrupted in the mouse dactylaplasia mutant. Nat Genet. 1999;23:104–107. doi: 10.1038/12709. [DOI] [PubMed] [Google Scholar]
- Chuang TH, Xu X, Kaartinen V, Heisterkamp N, Groffen J, Bokoch GM. Abr and bcr are multifunctional regulators of the rho GTP-binding protein family. Proc Natl Acad Sci USA. 1995;92:10282–10286. doi: 10.1073/pnas.92.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen V, Nagy A, Gonzalez-Gomez I, Groffen J, Heisterkamp N. Vestibular dysgenesis in mice lacking abr and bcr Cdc42/RacGAPs. Dev Dyn. 2002;223:517–525. doi: 10.1002/dvdy.10071. [DOI] [PubMed] [Google Scholar]
- Park TJ, Boyd K, Curran T. Cardiovascular and craniofacial defects in crk-null mice. Mol Cell Biol. 2006;26:6272–6282. doi: 10.1128/MCB.00472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato Y, Michiue T, Terabayashi T, et al. Nucleoredoxin regulates the Wnt/planar cell polarity pathway in xenopus. Genes Cells. 2008;13:965–975. doi: 10.1111/j.1365-2443.2008.01220.x. [DOI] [PubMed] [Google Scholar]
- Verma RS, Conte RA, Pitter JH. Tandem duplication of the terminal band of the long arm of chromosome 7 (dir dup (7)(q36----qter)) J Med Genet. 1992;29:344–345. doi: 10.1136/jmg.29.5.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, et al. The UCSC genome browser database: Update 2010. Nucleic Acids Res. 2010;38 (Database issue:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.