Abstract
The evolution of the nucleus, the defining feature of eukaryotic cells, was long shrouded in speculation and mystery. There is now strong evidence that nuclear pore complexes (NPCs) and nuclear membranes coevolved with the endomembrane system, and that the last eukaryotic common ancestor (LECA) had fully functional NPCs. Recent studies have identified many components of the nuclear envelope in living Opisthokonts, the eukaryotic supergroup that includes fungi and metazoan animals. These components include diverse chromatin-binding membrane proteins, and membrane proteins with adhesive lumenal domains that may have contributed to the evolution of nuclear membrane architecture. Further discoveries about the nucleoskeleton suggest that the evolution of nuclear structure was tightly coupled to genome partitioning during mitosis.
Introduction
The nucleus, a double membrane–bound compartment that contains the nuclear genome, is the quintessential morphological and functional feature of eukaryotes (Wilson and Berk, 2010). Besides chromatin, the most prominent structure of the nucleus is the nuclear envelope (NE): two bordering membranes with huge nuclear pore complexes (NPCs) that allow molecules to enter and exit the nucleus (Strambio-De-Castillia et al., 2010). Another obvious feature is the nucleolus, which is the site of rDNA gene expression and ribosome assembly (Németh and Längst, 2011). Less obvious, hence recognized only recently, is the dynamic and complex internal architecture of the nucleus, conceptually termed the nucleoskeleton, which includes intermediate filaments, actin and titin, and also functions during mitosis (Simon and Wilson, 2011). How did this structural complexity arise?
The small subunit RNA (SSU)–based phylogenetic “tree of life” points to three domains—Bacteria, Archaea, and Eucarya (Woese et al., 1990)—all of which have extremely deep origins (Pace, 2009), and share overlapping sets of genes. Did these three lineages arise independently from the precellular phase of biological evolution (Pace, 2009), or did the eukaryotic precursor arise by merger of bacterial and archaeal cells? The latter possibility is attractive given the compelling genomic evidence for two primary symbiotic events: the endosymbiosis of an alphaproteobacterium that ultimately gave rise to mitochondria, and the endosymbiosis of a cyanobacterium that gave rise to chloroplasts (Margulis, 1970; Pace, 2009). There is also strong phylogenetic and genomic evidence for secondary and tertiary endosymbiosis of plastids in some eukaryotic lineages (Palmer and Delwiche, 1996). However, in contrast to mitochondria, chloroplasts, and plastids, evidence for the involvement of endosymbiosis in the evolution of the nucleus is sparse or missing.
The early evolution of the eukaryotic lineage remains murky, in large part because the genetic diversity of extant—particularly single-celled—eukaryotes remains unclear (Dawson and Pace, 2002). Indeed, the greatest genetic diversity is seen among microbial (single-celled) eukaryotes (Sogin and Silberman, 1998). However as Pace (2009) pointed out, genome sequence comparisons of living eukaryotes provide “no evidence whatsoever” as to whether the earliest eukaryotes actually had nuclear membranes or NPCs as morphological features. This simple idea, that the first eukaryotic common ancestor (FECA) lacked nuclear morphology, frees one to consider how specific types of proteins in the FECA might have contributed to the subsequent incremental evolution of nuclear structure present in the last common eukaryotic ancestor (LECA; Fig. 1). As discussed in this review, new evidence based on the ancestral nature of endomembrane proteins suggests that the eukaryotic endomembrane system coevolved with, or spawned, the nuclear membranes and NPCs, which appear to have been fully functional in the LECA (Neumann et al., 2010).
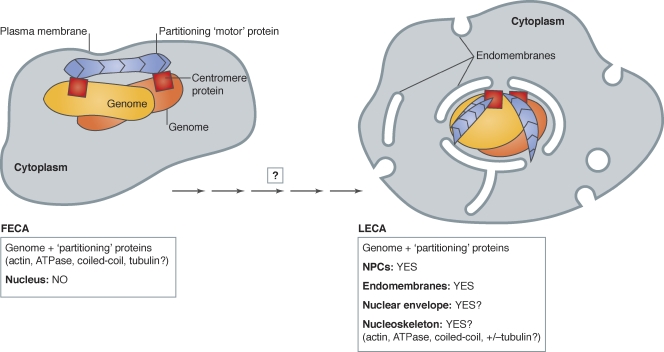
Figure 1.
Proposed incremental transition from FECA (no nuclear structure) to LECA (nucleus). The first eukaryotic common ancestor (FECA) is proposed to have lacked nuclear structure. Partitioning of the duplicated genome (yellow/orange) is proposed to be mediated by the polymerization of protein(s) related to bacterial par “motors” (blue; e.g., actin; ATPase; tubulin; DNA-binding coiled-coil protein), bound to centromere proteins (red squares). Over significant time, the FECA is proposed to have given rise to the last eukaryotic common ancestor (LECA), a cell with fully functional NPCs (not depicted) and endomembranes (Neumann et al., 2010) and, we suggest, a nucleoskeleton that included components involved in genome partitioning. After the LECA, further evolution of nuclear structure followed different pathways as seen in the six living eukaryotic supergroups (Hampl et al., 2009; see Fig. 3).
The LECA subsequently gave rise to six major eukaryotic supergroups, each of which includes microbial eukaryotes: Opisthokonts (e.g., fungi, animals, protists), Amoebozoa (e.g., Dictyostelium), Excavates (e.g., Trypanosomes, Giardia), Chromoalveolates (e.g., Plasmodium), Archaeplastids (e.g., plants), and Rhizaria (Hampl et al., 2009). Though the most basal branches are somewhat controversial (Rogozin et al., 2009; Parfrey et al., 2010), this classification system allows one to compare the genomes of diverse eukaryotes within each supergroup and create inventories of genes encoding known nuclear structure proteins. Comparisons between supergroups can then, in theory, identify core genes inferred as present in the LECA (Keeling, 2007). However, this approach is currently limited by the lack of annotation of most nucleoskeletal proteins (Simon and Wilson, 2011), and by lack of knowledge about the nuclear membrane protein components in most eukaryotes.
The immense diversity of microbial eukaryotes is not yet reflected in completed genome projects (Dawson and Fritz-Laylin 2009), which focused overwhelmingly (over 80%) on the Opisthokont (particularly animals and fungi, which lack many genes [“secondarily reduced” genomes]) and Archaeplastid (plant) lineages. Similarly, most functional knowledge about nuclear structure comes from model systems (animals, fungi, plants) that represent only two of the six eukaryotic supergroups. More sequenced genomes, and nuclear envelope proteomes, from other eukaryotic supergroups will be crucial to understand how nuclei evolved. All eukaryotic lineages are characterized by the loss, gain, expansion, and diversification of gene families (Fritz-Laylin et al., 2010). Thus, the history of nuclear structure after the LECA undoubtedly followed many different paths in the six major eukaryotic lineages. Understanding these differences, and shared features, would give unprecedented insight into the most fundamental aspects of nuclear structure and genome organization, and might also suggest therapeutic molecular targets in parasitic eukaryotes.
Co-evolution of NPCs and endomembranes: The proto-coatomer hypothesis
Two decades of intensive investigation have yielded a wealth of information about the NPC including its ∼30 constituent proteins (nucleoporins) and their stoichiometry, biochemistry, assembly, and three-dimensional positions within the NPC (Doucet and Hetzer, 2010; Fichtman et al., 2010; Wente and Rout, 2010). This knowledge includes the functions of specific folded domains within each nucleoporin (Devos et al., 2006). Remarkably, the components and structure of one NPC subcomplex (vertebrate Nup107-160 complex) resemble the membrane-bending protein coats that generate vesicles in the secretory and endomembrane pathways (Fig. 2; Devos et al., 2004). This stunning finding led to the proto-coatomer hypothesis, which suggests that both structures evolved from an ancestral membrane-curving protein(s) (Fig. 2; Devos et al., 2004; Hsia et al., 2007; Debler et al., 2008; Leksa and Schwartz, 2010).
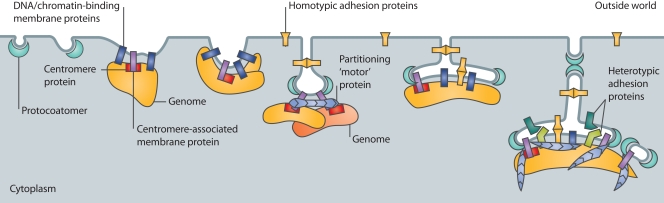
Figure 2.
Proposed contributions of membrane proteins to the evolution of nuclear structure. Different types of proteins are proposed to have contributed to the incremental evolution of nuclear structure including soluble proto-coatomer proteins (aqua), DNA- or chromatin-binding membrane proteins (navy blue), centromere- or par system–associated membrane proteins (light purple), homotypic membrane “adhesion” proteins (yellow), and heterotypic membrane adhesion proteins (teal and light green). Blue indicates partitioning proteins (actin, ATPase, DNA-binding coiled-coil protein, tubulin), many of which are components of the nucleoskeleton in living Opisthokonts (see text).
To test the proto-coatomer hypothesis, NPC proteins were purified from the divergent basal Excavate eukaryote Trypanosoma brucei, a major human pathogen. Detailed functional knowledge about specific folded polypeptide domains was crucial to identifying Trypanosome nucleoporins because the corresponding Trypanosome genes were unrecognizable by DNA sequence and amino acid comparisons alone (DeGrasse et al., 2009). The Trypanosome NPC proteome suggests conservation of NPC proteins and NPC architecture in the LECA, and supports the proto-coatomer hypothesis (DeGrasse et al., 2009). This hypothesis was significantly extended by an analysis of 60 eukaryotic genomes representing five supergroups, which placed at least 23 and as many as 26 (out of 30) nucleoporins in the LECA (Neumann et al., 2010). This conclusion was not affected by the position of the eukaryotic root. Among five known transmembrane nucleoporins, two (gp210, Ndc1) were identified as key components that anchor NPCs to the membrane in all five supergroups. Also conserved in all five supergroups were NPC “basket” nucleoporins Tpr and Nup50; the third basket protein, Nup153, which in vertebrates binds lamins directly (Smythe et al., 2000), was conserved in four out of five supergroups (Neumann et al., 2010). These conserved proteins, which were likely present in the LECA, have implications beyond NPC structure and function; as discussed in the next section, Tpr and Nup153 also have functions related to chromatin and gene expression.
Which other nuclear structural proteins were present in the LECA?
The presence of apparently functional NPCs in the LECA raises an intriguing question: did this ancestral nucleus have other nuclear structural proteins, and if so, which ones? To answer this question one needs clues about which proteins to look for in diverse genomes. Fortunately, many proteins that might have contributed to the evolution of nuclear structure have emerged from functional studies in Opisthokonts and plants. These proteins of interest include growing numbers of nuclear membrane proteins, which are discussed next, and functionally diverse nucleoskeletal proteins including actin, molecular motors, spectrin repeat proteins, coiled-coil proteins, and nuclear pore complex linked filaments (Simon and Wilson, 2011), which are discussed later in this review.
Nuclear membrane proteins: Uncharted territory
Mammals encode large repertoires (likely hundreds) of uncharacterized nuclear envelope transmembrane (NET) proteins (Wilson and Berk, 2010). This unexpected complexity was first revealed in a landmark proteomic study that identified over 60 different NET proteins in purified rat liver cell nuclear envelopes (Schirmer et al., 2003), and was confirmed and extended by studies in other mammalian cells (Wilkie et al., 2011). Most nuclear membrane proteins in Opisthokonts are either uncharacterized, or are not yet understood at the level of structural or functional detail that may be needed to identify orthologous genes in diverse eukaryotes. To bypass this problem, at least in part, one could determine the NET proteomes of diverse eukaryotes, and thereby identify specific relevant genes. Further analysis of conserved NET proteins, even in other Opisthokonts, is yielding surprises about nuclear structure. For example, the fungus Schizosaccharomyces pombe encodes a nuclear inner membrane protein named Ima1, identified as a potential analogue of mammalian NET5 (King et al., 2008). Functional studies show that Ima1 attaches heterochromatin to the NE and (through unknown connections) to microtubules (King et al., 2008). The Ima1 protein binds directly to centromeres and telomeres, and its properties suggest that heterochromatin provides a mechanical “nut” that reinforces the NE against forces generated by microtubules (King et al., 2008). This is consistent with biomechanical evidence that heterochromatin itself functions as a force-bearing structure (Dahl et al., 2005). Nuclear proteins that either mechanically reinforced chromatin, or protected chromatin from force, might have influenced the capacity of cells not only to survive external mechanical challenges, but also to exert force on the outside world (Dahl et al., 2008). Thus, the future characterization of conserved NET proteins has the potential to reveal new aspects of nuclear structure in living eukaryotes, as well as new principles about how this structure evolved.
Sticky membrane proteins help stabilize the envelope.
We propose that membrane proteins with “adhesive” extracellular domains contributed to the evolution of nuclear structure by stabilizing the parallel organization of curved or infolded plasma membranes (Fig. 2). This idea is based on the discovery of Opisthokont nuclear membrane proteins that mediate adhesion either homotypically (between two or more copies of one protein) or heterotypically (between distinct proteins). Note that the lumenal domains of membrane proteins face a compartment that is topologically equivalent to the outside of the cell. The lumenal domains of yeast transmembrane protein Pom152 (potential vertebrate orthologue: gp210) are predicted to self-interact via a cadherin fold (Devos et al., 2006). This fold is characteristic of the extracellular domains of certain cell surface proteins (e.g., cadherins) and mediates homotypic adhesion to neighboring cells (Franke, 2009). Although cadherin fold–containing nuclear membrane proteins have so far been seen only in Opisthokonts, other types of adhesive domains are widely conserved. For example, Pom121, a conserved NPC membrane protein, is recruited to chromatin by the DNA-binding nucleoporin ELYS and the Nup107-160 complex (Lau et al., 2009). These interactions somehow trigger adhesive Pom121-mediated fusion of parallel membranes to create new pores (Fichtman et al., 2010). In addition to this evidence for homotypic adhesion, there is growing evidence that certain nuclear membrane proteins mediate adhesion heterotypically.
At least three types of NE membrane proteins are required for the parallel organization of the inner and outer nuclear membranes. One family consists of the inner membrane protein lamina-associated polypeptide 1 (LAP1) and a related outer membrane protein, LULL1, which interact via their large lumenal domains in conjunction with soluble lumenal proteins named torsinA and torsinB (Nery et al., 2008; Vander Heyden et al., 2009; Kim et al., 2010). Because LAP1 and LULL1 are related, their adhesion might be considered homotypic. By contrast, two other adhesive families, consisting of KASH domain proteins and SUN domain proteins, interact heterotypically via their lumenal domains (Crisp et al., 2006; Starr and Fridolfsson, 2010). KASH and SUN domain proteins also have additional domains that mediate either self-interaction or direct binding to specific cytoskeletal proteins, nucleoskeletal proteins, or NE membrane proteins (Wilson and Berk, 2010). Although the experimental picture is far from complete, current evidence suggests KASH domain and SUN domain proteins form a variety of mechanically robust adhesion complexes at the NE, termed LINC complexes (links the nucleoskeleton and cytoskeleton; Crisp et al., 2006), some of which are essential for chromosome pairing during meiosis and sexual recombination (Fridkin et al., 2009; Hiraoka and Dernburg, 2009). In our limited search, the SUN domain was detected in every tested supergroup (Fig. 3, Table I). This strongly suggests the LECA had a SUN domain protein. By contrast, we found the KASH domain only in Opisthokonts (Fig. 3, Table I). The potential presence of a KASH domain protein in the LECA cannot presently be ruled out. However, if SUN domain proteins are indeed more ancient, they might also serve ancient (KASH-independent) roles. These roles might involve NPCs (Liu et al., 2007) because SUN1 has an early role in NPC assembly (Talamas and Hetzer, 2011). SUN domain proteins also interact with meiotic telomeres, histone H2A.Z, and couple the nucleus to the microtubule-organizing center (Hiraoka and Dernburg, 2009; Gardner et al., 2011). These findings support the idea that membrane proteins, including adhesive membrane proteins, influenced the evolution of nuclear structure.
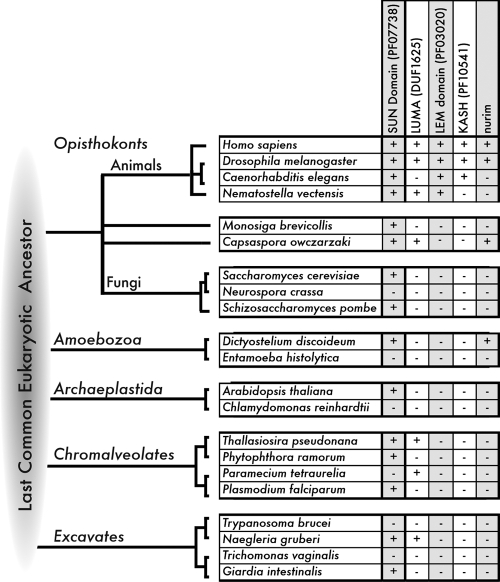
Figure 3.
Identifying nuclear membrane proteins in diverse eukaryotes. Representatives of five eukaryotic supergroup lineages, queried by BLAST search for potential conservation of open reading frames (ORFs) related to Opisthokont nuclear membrane proteins. Chart indicates whether an ORF(s) homologous to each queried polypeptide was detected (+) or not detected (−) in completed genomes of representative supergroup lineages (Opisthokonts, Amoebozoa, Archaeplastida, Chromalveolates, and Excavates) in GenBank using reciprocal BLAST with e-values > 0.01, as detailed in Table I. ORFs homologous to LUMA and nurim are also present in various bacterial lineages (not depicted). The specific evolutionary relationships between each supergroup and the last eukaryotic common ancestor (LECA) are controversial, as reflected by the lack of specific branching order between these groups. However, the detection of a nuclear-associated homologue (e.g., SUN domain, or LUMA) in more than two supergroups might suggest the homologue was present in the LECA. Negative results are not definitive; for example, we did not recover the S. cerevisiae LEM (“HEH”) domain protein Src1 (see next page). The species M. brevicollis and C. owczarsaki represent the Protist lineage within Opisthokonts and are distantly related to Fungi and Animals.
Table I.
ORFs in diverse eukaryotes that possess conserved nuclear membrane protein domains
Search results for selected nuclear protein ORFs in representatives of five eukaryotic supergroup lineages, listing genes or ORFs homologous to each queried polypeptide that were detected in completed genomes of representative supergroup lineages (Opisthokonts, Amoebozoa, Archaeplastida, Chromalveolates, and Excavates) in GenBank using reciprocal BLAST with e-values > 0.01. This method detected ORFs representing four of the seven known human LEM domain genes, and is therefore quite stringent. Multiple ORFs, for example LUMA-related ORFs in Paramecium, were not annotated so the number of encoding gene(s) remains to be determined.
Membrane proteins that bind chromatin or nucleoskeletal partner(s).
The evolution of nuclear structure may have been strongly influenced by membrane proteins capable of binding to DNA or chromatin proteins (Wilson and Foisner, 2010). One such family of proteins in metazoans has a characteristic “LEM domain” fold, first identified in LAP2, emerin, and MAN1 (Lin et al., 2000; Laguri et al., 2001; Wagner and Krohne, 2007). In fact, the first of two LEM domains in LAP2 confers direct binding to dsDNA (Cai et al., 2001). Other tested LEM domains confer binding to barrier-to-autointegration factor (BAF), a conserved metazoan protein that also binds directly to dsDNA, histone H3, and lamins (Margalit et al., 2007; Montes de Oca et al., 2009), and influences histone posttranslational modifications (Montes de Oca et al., 2011). LEM domain proteins have other domains that bind one or more components of the nucleoskeleton, or various signaling or gene-regulatory proteins (Wagner and Krohne, 2007; Wilson and Berk, 2010). The LEM domain protein emerin also binds KASH domain and SUN domain proteins directly (Simon and Wilson, 2011), and somehow couples mechanical force to downstream changes in gene expression, a phenomenon known as mechanotransduction (Lammerding et al., 2005). Vertebrate nuclei also express a specialized nonmembrane LEM domain protein named LAP2α that interacts with itself (as trimers), lamin A, chromatin, and telomeres, and is required to organize A-type lamins in the nuclear interior (Snyers et al., 2007; Gotic and Foisner, 2010; Dechat et al., 2011). Yeast, which lack lamins and BAF, nevertheless encode a LEM domain (“HEH” domain) inner nuclear membrane protein named Src1, which associates with and represses telomeric, subtelomeric, and rDNA genes (Grund et al., 2008). Interestingly, Src1 also functions during mitosis; cells that lack Src1 have shorter anaphase and longer telophase (Rodríguez-Navarro et al., 2002). Both the LEM domain fold (Cai et al., 2001) and a conserved C-terminal “MSC” (MAN1–Src1p–C-terminal) domain shared by Src1 and human MAN1 (Mans et al., 2004) are conserved in bacteria and may function to bind nucleic acids. Among eukaryotes, our limited search detected LEM domain–related ORFs only in Opisthokonts (Fig. 3, Table I). However, previous more extensive alignments found proteins with both features (LEM/HEH domain; MSC domain) in all tested eukaryotic supergroups, suggesting the LECA had a LEM domain protein (Mans et al., 2004).
The nurim protein has four membrane-spanning domains (and little else). This protein localizes at the nuclear inner membrane through unknown mechanisms because it shows no detectable binding to NPCs, lamins, or other intranuclear components (Rolls et al., 1999). Nurim is proposed to function in a pathway that sorts newly synthesized nuclear membrane proteins past the NPC to the inner membrane (King et al., 2006; Braunagel et al., 2007). Our search revealed nurim-related ORFs in two eukaryotic supergroups (Fig. 3). Suggesting potentially ancient origins, a previous study (Mans et al., 2004) grouped nurim in a protein superfamily that includes the mammalian inner nuclear membrane protein LBR (a sterol reductase; Holmer et al., 1998) and related enzymes in bacteria.
The LUMA protein (encoded by TMEM43) crosses the nuclear inner membrane four times, has a large uncharacterized lumenal domain, associates with SUN2, lamins, and emerin, and forms homo-oligomers (Bengtsson and Otto, 2008; Liang et al., 2011). LUMA oligomerization is disrupted by a mutation that causes Emery-Dreifuss muscular dystrophy (Liang et al., 2011). LUMA is conserved in three eukaryotic supergroups (Fig. 3) and notably also in bacteria (Bengtsson and Otto, 2008), suggesting LUMA might have been present in the LECA. The idea that LUMA, LEM domain proteins, nurim, and likely other nuclear membrane proteins have potentially ancient roles in nuclear structure and function will make it even more interesting to decipher their roles in living eukaryotes.
Actin and myosins: Ancient components of nuclear structure?
Eukaryotic actin and actin-dependent motors (myosins) are well known cytoskeletal components relevant to cell motility, and their evolutionary significance is nearly always discussed exclusively in these terms (Fritz-Laylin et al., 2010). Less appreciated are their fundamental and potentially ancient roles in nuclear structure and genome function. Polymerizable actin and myosins are involved in transcription by all three DNA-dependent RNA polymerases, mediate RNA export from the nucleus, and are required for the long-range movement of specific loci within the nucleus (Gieni and Hendzel, 2009; Hofmann, 2009; Mekhail and Moazed, 2010; Skarp and Vartiainen, 2010). At least six different myosin motors (Pestic-Dragovich et al., 2000; Salamon et al., 2003; Hofmann et al., 2006, 2009; Vreugde et al., 2006; Cameron et al., 2007; Pranchevicius et al., 2008; Lindsay and McCaffrey, 2009) and four different kinesin motors (Macho et al., 2002; Levesque et al., 2003; Mazumdar et al., 2004; Wu et al., 2008; Cross and Powers, 2011; Zhang et al., 2011) are present in animal nuclei, with roles that include transcription, intranuclear movement of chromatin, or export along pore-linked filament networks that connect the nucleolus to NPCs (Simon and Wilson, 2011). Myosin I motors are conserved in diverse eukaryotes (Foth et al., 2006; Hofmann et al., 2009) including the basal Excavate Naegleria gruberi (Goodson and Dawson, 2006), which has six myosin I homologues (Fritz-Laylin et al., 2010). Which (if any) Naegleria myosins actually function in the nucleus is unknown.
Other nucleoskeletal proteins include nuclear mitotic apparatus (NuMA; interphase roles unclear, but self-assembles into 3D space-filling structures; Harborth et al., 1999; Radulescu and Cleveland, 2010), nuclear spectrins (e.g., αII-spectrin scaffolds DNA repair complexes; Young and Kothary, 2005; Zhang et al., 2010), and nuclear protein 4.1 (binds NuMA; associates with pore-linked filaments; helps organize the nucleoskeleton and several NE membrane proteins; Meyer et al., 2011; Simon and Wilson, 2011). Indeed, the ancestral spectrin-repeat protein is proposed to have functioned in the nucleus (Young and Kothary, 2005). Nuclear titin can bind directly to nuclear intermediate filament proteins (Zastrow et al., 2006), and is essential for chromosome condensation during mitosis (Machado et al., 1998; Machado and Andrew, 2000; Zhong et al., 2010). Proteins such as actin, myosins, NuMA, spectrins, and titin were only recently recognized as having fundamental roles in nuclear structure and genome function in living eukaryotes (Simon and Wilson, 2011), and for historical reasons their roles in the nucleoskeleton remain largely un-annotated. Kinesins, of which the LECA is proposed to have possessed ∼11 (Wickstead et al., 2010), are also present in the nucleus (Simon and Wilson, 2011). Nuclear kinesins associate with chromatin, and one (Kif4A) is involved in the response to DNA damage (Wu et al., 2008); otherwise, little is known about their nuclear functions.
Coiled-coil nucleoskeletal proteins
DNA-binding coiled-coil nucleoskeletal proteins are present in two multicellular lineages, animals (Opisthokonts) and plants (Archaeplastids), but evolved independently. In the case of animals, these proteins (lamins) comprise the ancestral intermediate filament (Prokocimer et al., 2009) from which cytoplasmic intermediate filaments later evolved. Nuclear intermediate filaments (lamin filaments) are major structural components of the animal nucleoskeleton, with genetic links to a variety of human diseases (Dittmer and Misteli, 2011). Lamins support or influence nearly every aspect of genome biology, including replication, transcription, signaling, development, and chromosome organization (Prokocimer et al., 2009; Dechat et al., 2010, 2011; Wilson and Berk, 2010). All metazoans have one or two genes encoding “B-type” lamins, whereas complex animals (insects, vertebrates) have an additional gene encoding an independent network of “A-type” lamin filaments required for the physiology of many exquisitely mechanosensitive cell types such as muscle and bone (Dahl et al., 2008). Other known coiled-coil structural proteins in Opisthokonts include the conserved NPC basket protein Tpr (Krull et al., 2004) and Smc (structural maintenance of chromosomes; Wong, 2010). In multicellular animals, where active genes can be located deep within the nucleus, Tpr and related proteins are proposed components of nucleoskeletal pore-linked filaments that connect NPCs to active genes and the nucleolus, and facilitate actin- and myosin-dependent export from the nucleus (Simon and Wilson, 2011). In yeast, which have smaller nuclei, active genes are tethered to the NPC via direct binding of conserved promoter elements (e.g., “DNA zip codes”; Ahmed et al., 2010) to specific nucleoporins (Casolari et al., 2004; Kalverda and Fornerod, 2010). Whether other eukaryotic supergroups have pore-linked filament networks is unknown.
Higher plants lack intermediate filaments (Rose et al., 2005), but do have functionally analogous filaments including those formed by the dsDNA-binding coiled-coil protein MPF1 (Samaniego et al., 2006; Fiserova et al., 2009; Meier and Brkljacic, 2009). Thus, genes encoding different major coiled-coil nucleoskeletal proteins are proposed to have arisen independently, after the LECA, in the Opisthokont and Archaeplastid lineages. These genes may have profoundly influenced genome organization and nuclear structure because they correlate with the emergence of multicellular organisms.
Proposed impact of genome tethering to the cell membrane
Evolution clearly favored the stabilization of positively curved and negatively curved membranes by proto-coatomer proteins (Devos et al., 2004) and ESCRT proteins (Samson and Bell, 2009), respectively. However, these molecules do not account for the fundamentally genome-associated nature of NPCs, or their intimate roles in mitosis and chromosome segregation. For this reason we suggest that the transition from FECA to LECA was driven, in part, by several types of membrane proteins. Among the earliest, we propose, were those that bound DNA or chromatin and thereby tethered the genome to the cell membrane. Stable tethering of relatively dense chromatin would have imposed a mechanical load on the cell membrane (Fig. 2). This load might have had little effect on membrane curvature in cells with strong membrane attachments to an external cell wall. By contrast, in cells with weaker external reinforcement (e.g., FECA and perhaps the precursor of planctomycetes bacteria, which enclose their genome within a nucleus-like structure; Fuerst and Sagulenko, 2011), the chromatin-loaded membrane might have become extensively infolded, threatening cell structure and potentially interfering with chromosome segregation or cell division. We therefore suggest that the incremental molecular evolution of nuclear envelope and nucleoskeletal structure was tightly coupled to the evolution of chromosome segregation and mitosis. This idea is supported by evidence from living eukaryotes that many NPC and nucleoskeletal proteins are essential for chromosome segregation and mitosis, as discussed in the next section. In parallel, we suggest there was strong positive selection, both for adhesive proteins that stabilized and mechanically reinforced the structure of infolded membranes (Fig. 2) and for fusogenic proteins that prevented infolded membranes from interfering with mitosis.
Was evolution of the NPC/nucleoskeleton coupled to chromosome segregation and mitosis?
There is growing evidence that nucleoporins contact active genes, organize heterochromatin, and couple mRNA synthesis to nuclear export (Strambio-De-Castillia et al., 2010; Liang and Hetzer, 2011). Opisthokont nucleoporins are also emerging as central players in mitosis, involved in chromosome condensation and sister chromatid cohesion (Nakano et al., 2011), kinetochore assembly (Salina et al., 2003; Rasala et al., 2006; Roux and Burke, 2006), regulation of microtubule-dependent motors (basket protein Tpr; Nakano et al., 2010), regulation of microtubule polymerization at kinetochores (Mishra et al., 2010), mitotic checkpoint regulation (De Souza and Osmani, 2009; Lussi et al., 2010; Wozniak et al., 2010), and spindle assembly (Nakano et al., 2011). These findings strongly suggest NPCs evolved not merely as “portals”, but as membrane-tethered structural hubs for the genome and mitosis.
Similarly, the textbook picture of mitosis, in which chromosomes are segregated primarily by spindle microtubules, is incomplete. A separate space-filling structure, the spindle “matrix” with proposed elastic hydro-gel properties, is now known to support the spindle (Zheng, 2010; Johansen et al., 2011). This matrix includes both mitotically reorganized nucleoporins (e.g., Tpr; Ding et al., 2009; Lince-Faria et al., 2009) and nucleoskeletal proteins (e.g., NuMA, B-type lamins; Simon and Wilson, 2011). Intriguingly, chromosome segregation in oocytes is driven by the contraction of a nuclear actin network (Lénárt et al., 2005). Similarly in yeast, chromosome segregation can occur in the absence of spindle microtubules through a nuclear fission process that requires actin (Castagnetti et al., 2010). These findings reveal actin and potentially other components of the Opisthokont nucleoskeleton in a new light: as genome-partitioning proteins. Was chromosome segregation (mitosis) a driving force in the evolution of the nucleoskeleton?
The idea that the nucleoskeleton evolved from ancient genome-segregating proteins is supported by the conservation of many related proteins as components of genome partitioning (“par”) systems in bacteria. Bacterial partitioning is best understood for plasmids, and involves three simple components: a repeated DNA sequence (centromeric DNA), a centromere-binding protein, and an associated force-generating (“motor”) protein that separates the two centromeres by forming polymers (Schumacher, 2008). Of these components, only the motors—four kinds—are significantly conserved. Most bacteria use a motor with a Walker-type ATPase motif (type I par system) or an actin/hsp70 superfamily protein (type II; Schumacher, 2008). Other bacteria use a tubulin/FtsZ GTPase superfamily protein (type III) or an unusual (type IV) protein that is predicted to form coiled-coil polymers and also has a predicted DNA-binding domain, potentially uniting both the centromere-binding and motor functions in a single polypeptide (Simpson et al., 2003; Schumacher, 2008). Eukaryotes express proteins related to potentially all four bacterial par motors. Actin is both a major component of the interphase nucleoskeleton (as discussed earlier) and essential for chromosome segregation (Castagnetti et al., 2010). The Smc family of Walker-type ATPases are conserved in all living cells (Hirano, 2005). Tubulin forms intranuclear microtubules in eukaryotes with “closed” mitosis, or “spindle matrix-associated” microtubules in eukaryotes with open mitosis, and is mitotically regulated by nucleoporins. Less clear is whether any nucleoskeletal protein(s) are related to coiled-coil (type IV par) proteins, but candidates include lamins, Tpr, and Smc. Interestingly, certain nuclear membrane proteins also appear to function during mitosis: Samp1, a nuclear inner membrane protein, colocalizes with the mitotic spindle (Buch et al., 2009).
Concluding remarks
The “conserved protein fold” strategy, coupled to purification of NPCs from diverse eukaryotes, yielded brilliant insight into the early coevolution of NPCs with endomembranes (DeGrasse et al., 2009). Further explorations of nuclear transmembrane and nucleoskeletal proteins purified from diverse eukaryotes may yield fascinating insights into the LECA nucleus, and the human cell nucleus. Current limitations include lack of knowledge about most Opisthokont nuclear membrane proteins, and a paucity of sequenced genomes from diverse eukaryotes. Genome analysis of the free-living predatory amoebo-flagellate Naegleria gruberi, which diverged from other eukaryotic lineages over a billion years ago, reveals a rich repertoire of proteins involved in cell structure, signaling, metabolism, and sexual recombination (Fritz-Laylin et al., 2010). This organism has a typical-appearing nucleus and can be cultured in the laboratory (Fulton et al., 1984; Fritz-Laylin et al., 2011). Naegleria, other diverse Excavates including Giardia and Trypanosoma, and laboratory-friendly members of other eukaryotic supergroups including Amoeba (Dictyostelium) and Archaeplastids/Plants (Chlamydomonas) are all available to explore the evolution of nuclear structure. Additional clues to the early evolution of nuclear structure, whether independent or based on shared genes, may come from an unlikely source: planctomycetes bacteria, which enclose their genome within a double membrane, express a clathrin-related protein, and have an endocytosis-like pathway (Fuerst and Webb, 1991; Fuerst and Sagulenko, 2011). It will be exciting to understand how different kinds of proteins and possibly other types of molecules including noncoding RNAs (Pauli et al., 2011) and ADP-ribose chains (Chang et al., 2005) might have shaped the evolution of nuclear structure before the LECA, and to this day.
Acknowledgments
Illustrations provided by Neil Smith (neil@neilsmithillustration.co.uk), based on collage originals by K.L. Wilson.
The authors gratefully acknowledge research funding from the National Institutes of Health (RO1 GM048646 to K.L. Wilson, and R01 AI77571 to S.C. Dawson).
The authors declare no conflicts of interest.
Footnotes
Abbreviations used in this paper:
- FECA
- first eukaryotic common ancestor
- LAP
- lamina-associated polypeptide
- LECA
- last eukaryotic common ancestor
- LEM
- LAP2, emerin and MAN1
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- NuMA
- nuclear mitotic apparatus
- Smc
- structural maintenance of chromosomes
References
- Ahmed S., Brickner D.G., Light W.H., Cajigas I., McDonough M., Froyshteter A.B., Volpe T., Brickner J.H.. 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 12:111–118. 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson L., Otto H.. 2008. LUMA interacts with emerin and influences its distribution at the inner nuclear membrane. J. Cell Sci. 121:536–548. 10.1242/jcs.019281 [DOI] [PubMed] [Google Scholar]
- Braunagel S.C., Williamson S.T., Ding Q., Wu X., Summers M.D.. 2007. Early sorting of inner nuclear membrane proteins is conserved. Proc. Natl. Acad. Sci. USA. 104:9307–9312. 10.1073/pnas.0703186104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch C., Lindberg R., Figueroa R., Gudise S., Onischenko E., Hallberg E.. 2009. An integral protein of the inner nuclear membrane localizes to the mitotic spindle in mammalian cells. J. Cell Sci. 122:2100–2107. 10.1242/jcs.047373 [DOI] [PubMed] [Google Scholar]
- Cai M., Huang Y., Ghirlando R., Wilson K.L., Craigie R., Clore G.M.. 2001. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 20:4399–4407. 10.1093/emboj/20.16.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron R.S., Liu C., Mixon A.S., Pihkala J.P., Rahn R.J., Cameron P.L.. 2007. Myosin16b: The COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression. Cell Motil. Cytoskeleton. 64:19–48. 10.1002/cm.20162 [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A.. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439. 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Castagnetti S., Oliferenko S., Nurse P.. 2010. Fission yeast cells undergo nuclear division in the absence of spindle microtubules. PLoS Biol. 8:e1000512. 10.1371/journal.pbio.1000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P., Coughlin M., Mitchison T.J.. 2005. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 7:1133–1139. 10.1038/ncb1322 [DOI] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D.. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53. 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M.K., Powers M.A.. 2011. Nup98 regulates bipolar spindle assembly through association with microtubules and opposition of MCAK. Mol. Biol. Cell. 22:661–672. 10.1091/mbc.E10-06-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K.N., Engler A.J., Pajerowski J.D., Discher D.E.. 2005. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J. 89:2855–2864. 10.1529/biophysj.105.062554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K.N., Ribeiro A.J., Lammerding J.. 2008. Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102:1307–1318. 10.1161/CIRCRESAHA.108.173989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S.C., Fritz-Laylin L.K.. 2009. Sequencing free-living protists: the case for metagenomics. Environ. Microbiol. 11:1627–1631. 10.1111/j.1462-2920.2009.01965.x [DOI] [PubMed] [Google Scholar]
- Dawson S.C., Pace N.R.. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA. 99:8324–8329. 10.1073/pnas.062169599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C.P., Osmani S.A.. 2009. Double duty for nuclear proteins—the price of more open forms of mitosis. Trends Genet. 25:545–554. 10.1016/j.tig.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler E.W., Ma Y., Seo H.S., Hsia K.C., Noriega T.R., Blobel G., Hoelz A.. 2008. A fence-like coat for the nuclear pore membrane. Mol. Cell. 32:815–826. 10.1016/j.molcel.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Dechat T., Adam S.A., Taimen P., Shimi T., Goldman R.D.. 2010. Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2:a000547. 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Gesson K., Foisner R.. 2011. Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harb. Symp. Quant. Biol. In press. [DOI] [PubMed] [Google Scholar]
- DeGrasse J.A., DuBois K.N., Devos D., Siegel T.N., Sali A., Field M.C., Rout M.P., Chait B.T.. 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics. 8:2119–2130. 10.1074/mcp.M900038-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Alber F., Williams R., Chait B.T., Sali A., Rout M.P.. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2:e380. 10.1371/journal.pbio.0020380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B.T., Rout M.P., Sali A.. 2006. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 103:2172–2177. 10.1073/pnas.0506345103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Yao C., Lince-Faria M., Rath U., Cai W., Maiato H., Girton J., Johansen K.M., Johansen J.. 2009. Chromator is required for proper microtubule spindle formation and mitosis in Drosophila. Dev. Biol. 334:253–263. 10.1016/j.ydbio.2009.07.027 [DOI] [PubMed] [Google Scholar]
- Dittmer T.A., Misteli T.. 2011. The lamin protein family. Genome Biol. 12:222. 10.1186/gb-2011-12-5-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet C.M., Hetzer M.W.. 2010. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 119:469–477. 10.1007/s00412-010-0289-2 [DOI] [PubMed] [Google Scholar]
- Fichtman B., Ramos C., Rasala B., Harel A., Forbes D.J.. 2010. Inner/Outer nuclear membrane fusion in nuclear pore assembly: biochemical demonstration and molecular analysis. Mol. Biol. Cell. 21:4197–4211. 10.1091/mbc.E10-04-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J., Kiseleva E., Goldberg M.W.. 2009. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 59:243–255. 10.1111/j.1365-313X.2009.03865.x [DOI] [PubMed] [Google Scholar]
- Foth B.J., Goedecke M.C., Soldati D.. 2006. New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. USA. 103:3681–3686. 10.1073/pnas.0506307103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke W.W. 2009. Discovering the molecular components of intercellular junctions—a historical view. Cold Spring Harb. Perspect. Biol. 1:a003061. 10.1101/cshperspect.a003061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin A., Penkner A., Jantsch V., Gruenbaum Y.. 2009. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell. Mol. Life Sci. 66:1518–1533. 10.1007/s00018-008-8713-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin L.K., Prochnik S.E., Ginger M.L., Dacks J.B., Carpenter M.L., Field M.C., Kuo A., Paredez A., Chapman J., Pham J., et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 140:631–642. 10.1016/j.cell.2010.01.032 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin L.K., Ginger M.L., Walsh C., Dawson S.C., Fulton C.. 2011. The Naegleria genome: a free-living microbial eukaryote lends unique insights into core eukaryotic cell biology. Res. Microbiol. 162:607–618. 10.1016/j.resmic.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst J.A., Sagulenko E.. 2011. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9:403–413. 10.1038/nrmicro2578 [DOI] [PubMed] [Google Scholar]
- Fuerst J.A., Webb R.I.. 1991. Membrane-bounded nucleoid in the eubacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. USA. 88:8184–8188. 10.1073/pnas.88.18.8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton C., Webster C., Wu J.S.. 1984. Chemically defined media for cultivation of Naegleria gruberi. Proc. Natl. Acad. Sci. USA. 81:2406–2410. 10.1073/pnas.81.8.2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J.M., Smoyer C.J., Stensrud E.S., Alexander R., Gogol M., Wiegraebe W., Jaspersen S.L.. 2011. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J. Cell Biol. 193:489–507. 10.1083/jcb.201011017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieni R.S., Hendzel M.J.. 2009. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem. Cell Biol. 87:283–306. 10.1139/O08-133 [DOI] [PubMed] [Google Scholar]
- Goodson H.V., Dawson S.C.. 2006. Multiplying myosins. Proc. Natl. Acad. Sci. USA. 103:3498–3499. 10.1073/pnas.0600045103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotic I., Foisner R.. 2010. Multiple novel functions of lamina associated polypeptide 2α in striated muscle. Nucleus. 1:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund S.E., Fischer T., Cabal G.G., Antúnez O., Pérez-Ortín J.E., Hurt E.. 2008. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J. Cell Biol. 182:897–910. 10.1083/jcb.200803098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampl V., Hug L., Leigh J.W., Dacks J.B., Lang B.F., Simpson A.G., Roger A.J.. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc. Natl. Acad. Sci. USA. 106:3859–3864. 10.1073/pnas.0807880106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Wang J., Gueth-Hallonet C., Weber K., Osborn M.. 1999. Self assembly of NuMA: multiarm oligomers as structural units of a nuclear lattice. EMBO J. 18:1689–1700. 10.1093/emboj/18.6.1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. 2005. SMC proteins and chromosome mechanics: from bacteria to humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:507–514. 10.1098/rstb.2004.1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Dernburg A.F.. 2009. The SUN rises on meiotic chromosome dynamics. Dev. Cell. 17:598–605. 10.1016/j.devcel.2009.10.014 [DOI] [PubMed] [Google Scholar]
- Hofmann W.A. 2009. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol. 273:219–263. 10.1016/S1937-6448(08)01806-6 [DOI] [PubMed] [Google Scholar]
- Hofmann W.A., Johnson T., Klapczynski M., Fan J.L., de Lanerolle P.. 2006. From transcription to transport: emerging roles for nuclear myosin I. Biochem. Cell Biol. 84:418–426. 10.1139/o06-069 [DOI] [PubMed] [Google Scholar]
- Hofmann W.A., Richards T.A., de Lanerolle P.. 2009. Ancient animal ancestry for nuclear myosin. J. Cell Sci. 122:636–643. 10.1242/jcs.030205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmer L., Pezhman A., Worman H.J.. 1998. The human lamin B receptor/sterol reductase multigene family. Genomics. 54:469–476. 10.1006/geno.1998.5615 [DOI] [PubMed] [Google Scholar]
- Hsia K.C., Stavropoulos P., Blobel G., Hoelz A.. 2007. Architecture of a coat for the nuclear pore membrane. Cell. 131:1313–1326. 10.1016/j.cell.2007.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen K.M., Forer A., Yao C., Girton J., Johansen J.. 2011. Do nuclear envelope and intranuclear proteins reorganize during mitosis to form an elastic, hydrogel-like spindle matrix? Chromosome Res. 19:345–365. 10.1007/s10577-011-9187-6 [DOI] [PubMed] [Google Scholar]
- Kalverda B., Fornerod M.. 2010. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle. 9:4812–4817. 10.4161/cc.9.24.14328 [DOI] [PubMed] [Google Scholar]
- Keeling P.J. 2007. Genomics. Deep questions in the tree of life. Science. 317:1875–1876. 10.1126/science.1149593 [DOI] [PubMed] [Google Scholar]
- Kim C.E., Perez A., Perkins G., Ellisman M.H., Dauer W.T.. 2010. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci. USA. 107:9861–9866. 10.1073/pnas.0912877107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M.C., Lusk C.P., Blobel G.. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007. 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- King M.C., Drivas T.G., Blobel G.. 2008. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 134:427–438. 10.1016/j.cell.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S., Thyberg J., Björkroth B., Rackwitz H.R., Cordes V.C.. 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell. 15:4261–4277. 10.1091/mbc.E04-03-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguri C., Gilquin B., Wolff N., Romi-Lebrun R., Courchay K., Callebaut I., Worman H.J., Zinn-Justin S.. 2001. Structural characterization of the LEM motif common to three human inner nuclear membrane proteins. Structure. 9:503–511. 10.1016/S0969-2126(01)00611-6 [DOI] [PubMed] [Google Scholar]
- Lammerding J., Hsiao J., Schulze P.C., Kozlov S., Stewart C.L., Lee R.T.. 2005. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 170:781–791. 10.1083/jcb.200502148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.K., Delmar V.A., Chan R.C., Phung Q., Bernis C., Fichtman B., Rasala B.A., Forbes D.J.. 2009. Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol. Biol. Cell. 20:4043–4058. 10.1091/mbc.E09-02-0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leksa N.C., Schwartz T.U.. 2010. Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: Commonalities, differences, challenges. Nucleus. 1:314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P., Bacher C.P., Daigle N., Hand A.R., Eils R., Terasaki M., Ellenberg J.. 2005. A contractile nuclear actin network drives chromosome congression in oocytes. Nature. 436:812–818. 10.1038/nature03810 [DOI] [PubMed] [Google Scholar]
- Levesque A.A., Howard L., Gordon M.B., Compton D.A.. 2003. A functional relationship between NuMA and kid is involved in both spindle organization and chromosome alignment in vertebrate cells. Mol. Biol. Cell. 14:3541–3552. 10.1091/mbc.E03-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Hetzer M.W.. 2011. Functional interactions between nucleoporins and chromatin. Curr. Opin. Cell Biol. 23:65–70. 10.1016/j.ceb.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W.C., Mitsuhashi H., Keduka E., Nonaka I., Noguchi S., Nishino I., Hayashi Y.K.. 2011. TMEM43 mutations in Emery-Dreifuss muscular dystrophy-related myopathy. Ann. Neurol. 69:1005–1013. 10.1002/ana.22338 [DOI] [PubMed] [Google Scholar]
- Lin F., Blake D.L., Callebaut I., Skerjanc I.S., Holmer L., McBurney M.W., Paulin-Levasseur M., Worman H.J.. 2000. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 275:4840–4847. 10.1074/jbc.275.7.4840 [DOI] [PubMed] [Google Scholar]
- Lince-Faria M., Maffini S., Orr B., Ding Y., Cláudia Florindo, Sunkel C.E., Tavares A., Johansen J., Johansen K.M., Maiato H.. 2009. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184:647–657. 10.1083/jcb.200811012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay A.J., McCaffrey M.W.. 2009. Myosin Vb localises to nucleoli and associates with the RNA polymerase I transcription complex. Cell Motil. Cytoskeleton. 66:1057–1072. 10.1002/cm.20408 [DOI] [PubMed] [Google Scholar]
- Liu Q., Pante N., Misteli T., Elsagga M., Crisp M., Hodzic D., Burke B., Roux K.J.. 2007. Functional association of Sun1 with nuclear pore complexes. J. Cell Biol. 178:785–798. 10.1083/jcb.200704108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussi Y.C., Shumaker D.K., Shimi T., Fahrenkrog B.. 2010. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus. 1:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C., Andrew D.J.. 2000. D-Titin: a giant protein with dual roles in chromosomes and muscles. J. Cell Biol. 151:639–652. 10.1083/jcb.151.3.639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C., Sunkel C.E., Andrew D.J.. 1998. Human autoantibodies reveal titin as a chromosomal protein. J. Cell Biol. 141:321–333. 10.1083/jcb.141.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho B., Brancorsini S., Fimia G.M., Setou M., Hirokawa N., Sassone-Corsi P.. 2002. CREM-dependent transcription in male germ cells controlled by a kinesin. Science. 298:2388–2390. 10.1126/science.1077265 [DOI] [PubMed] [Google Scholar]
- Mans B.J., Anantharaman V., Aravind L., Koonin E.V.. 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 3:1612–1637. 10.4161/cc.3.12.1316 [DOI] [PubMed] [Google Scholar]
- Margalit A., Brachner A., Gotzmann J., Foisner R., Gruenbaum Y.. 2007. Barrier-to-autointegration factor—a BAFfling little protein. Trends Cell Biol. 17:202–208. 10.1016/j.tcb.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Margulis L. 1970. Origin of Eukaryotic Cells. New Haven, CT, Yale University Press. [Google Scholar]
- Mazumdar M., Sundareshan S., Misteli T.. 2004. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J. Cell Biol. 166:613–620. 10.1083/jcb.200401142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I., Brkljacic J.. 2009. Adding pieces to the puzzling plant nuclear envelope. Curr. Opin. Plant Biol. 12:752–759. 10.1016/j.pbi.2009.09.016 [DOI] [PubMed] [Google Scholar]
- Mekhail K., Moazed D.. 2010. The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 11:317–328. 10.1038/nrm2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A.J., Almendrala D.K., Go M.M., Krauss S.W.. 2011. Structural protein 4.1R is integrally involved in nuclear envelope protein localization, centrosome-nucleus association and transcriptional signaling. J. Cell Sci. 124:1433–1444. 10.1242/jcs.077883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R.K., Chakraborty P., Arnaoutov A., Fontoura B.M., Dasso M.. 2010. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12:164–169. 10.1038/ncb2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R., Shoemaker C.J., Gucek M., Cole R.N., Wilson K.L.. 2009. Barrier-to-autointegration factor proteome reveals chromatin-regulatory partners. PLoS ONE. 4:e7050. 10.1371/journal.pone.0007050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R., Andreassen P.R., Wilson K.L.. 2011. Barrier-to-Autointegration Factor influences specific histone modifications. Nucleus. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Funasaka T., Hashizume C., Wong R.W.. 2010. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J. Biol. Chem. 285:10841–10849. 10.1074/jbc.M110.105890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Wang W., Hashizume C., Funasaka T., Sato H., Wong R.W.. 2011. Unexpected role of nucleoporins in coordination of cell cycle progression. Cell Cycle. 10:425–433. 10.4161/cc.10.3.14721 [DOI] [PubMed] [Google Scholar]
- Németh A., Längst G.. 2011. Genome organization in and around the nucleolus. Trends Genet. 27:149–156. 10.1016/j.tig.2011.01.002 [DOI] [PubMed] [Google Scholar]
- Nery F.C., Zeng J., Niland B.P., Hewett J., Farley J., Irimia D., Li Y., Wiche G., Sonnenberg A., Breakefield X.O.. 2008. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J. Cell Sci. 121:3476–3486. 10.1242/jcs.029454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann N., Lundin D., Poole A.M.. 2010. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE. 5:e13241. 10.1371/journal.pone.0013241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N.R. 2009. Mapping the tree of life: progress and prospects. Microbiol. Mol. Biol. Rev. 73:565–576. 10.1128/MMBR.00033-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.D., Delwiche C.F.. 1996. Second-hand chloroplasts and the case of the disappearing nucleus. Proc. Natl. Acad. Sci. USA. 93:7432–7435. 10.1073/pnas.93.15.7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey L.W., Grant J., Tekle Y.I., Lasek-Nesselquist E., Morrison H.G., Sogin M.L., Patterson D.J., Katz L.A.. 2010. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst. Biol. 59:518–533. 10.1093/sysbio/syq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A., Rinn J.L., Schier A.F.. 2011. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12:136–149. 10.1038/nrg2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestic-Dragovich L., Stojiljkovic L., Philimonenko A.A., Nowak G., Ke Y., Settlage R.E., Shabanowitz J., Hunt D.F., Hozak P., de Lanerolle P.. 2000. A myosin I isoform in the nucleus. Science. 290:337–341. 10.1126/science.290.5490.337 [DOI] [PubMed] [Google Scholar]
- Pranchevicius M.C., Baqui M.M., Ishikawa-Ankerhold H.C., Lourenço E.V., Leão R.M., Banzi S.R., dos Santos C.T., Roque-Barreira M.C., Espreafico E.M., Larson R.E.. 2008. Myosin Va phosphorylated on Ser1650 is found in nuclear speckles and redistributes to nucleoli upon inhibition of transcription. Cell Motil. Cytoskeleton. 65:441–456. 10.1002/cm.20269 [DOI] [PubMed] [Google Scholar]
- Prokocimer M., Davidovich M., Nissim-Rafinia M., Wiesel-Motiuk N., Bar D.Z., Barkan R., Meshorer E., Gruenbaum Y.. 2009. Nuclear lamins: key regulators of nuclear structure and activities. J. Cell. Mol. Med. 13:1059–1085. 10.1111/j.1582-4934.2008.00676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu A.E., Cleveland D.W.. 2010. NuMA after 30 years: the matrix revisited. Trends Cell Biol. 20:214–222. 10.1016/j.tcb.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B.A., Orjalo A.V., Shen Z., Briggs S., Forbes D.J.. 2006. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 103:17801–17806. 10.1073/pnas.0608484103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro S., Igual J.C., Pérez-Ortín J.E.. 2002. SRC1: an intron-containing yeast gene involved in sister chromatid segregation. Yeast. 19:43–54. 10.1002/yea.803 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Basu M.K., Csürös M., Koonin E.V.. 2009. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol. Evol. 1:99–113. 10.1093/gbe/evp011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M.M., Stein P.A., Taylor S.S., Ha E., McKeon F., Rapoport T.A.. 1999. A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein. J. Cell Biol. 146:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A., Schraegle S.J., Stahlberg E.A., Meier I.. 2005. Coiled-coil protein composition of 22 proteomes—differences and common themes in subcellular infrastructure and traffic control. BMC Evol. Biol. 5:66. 10.1186/1471-2148-5-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Burke B.. 2006. From pore to kinetochore and back: regulating envelope assembly. Dev. Cell. 11:276–278. 10.1016/j.devcel.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Salamon M., Millino C., Raffaello A., Mongillo M., Sandri C., Bean C., Negrisolo E., Pallavicini A., Valle G., Zaccolo M., et al. 2003. Human MYO18B, a novel unconventional myosin heavy chain expressed in striated muscles moves into the myonuclei upon differentiation. J. Mol. Biol. 326:137–149. 10.1016/S0022-2836(02)01335-9 [DOI] [PubMed] [Google Scholar]
- Salina D., Enarson P., Rattner J.B., Burke B.. 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162:991–1001. 10.1083/jcb.200304080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaniego R., Jeong S.Y., Meier I., de la Espina S.M.. 2006. Dual location of MAR-binding, filament-like protein 1 in Arabidopsis, tobacco, and tomato. Planta. 223:1201–1206. 10.1007/s00425-005-0168-x [DOI] [PubMed] [Google Scholar]
- Samson R.Y., Bell S.D.. 2009. Ancient ESCRTs and the evolution of binary fission. Trends Microbiol. 17:507–513. 10.1016/j.tim.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Schirmer E.C., Florens L., Guan T., Yates J.R., III, Gerace L.. 2003. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 301:1380–1382. 10.1126/science.1088176 [DOI] [PubMed] [Google Scholar]
- Schumacher M.A. 2008. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 412:1–18. 10.1042/BJ20080359 [DOI] [PubMed] [Google Scholar]
- Simon D.N., Wilson K.L.. 2011. The nucleoskeleton as a genome-associated dynamic network of networks. Nat. Rev. Mol. Cell Biol. In press. [DOI] [PubMed] [Google Scholar]
- Simpson A.E., Skurray R.A., Firth N.. 2003. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J. Bacteriol. 185:2143–2152. 10.1128/JB.185.7.2143-2152.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarp K.P., Vartiainen M.K.. 2010. Actin on DNA-an ancient and dynamic relationship. Cytoskeleton (Hoboken). 67:487–495. [DOI] [PubMed] [Google Scholar]
- Smythe C., Jenkins H.E., Hutchison C.J.. 2000. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 19:3918–3931. 10.1093/emboj/19.15.3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyers L., Vlcek S., Dechat T., Skegro D., Korbei B., Gajewski A., Mayans O., Schöfer C., Foisner R.. 2007. Lamina-associated polypeptide 2-alpha forms homo-trimers via its C terminus, and oligomerization is unaffected by a disease-causing mutation. J. Biol. Chem. 282:6308–6315. 10.1074/jbc.M605782200 [DOI] [PubMed] [Google Scholar]
- Sogin M.L., Silberman J.D.. 1998. Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int. J. Parasitol. 28:11–20. 10.1016/S0020-7519(97)00181-1 [DOI] [PubMed] [Google Scholar]
- Starr D.A., Fridolfsson H.N.. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26:421–444. 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P.. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501. 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Talamas J.A., Hetzer M.W.. 2011. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J. Cell Biol. 194:27–37. 10.1083/jcb.201012154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heyden A.B., Naismith T.V., Snapp E.L., Hodzic D., Hanson P.I.. 2009. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell. 20:2661–2672. 10.1091/mbc.E09-01-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreugde S., Ferrai C., Miluzio A., Hauben E., Marchisio P.C., Crippa M.P., Bussi M., Biffo S.. 2006. Nuclear myosin VI enhances RNA polymerase II-dependent transcription. Mol. Cell. 23:749–755. 10.1016/j.molcel.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Wagner N., Krohne G.. 2007. LEM-Domain proteins: new insights into lamin-interacting proteins. Int. Rev. Cytol. 261:1–46. 10.1016/S0074-7696(07)61001-8 [DOI] [PubMed] [Google Scholar]
- Wente S.R., Rout M.P.. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2:a000562. 10.1101/cshperspect.a000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B., Gull K., Richards T.A.. 2010. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol. Biol. 10:110. 10.1186/1471-2148-10-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie G.S., Korfali N., Swanson S.K., Malik P., Srsen V., Batrakou D.G., de las Heras J., Zuleger N., Kerr A.R., Florens L., Schirmer E.C.. 2011. Several novel nuclear envelope transmembrane proteins identified in skeletal muscle have cytoskeletal associations. Mol. Cell. Proteomics. 10:M110: 003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.L., Berk J.M.. 2010. The nuclear envelope at a glance. J. Cell Sci. 123:1973–1978. 10.1242/jcs.019042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.L., Foisner R.. 2010. Lamin-binding proteins. Cold Spring Harb. Perspect. Biol. 2:a000554. 10.1101/cshperspect.a000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R., Kandler O., Wheelis M.L.. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA. 87:4576–4579. 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.W. 2010. An update on cohesin function as a ‘molecular glue’ on chromosomes and spindles. Cell Cycle. 9:1754–1758. 10.4161/cc.9.9.11806 [DOI] [PubMed] [Google Scholar]
- Wozniak R., Burke B., Doye V.. 2010. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell. Mol. Life Sci. 67:2215–2230. 10.1007/s00018-010-0325-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Zhou L., Khidr L., Guo X.E., Kim W., Lee Y.M., Krasieva T., Chen P.L.. 2008. A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle. 7:2013–2020. 10.4161/cc.7.13.6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.G., Kothary R.. 2005. Spectrin repeat proteins in the nucleus. Bioessays. 27:144–152. 10.1002/bies.20177 [DOI] [PubMed] [Google Scholar]
- Zastrow M.S., Flaherty D.B., Benian G.M., Wilson K.L.. 2006. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J. Cell Sci. 119:239–249. 10.1242/jcs.02728 [DOI] [PubMed] [Google Scholar]
- Zhang P., Sridharan D., Lambert M.W.. 2010. Knockdown of mu-calpain in Fanconi anemia, FA-A, cells by siRNA restores alphaII spectrin levels and corrects chromosomal instability and defective DNA interstrand cross-link repair. Biochemistry. 49:5570–5581. 10.1021/bi100656j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shao H., Huang Y., Yan F., Chu Y., Hou H., Zhu M., Fu C., Aikhionbare F., Fang G., et al. 2011. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 286:3033–3046. 10.1074/jbc.M110.165340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. 2010. A membranous spindle matrix orchestrates cell division. Nat. Rev. Mol. Cell Biol. 11:529–535. 10.1038/nrm2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Wilson K.L., Dahl K.N.. 2010. Beyond lamins other structural components of the nucleoskeleton. Methods Cell Biol. 98:97–119. 10.1016/S0091-679X(10)98005-9 [DOI] [PMC free article] [PubMed] [Google Scholar]