Abstract
Contact of Mycobacterium tuberculosis (M.tb) with the immune system requires interactions between microbial surface molecules and host pattern recognition receptors. Major M.tb-exposed cell envelope molecules, such as lipomannan (LM), contain subtle structural variations that affect the nature of the immune response. Here we show that LM from virulent M.tb (TB-LM), but not from avirulent Myocobacterium smegmatis (SmegLM), is a potent inhibitor of TNF biosynthesis in human macrophages. This difference in response is not because of variation in Toll-like receptor 2-dependent activation of the signaling kinase MAPK p38. Rather, TB-LM stimulation leads to destabilization of TNF mRNA transcripts and subsequent failure to produce TNF protein. In contrast, SmegLM enhances MAPK-activated protein kinase 2 phosphorylation, which is critical for maintaining TNF mRNA stability in part by contributing microRNAs (miRNAs). In this context, human miRNA miR-125b binds to the 3′ UTR region of TNF mRNA and destabilizes the transcript, whereas miR-155 enhances TNF production by increasing TNF mRNA half-life and limiting expression of SHIP1, a negative regulator of the PI3K/Akt pathway. We show that macrophages incubated with TB-LM and live M.tb induce high miR-125b expression and low miR-155 expression with correspondingly low TNF production. In contrast, SmegLM and live M. smegmatis induce high miR-155 expression and low miR-125b expression with high TNF production. Thus, we identify a unique cellular mechanism underlying the ability of a major M.tb cell wall component, TB-LM, to block TNF biosynthesis in human macrophages, thereby allowing M.tb to subvert host immunity and potentially increase its virulence.
Keywords: lipoglycans, innate immunity, cell signaling, intracellular pathogen
Mycobacterium tuberculosis (M.tb) subverts host innate immune responses and survives within macrophages by various mechanisms, particularly by using its unique outer cell wall molecules including lipoglycoconjugates, such as mannose-capped lipoarabinomannan, lipomannan (LM), and phosphatidyl-myo-inositol mannosides (PIMs), in addition to mannosylated lipopeptides (1). Whereas much attention has been given to the biological properties of mannose-capped lipoarabinomannan and PIMs, there has been less interest in studying LM. Mycobacterial LM is structurally defined by a GPI anchor linked to a linear α(1→6)- mannan containing some α(1→2)-mannose branches (2). In particular, LMs from different mycobacterial species exhibit both proinflammatory and anti-inflammatory responses through Toll-like receptor (TLR) 2-dependent and -independent pathways (3, 4). Although LMs from different mycobacterial strains and species clearly activate several immune responses in macrophages, the molecular and cellular mechanisms underlying this activation remain unclear, particularly in human macrophages.
TNF plays a central role in the establishment of M.tb infection and maintenance of latent tuberculosis (5). The relative production of TNF varies among pathogenic and nonpathogenic mycobacterial species (6). Regulation of TNF biosynthesis is complex. Like many eukaryotic proteins, TNF contains an adenylate/uridylate-rich element (ARE) in its mRNA 3′ UTR that is frequently targeted by RNA binding proteins for degradation (7). The initiation or stabilization of TNF transcription is thought to be controlled by various proteins, including tristetraprolin (TTP), human antigen R, T-cell intracytoplasmic antigen-1, and TIA-1–related protein (8). In addition to mRNA stability, posttranscriptional regulation of many inflammatory genes occurs through p38 MAPK-mediated activation of MAPK-activated protein kinase 2 (MK2) (9). Activated MK2 stabilizes TNF mRNA through TTP phosphorylation (10). Nonphosphorylated TTP binds to the ARE region of target mRNAs and induces rapid degradation through various mechanisms (11–13).
Another means of eukaryotic control of gene expression is through microRNAs (miRNAs), which function as posttranscriptional regulators of many genes. MicroRNAs mediate their effect by binding mRNA 3′ UTR regions usually resulting in mRNA degradation. MiR-125b targets the 3′ UTR region of TNF mRNA transcript destabilizing it (14). MiR-155 targets the 3′ UTR region of the inositol phosphatase SHIP1 mRNA, leading to its degradation. SHIP1 functions a negative regulator of TNF production (15). It was recently recognized that the pathogenic bacterium Francisella can regulate miRNAs, thereby modulating host immunologic responses (16). Despite the apparent differences in TNF production in response to mycobacterial species and their cell wall products, no studies to date have analyzed their effects on miRNAs.
In the present study, we examined the effects of LMs from virulent (M.tb H37Rv) and avirulent (Mycobacterium smegmatis) mycobacteria on TNF production in human macrophages. Although both SmegLM and TB-LM activate TLR2-dependent MAPK p38 and the PI3K/Akt pathway with production of steady-state TNF mRNA, TB-LM treated macrophages fail to produce TNF protein. We found that this deficiency is due to increased turnover of TNF mRNA and decreased levels of polysome-bound TNF mRNA in response to TB-LM compared with SmegLM as a result of differential activation and expression of MK2, miR-125b, and miR-155.
Results
Differential Production of TNF by LM-Stimulated Human Macrophages.
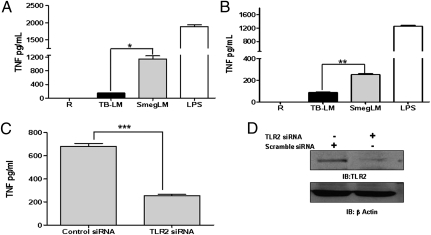
We compared the levels of TNF production induced by TB-LM and by SmegLM in human monocyte-derived macrophages (MDMs) (17) after 24 h of exposure. MDMs incubated with TB-LM produced significantly less TNF than those incubated with SmegLM [14.11% ± 1.72% that of SmegLM (mean ± SEM; n = 5)] (Fig. 1A). A similar response was observed using the human monocytic cell line THP-1 (Fig. 1B). Given that certain LMs serve as ligands for TLR2 (3, 4), we next investigated the involvement of TLR2 in SmegLM-induced TNF production. In contrast to the control MDMs, TNF production in response to SmegLM was significantly reduced in cells transfected with TLR2 siRNA (Fig. 1 C and D), demonstrating that TLR2 is an important receptor for SmegLM-mediated TNF production in human macrophages.
Fig. 1.
LM from M.tb stimulates minimal TNF production, whereas LM from M. smegmatis stimulates robust TNF production depending on TLR2. (A and B) MDM monolayers (A) or THP-1 cells (B) were incubated with TB-LM (5 μg/mL), SmegLM (5 μg/mL), or LPS as a positive control for 24 h. Cell-free culture supernatants were analyzed for TNF production by ELISA. The results in A and B are cumulative data from five and three experiments, respectively, each performed in triplicate (*P < 0.01; **P < 0.001). (C) MDMs were transfected with TLR2 siRNA or scramble siRNA (control) and plated in RPMI containing 20% autologous serum. After 24 h, cells were washed and stimulated with SmegLM (5 μg/mL) for 24 h. Cell culture supernatants were collected and analyzed for TNF production by ELISA (***P < 0.0001). (D) The cell lysates were examined for TLR2 and β−actin expression by Western blot analysis. The graph and Western blot are representative of three experiments.
TB-LM and SmegLM Activate MAPK p38 and Akt in Macrophages.
Mycobacterial cell wall components, such as lipoarabinomannan (LAM), LM, and PIMs, can activate the MAPK/Akt pathway, leading to the production of proinflammatory mediators (18, 19). We next examined whether TB-LM and SmegLM differentially activate these upstream kinases involved in TNF biosynthesis in human macrophages. Our results show that both LMs were able to activate these kinases, equivalently in the case of MAPK p38 and quantitatively greater in the case of Akt with SmegLM (Fig. S1). In addition, TLR2 knockdown in MDMs confirmed that the activation of Akt and MAPK p38 by TB-LM occurs through TLR2 (Fig. S2 A and B). Thus, the differences in activation of MAPK p38 and Akt by TB-LM and SmegLM do not fully account for the marked difference in TNF protein production observed for these LMs; however, these kinases likely are important in the initiation of TNF transcription.
TNF Promoter Activity and Expression in Macrophages Stimulated with LMs.
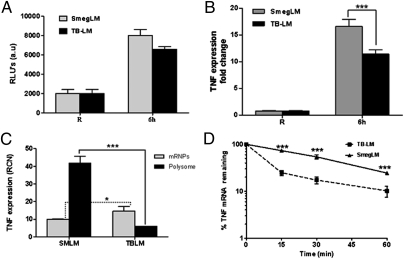
We next investigated whether TB-LM and SmegLM differentially induce TNF promoter activity and steady-state mRNA. Our results show that TB-LM–stimulated macrophages did not significantly decrease promoter activity compared with SmegLM-stimulated cells (Fig. 2A; P = not significant). However, significantly less TNF mRNA was present after 6 h of stimulation with TB-LM than after comparable stimulation with SmegLM (Fig. 2B), suggesting that one or more posttranscriptional processes may be responsible for the lower protein levels shown in Fig. 1.
Fig. 2.
TB-LM and SmegLM enhance TNF promoter activity and expression in human macrophages. (A) MDMs were transfected with a luciferase reporter plasmid (K-13 long) by nucleofection and plated in RPMI containing 20% autologous serum. After overnight incubation, the cells were washed and stimulated with TB-LM or SmegLM (5 μg/mL) or were left unstimulated. After 6 h of incubation, luciferase activity was measured with a luminometer. The graph represents three experiments (P = not significant for TB-LM vs. SmegLM). (B) MDMs were stimulated with TB-LM or SmegLM or were left unstimulated for 6 h, and total RNA was extracted and analyzed for TNF mRNA transcripts by qRT-PCR. The graph is representative of three experiments (***P < 0.0001). (C) Polysome analysis was performed in MDMs after LM stimulation as in B. Cell lysates devoid of nuclei were fractionated on sucrose step gradients, and total RNA was extracted from the polysome and mRNP fractions and analyzed for TNF mRNA transcripts by qRT-PCR. The graph is representative of three experiments (*P < 0.01; ***P < 0.0001). (D) A TNF mRNA stability assay was performed in MDMs by incubating cells with actinomycin D (5 μg/mL) for different time points (15, 30, and 60 min) after stimulation with LMs for 6 h and washing. Total RNA was extracted and analyzed for TNF mRNA transcript levels by qRT-PCR. The graph shows cumulative data from three experiments (***P < 0.0001).
TB-LM Leads to Reduced Translation and Stability of TNF mRNA in Macrophages.
Posttranscriptional control of TNF expression is exerted at the initiation of translation and at the level of mRNA turnover (20, 21). The efficiency of translation initiation is generally assayed by the amount of a given mRNA on polysomes; given our small sample size, this was determined using sucrose step gradients, a rapid technique with yields comparable to those obtained with linear gradients (22). MDMs were incubated with SmegLM or TB-LM for 6 h, lysed, and loaded onto step gradients. Then the levels of TNF mRNA in the pellet (polysomes) and interface [nontranslating mRNA–protein complexes (mRNPs)] were determined by quantitative RT-PCR (qRT-PCR). Overall, our results show that 80.04% ± 1.36% (n = 3) of the TNF transcripts were associated with polysomes in SmegLM-stimulated MDMs (Fig. 2C). In sharp contrast, 71.95% ± 2.78% (n = 3) of the TNF transcripts were associated with the mRNP fraction in TB-LM–stimulated cells. These results indicate that treatment with TB-LM affects the translation of TNF mRNA, most likely by inhibiting initiation, and that this is a contributing factor to the significantly lower amount of TNF elaborated from TB-LM–treated cells compared with SmegLM-treated cells.
Given that the amount of TNF mRNA was also lower in TB-LM–treated cells, we next examined the impact of LM stimulation on TNF mRNA stability. TNF mRNA decayed with biphasic kinetics in TB-LM–treated cells, with 73.51% ± 2.13% of TNF mRNA lost within 15 min of actinomycin D treatment (Fig. 2D). In contrast, the TNF mRNA half-life was 40 min in SmegLM-treated cells, and decay followed linear kinetics. The difference in decay curves suggests differences in the mechanism through which TNF mRNA is degraded in TB-LM–treated cells vs. SmegLM-treated cells. The data shown in Fig. 2 C and D indicate that TB-LM suppresses TNF production by acting on both the translation and stability of TNF mRNA.
SmegLM and M. smegmatis, but Not TB-LM and M.tb, Are Potent MK2 Activators in Macrophages.
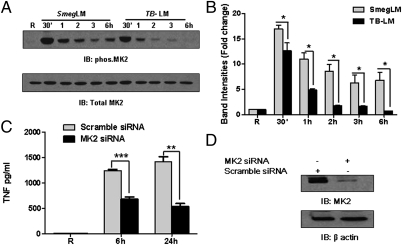
To understand the molecular mechanism(s) underlying enhanced TNF mRNA stability in SmegLM-stimulated macrophages, we analyzed the activation of MK2, a key molecule for TNF mRNA stability (23). Activated MK2 translocates to the cytoplasm and maintains TNF mRNA stability through TTP phosphorylation, leading to its removal from the TNF transcript ARE (10). We found that SmegLM stimulation for different time periods led to a more robust and prolonged MK2 phosphorylation compared with that achieved by TB-LM stimulation (Fig. 3 A and B), and that this was associated with increased TTP phosphorylation after SmegLM stimulation (Fig. S3A). Thus, the relative lack of MK2 activation in macrophages provides a mechanism for the reduction of TNF mRNA stability after TB-LM stimulation. In concert with this finding, MK2 knockdown in MDMs significantly reduced TNF production in response to SmegLM (Fig. 3 C and D). Finally, like SmegLM stimulation, M. smegmatis stimulation led to a more robust MK2 phosphorylation compared with that of M.tb H37Rv stimulation (Fig. S3B). These findings demonstrate the importance of MK2 in TNF mRNA stability and TNF production in response to LMs and their associated mycobacteria.
Fig. 3.
TNF production is regulated by MK2 in human macrophages. (A and B) MDMs were stimulated with TB-LM (5 μg/mL) or SmegLM (5 μg/mL) or left untreated for different time periods (30 min, 1 , 2 , 3 , and 6 h). Cell lysates were examined by Western blot analysis using phospho-MK2, followed by total MK2 Abs. (A) A representative Western blot from three experiments. (B) Cumulative data of band intensities from three experiments (*P < 0.01). (C) MDMs were transfected with MK2 siRNA or scramble siRNA, and after 48 h cells were washed and stimulated with SmegLM (5 μg/mL) for 6 h and 24 h. Cell culture supernatants were collected and analyzed for TNF production by ELISA. The graph is representative of three experiments (**P < 0.001; ***P < 0.0001). (D) The cell lysates in C were examined for MK2 and β-actin expression by Western blot analysis.
TB-LM Stimulation Markedly Up-Regulates hsa-miR-125b Expression, Whereas SmegLM Markedly Up-Regulates hsa-miR-155 Expression in Macrophages.
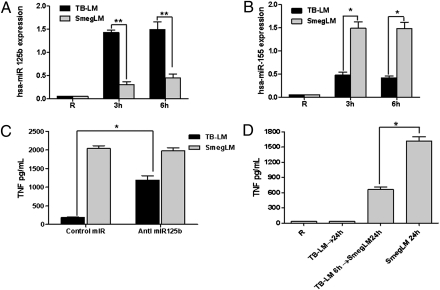
miRNAs represent another potential important class of molecules for regulating TNF biosynthesis in response to LMs. hsa-miR-125b targets the 3′ UTR region of TNF mRNA down-regulating TNF expression in response to LPS in mouse macrophages (14). In contrast, hsa-miR-155 targets SHIP1, a negative regulator of the PI3K/Akt pathway involved in TNF biosynthesis (15). In addition, expression of miR-155 was recently shown to increase TNF production through increasing mRNA stability and half-life (24). Thus, we evaluated whether hsa-miR-125b expression is up-regulated in MDMs stimulated with TB-LM compared with those stimulated with SmegLM. The results show that TB-LM–stimulated MDMs have enhanced hsa-miR-125b expression (a 33.0 ± 6.98-fold increase at 3 h and a 35.46 ± 9.56-fold increase at 6 h; n = 3) (Fig. 4A), whereas SmegLM-stimulated MDMs have enhanced hsa-miR-155 expression (a 29.75 ± 3.90-fold increase at 3 h and a 28.67 ± 3.42-fold increase at 6 h; n = 3) (Fig. 4B). These results suggest that TB-LM–induced hsa-miR-125b binds to the 3′ UTR region of TNF mRNA, contributing to the inhibition of translation in Fig. 2C and possibly its accelerated degradation in Fig. 2D, whereas SmegLM-induced hsa-miR-155 increases TNF mRNA stability and targets SHIP1 for destruction, thereby enhancing PI3K/Akt-mediated signaling and TNF mRNA stability. In concert with this, SmegLM induced greater Akt activation than TB-LM (Fig. S1B) and demonstrated reduced expression of SHIP1 at later time points (Fig. S4). In a similar fashion, live M. smegmatis had reduced SHIP1 expression at later time points compared with live M.tb (Fig. S5). Together, these findings provide evidence that differential regulation of miRNAs by TB-LM and SmegLM is also an important contributor to the differences in TNF production between TB-LM and SmegLM.
Fig. 4.
LM stimulation alters hsa-miR-125b and hsa-miR-155 expression, and regulation of has-miR-125b expression alters TNF production in human macrophages. (A and B) TB-LM, SmegLM, and unstimulated cells were analyzed for miRNA expression by qRT-PCR. (A) hsa- miR-125b expression. (B) hsa-miR-155 expression. The graphs are representative of three experiments (*P < 0.01; **P < 0.001). (C) For blocking hsa-miR-125b expression, MDMs were transfected with anti-miR miRNA inhibitor specific for hsa-miR-125b and then stimulated with TB-LM or SmegLM (5 μg/mL). Cell-free supernatants were harvested and analyzed for TNF production by ELISA. The graph is representative of three experiments (*P < 0.01). (D) MDMs were stimulated with TB-LM (5 μg/mL) for 6 h, washed with warm media, and resuspended in RPMI medium supplemented with 10 mM Hepes and 0.4% human serum albumin containing SmegLM (5 μg/mL) for 24 h. Cell-free culture supernatants were harvested and analyzed for TNF production by ELISA. The graph is representative of three experiments (*P < 0.01).
Up-regulation of hsa-miR-155 has been linked to TLR2 activation (16), and we determined that TLR2 plays an important role in SmegLM-mediated macrophage TNF production (Fig. 1C). Thus, we elected to knock down TLR2 and assess miRNA expression after LM stimulation. Our results show that TLR2 knockdown significantly reduced hsa-miR-155 expression in response to SmegLM, but had no effect on the low level of TB-LM–mediated hsa-miR-155 expression (Fig. S6A). In addition, TLR2 knockdown had no effect on hsa-miR-125b expression in response to either LM (Fig. S6B), indicating that hsa-miR-125b expression is regulated through another pathway.
Modulation of hsa-miR-125b Expression and TB-LM Pretreatment of Macrophages Alter TNF Production in Response to LMs.
To confirm the involvement of hsa-miR-125b in TNF production in response to LMs, MDMs were transfected with an anti-miR miRNA inhibitor specific for hsa-miR-125b, stimulated with TB-LM or SmegLM and supernatants, and then analyzed for TNF. Inhibition of hsa-miR-125b expression in macrophages enhanced TNF production (by 5.5 ± 0.87-fold; n = 3) in response to TB-LM, but had no effect on SmegLM-induced TNF (Fig. 4C). Next, MDMs were prestimulated with TB-LM (to induce hsa-miR-125b) for 6 h, washed, and treated with SmegLM for 24 h. TB-LM pretreatment of macrophages inhibited SmegLM-mediated TNF production (a 44.53% ± 7.17% decrease, n = 3) (Fig. 4D). These results support the role of hsa-miR-125b expression in regulating TNF production in macrophages after LM stimulation.
Differential Expression of hsa-miR-125b and hsa-miR-155 Influences TNF Production in M.tb- and M. smegmatis-Infected Macrophages.
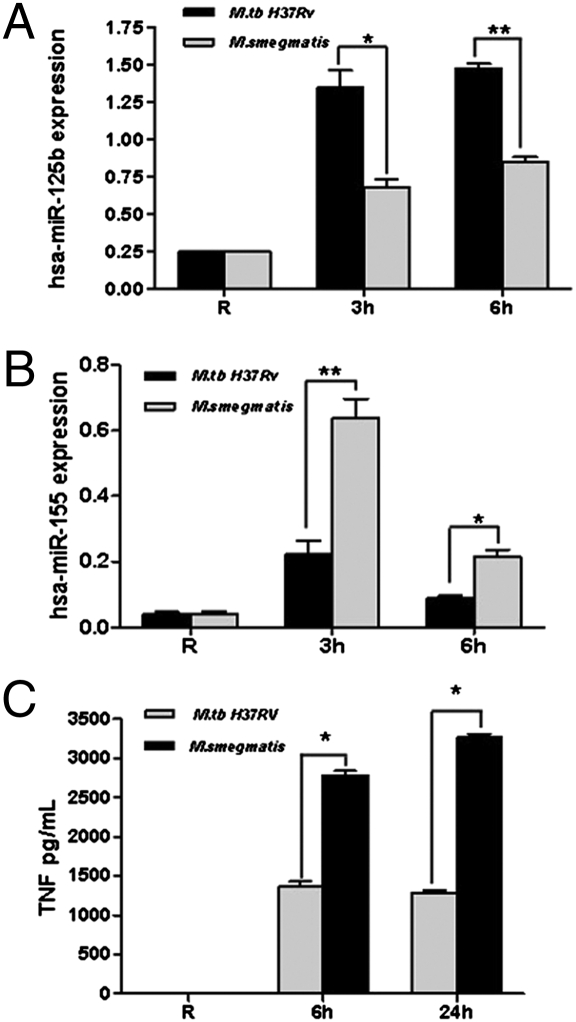
To ascertain whether the miRNA results obtained with purified LMs are applicable to live bacteria, MDMs were infected with M.tb H37Rv and M. smegmatis, and miRNA expression was analyzed. The results show that M.tb infection enhanced hsa-miR-125b expression (Fig. 5A) and limited hsa-miR-155 expression (Fig. 5B) relative to M. smegmatis infection. Furthermore, consistent with our miRNA results with LMs, MDMs infected with M. smegmatis produced higher TNF levels compared with those infected with M.tb (Fig. 5C). These results provide evidence that after M. smegmatis infection of human macrophages, TNF mRNA stability is increased by down-regulation of hsa-miR-125b expression and up-regulation of hsa-miR-155 expression, which together promote the proinflammatory response.
Fig. 5.
Regulation of hsa-miR-125b and hsa-miR-155 expression and TNF production in human macrophages in response to mycobacteria. (A and B) MDMs were infected with M.tb H37Rv or M. smegmatis [multiplicity of infection (MOI) of 5:1] for 2 h. Cells were washed and incubated in 2% autologous serum for an additional 3 h or 6 h for hsa-miR-125b and hsa-miR-155 expression studies and for an additional 24 h for cytokine assays. (A) hsa-miR-125b expression. (B) hsa-miR-155 expression. The graphs are from three independent experiments, each performed in triplicate (*P < 0.01; **P < 0.001). (C) Cell-free supernatants were analyzed for TNF production by ELISA. The graph is representative of three experiments, each performed in triplicate (*P < 0.01).
Discussion
TNF plays a crucial role in the control of mycobacterial growth in macrophages through the activation of macrophage-mediated mycobactericidal mechanisms (25) and in protective immunity (26). Pathogenic mycobacteria have been shown to induce less TNF than nonpathogenic mycobacteria in human macrophages (27). The regulation of expression and secretion of TNF by pathogenic mycobacteria is likely a key factor in modulating the host cell immune response. Here we report previously unexplored regulatory pathways that target TNF mRNA expression and stability in M.tb-infected human macrophages and bring to light an important role for miRNAs in this process.
LAMs and LMs are major mycobacterial cell wall immunomodulatory molecules (2, 18). The bacillus Calmette–Guérin LM fatty acids can determine LMs’ ability to activate TLRs. Triacylated LM and tetra-acylated LM act as agonists of TLR2/TLR1 and TLR4, respectively, generating a proinflammatory response, whereas diacylated LM induces an anti-inflammatory response (28, 29). Mannan chain length also contributes to binding to TLR2, related in part to the avidity for the receptor (30). M. avium paratuberculosis and its LM type result in limited TNF expression in mouse macrophages (19). We performed detailed biochemical analyses on the SmegLM and TB-LM used in this study (monosaccharide, fatty acid, and succinate composition and linkage analysis by GC/MS; size and molecular composition by SDS/PAGE and MALDI-MS; and verification of chemical and structural composition by NMR). These analyses show significant compositional and structural differences between the LMs. Specifically, our results demonstrate that M.tb H37Rv produces a large, neutral LM, whereas M. smegmatis produces a smaller, linear, acidic LM. Compared with the TB-LM, the SmegLM has a linear and shorter mannan domain that is heavily substituted with acyl groups (succinates) and contains more arabinose residues (31). Thus, several unique structural motifs of the LMs likely contribute to our results.
We found markedly increased TNF production in macrophages by SmegLM compared with TB-LM. Consistent with a previous report (4), we determined that human macrophages recognize SmegLM through TLR2, leading to TNF production. Similarly, LAMs, and particularly LMs from the less pathogenic Mycobacterium chelonae and Mycobacterium kansasii, induce TNF production in the human macrophage-like cell line THP-1 by a TLR2-dependent mechanism (4). Our study provides evidence indicating that although TB-LM activates macrophages and induces signaling cascades required for TNF mRNA expression through TLR2, there is reduced translation and rapid degradation of TNF mRNA in TB-LM stimulated macrophages. Importantly, the central findings were recapitulated using live M.tb and M. smegmatis. Thus, our results reveal a unique mechanism by which M.tb is able to subvert the host cell immune response.
MAPK p38 and Akt activation contributes significantly to TNF expression in mycobacteria-infected macrophages (32). In this regard, pathogenic M. avium infection limits MAPK p38 activation in mouse macrophages compared with M. smegmatis (6, 19). Surprisingly, we found no significant difference in MAPK p38 activation between TB-LM and SmegLM after stimulation of human macrophages through TLR2, but a reproducible decrease in Akt activation in the case of TB-LM. In addition, we found a decrease in TNF expression in response to TB-LM as demonstrated by steady-state mRNA levels. This led us to examine the stability of TNF mRNA, focusing on MK2 (23) and miRNAs (14).
There is growing evidence that MK2, a MAPK p38-regulated kinase, effects TNF mRNA stability (33, 10). In particular, mice that lack MK2 show increased stress resistance and survive an LPS-induced endotoxic shock/cytokine storm due to decreased TNF production with no change in TNF receptor-mediated signaling (34). MK2 deficiency in mice results in increased susceptibility to Listeria monocytogenes infection (35), and MK2 differentially regulates Plasmodium falciparum glycosylphosphatidylinositol-induced production of TNF in mouse macrophages (36). These findings indicate an important role for MK2 in host defense against intracellular pathogens through the regulation of TNF production required for activation of antimicrobial effecter mechanisms. Our results demonstrate that SmegLM and live M. smegmatis phosphorylate MK2 more robustly and over a longer period compared with TB-LM. These results are correlated with the time course of TNF mRNA production and decay, indicating that regulation of MK2 is responsible in part for the decreased TNF production in TB-LM stimulated macrophages.
The critical role of miRNAs in various biological processes in health and disease, including the release of inflammatory mediators, is emerging (37, 38). An important discovery in the current study is the identification of specific miRNAs that link TNF protein repression with mRNA degradation in TB infection. Our findings reveal differential expression of hsa-miR-125b and hsa-miR-155 in TB-LM–stimulated macrophages. miR-125b targets the 3′ UTR of the TNF transcript and down-regulates TNF production (14). It also enhances the stability of κB-Ras2, an inhibitor of NFκB signaling in human macrophages, thereby decreasing the inflammatory response (39). Consistent with this, TB-LM induced increased hsa-miR-125b expression, which correlated with the decreased TNF production. In contrast, SmegLM induced increased hsa-miR-155 expression in macrophages, with a basal level of hsa-miR-125b expression that correlated with increased TNF production. We found that SmegLM induction of hsa-miR-155, but not of hsa-miR-125b, is mediated through TLR2. Anti–miR-125b significantly increased TNF production in response to TB-LM, and pretreatment of macrophages with TB-LM significantly decreased TNF production in SmegLM-stimulated cells, likely through hsa-miR-125b induced translational repression and/or degradation of TNF mRNA. We reproduced our results using live M.tb and M. smegmatis.
miRNA hsa-miR-155 regulates TNF production by targeting the inositol phosphatase SHIP1 for degradation through its 3′ UTR interaction (15) and enhances TNF mRNA stability (24). We found that SmegLM- and M. smegmatis-stimulated macrophages enhanced hsa-miR-155 expression, which led to reduced SHIP1 expression at later time points (accounting for greater Akt activation in the case of SmegLM), thereby enhancing TNF mRNA stability and TNF production. Taken together, our results provide evidence that miRNAs are important regulators of TNF production during mycobacterial infection. Apart from the current work, only some very recent publications have identified the fact that pathogenic microorganisms can induce miRNA expression in immune cells (16, 40).
In the present study, TLR2-dependent activation of MAPK p38 and Akt by TB-LM and SmegLM did not fully account for the difference in TNF protein production observed for these LMs. Rather, differential MK2 activation and miRNA expression were key to the regulation of translation and mRNA stability. MK2 is activated by MAPK p38, and it is possible that the differential activation of MK2 is associated with the level of nuclear translocation of activated MAPK p38, which is dependent on the type of stimulus given (41). Other reports demonstrate that changes to the cytoskeletal infrastructure, such as Rho/ROCK and actin cytoskeleton (42), microtubules, and dynein also regulate the translocation of MAPK p38 to the nucleus (43). We speculate that TB-LM–activated MAPK p38 may fail to translocate to the nucleus. Differential expression of miRNAs in response to TB-LM and SmegLM may be due to the activation level of transcription factors involved in induction of miRNA expression in human macrophages (16, 39).
Materials and Methods
Transfection and Luciferase Assays.
MDMs were transfected with 200 nM scramble siRNA or TLR2 siRNA (sense: CUGGUAGUUGUGGGUUGAAGCdTdT; antisense: GCUUCAACCCACAACUACCAGdTdT) for 48 h as described previously (44), with anti-miR miRNA inhibitors and plasmid-expressing TNF luciferase (11110:k3 long; Addgene) using an Amaxa Nucleofector for 16 h or with MK2 siRNA (smart pool) for 48 h. The transfection efficiency of MDMs was >95% as determined by transfecting a GFP-expressing plasmid into MDMs and then analyzing the percentage of GFP-expressing cells by fluorescence microscopy (Fig. S7). Transfected cells were used for subsequent experiments, including cytokine ELISA, luciferase assays, and Western blot analyses. For the promoter reporter assay, transfected cells were either left unstimulated or stimulated with TB-LM or SmegLM (5 μg/mL) for different time periods. Cells were lysed in 100 μL of Luciferase Cell Culture Lysis Reagent (Promega). Luciferase activity was measured using Promega Luciferase Assay Reagent. For blocking miR-125b, MDMs were transfected with anti-miR miRNA inhibitor for miR-125b or negative control (100 nM) using an Amaxa Nucleofector. The transfectants were stimulated with TB-LM or SmegLM, and TNF was measured by ELISA.
MicroRNA Assay.
MDMs were stimulated with LMs or infected with M.tb or M. smegmatis for 6 h in triplicate wells, and cells were lysed for total RNA extraction using the method described above. cDNA was synthesized using a Taqman MicroRNA reverse-transcription kit (Applied Biosystems) and the specific reverse-transcription primers for hsa-miR-125b, hsa-miR-155, and housekeeping small RNA RNU44 (Applied Biosystems). Real-time PCR reactions were prepared by mixing miRNA-specific cDNA, Taqman primers with Taqman Universal PCR master mix, and No AmpErase UNG (Applied Biosystems) and run in a BioRad CFX96 Real-Time System. Control reactions containing no reverse transcriptase and no cDNA template were performed as well. Triplicate samples were analyzed in duplicate by qRT-PCR.
Additional information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants AI059639 and AI052458 (to L.S.S.) and GM038277 (to D.R.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112660108/-/DCSupplemental.
References
- 1.Schlesinger LS. Phagocytosis and Toll-like receptors in tuberculosis. In: Rom WN, Garay SM, editors. Tuberculosis. 2nd Ed. Philadelphia: Lippincott, Williams, & Wilkins; 2004. pp. 203–214. [Google Scholar]
- 2.Torrelles JB, Schlesinger LS. Diversity in Mycobacterium tuberculosis mannosylated cell wall determinants impacts adaptation to the host. Tuberculosis (Edinb) 2010;90(2):84–93. doi: 10.1016/j.tube.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quesniaux VJ, et al. Toll-like receptor 2 (TLR2)-dependent–positive and TLR2-independent–negative regulation of proinflammatory cytokines by mycobacterial lipomannans. J Immunol. 2004;172:4425–4434. doi: 10.4049/jimmunol.172.7.4425. [DOI] [PubMed] [Google Scholar]
- 4.Vignal C, et al. Lipomannans, but not lipoarabinomannans, purified from Mycobacterium chelonae and Mycobacterium kansasii induce TNF-α and IL-8 secretion by a CD14 Toll-like receptor 2-dependent mechanism. J Immunol. 2003;171:2014–2023. doi: 10.4049/jimmunol.171.4.2014. [DOI] [PubMed] [Google Scholar]
- 5.Stenger S. Immunological control of tuberculosis: Role of tumour necrosis factor and more. Ann Rheum Dis. 2005;64(Suppl 4):iv24–iv28. doi: 10.1136/ard.2005.042531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yadav M, Roach SK, Schorey JS. Increased mitogen-activated protein kinase activity and TNF-α production associated with Mycobacterium smegmatis- but not Mycobacterium avium-infected macrophages requires prolonged stimulation of the calmodulin/calmodulin kinase and cyclic AMP/protein kinase A pathways. J Immunol. 2004;172:5588–5597. doi: 10.4049/jimmunol.172.9.5588. [DOI] [PubMed] [Google Scholar]
- 7.Chen YL, et al. Differential regulation of ARE-mediated TNFα and IL-1β mRNA stability by lipopolysaccharide in RAW264.7 cells. Biochem Biophys Res Commun. 2006;346(1):160–168. doi: 10.1016/j.bbrc.2006.05.093. [DOI] [PubMed] [Google Scholar]
- 8.Piecyk M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brook M, et al. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol Cell Biol. 2006;26:2408–2418. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitti E, et al. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol. 2006;26:2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: Non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 12.Chen CY, et al. AU-binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 13.Jing Q, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 14.Tili E, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 15.O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremer TJ, et al. MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS ONE. 2009;4:e8508. doi: 10.1371/journal.pone.0008508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlesinger LS. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J Immunol. 1993;150:2920–2930. [PubMed] [Google Scholar]
- 18.Briken V, Porcelli SA, Besra GS, Kremer L. Mycobacterial lipoarabinomannan and related lipoglycans: From biogenesis to modulation of the immune response. Mol Microbiol. 2004;53:391–403. doi: 10.1111/j.1365-2958.2004.04183.x. [DOI] [PubMed] [Google Scholar]
- 19.Basler T, et al. Reduced transcript stabilization restricts TNF-α expression in RAW264.7 macrophages infected with pathogenic mycobacteria: Evidence for an involvement of lipomannan. J Leukoc Biol. 2010;87(1):173–183. doi: 10.1189/jlb.0309207. [DOI] [PubMed] [Google Scholar]
- 20.Han J, Beutler B. The essential role of the UA-rich sequence in endotoxin-induced cachectin/TNF synthesis. Eur Cytokine Netw. 1990;1(2):71–75. [PubMed] [Google Scholar]
- 21.Cao Y, et al. IKKα provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell. 2001;107:763–775. doi: 10.1016/s0092-8674(01)00599-2. [DOI] [PubMed] [Google Scholar]
- 22.Otsuka Y, Schoenberg DR. Approaches for studying PMR1 endonuclease-mediated mRNA decay. Methods Enzymol. 2008;448:241–263. doi: 10.1016/S0076-6879(08)02613-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neininger A, et al. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J Biol Chem. 2002;277:3065–3068. doi: 10.1074/jbc.C100685200. [DOI] [PubMed] [Google Scholar]
- 24.Bala S, et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2010;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bermudez LE, Young LS. Tumor necrosis factor, alone or in combination with IL-2, but not IFN-γ, is associated with macrophage killing of Mycobacterium avium complex. J Immunol. 1988;140:3006–3013. [PubMed] [Google Scholar]
- 26.Flynn JL, et al. Tumor necrosis factor-α is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 27.Beltan E, Horgen L, Rastogi N. Secretion of cytokines by human macrophages upon infection by pathogenic and non-pathogenic mycobacteria. Microb Pathog. 2000;28:313–318. doi: 10.1006/mpat.1999.0345. [DOI] [PubMed] [Google Scholar]
- 28.Doz E, et al. Acylation determines the Toll-like receptor (TLR)-dependent positive versus TLR2-, mannose receptor-, and SIGNR1-independent negative regulation of pro-inflammatory cytokines by mycobacterial lipomannan. J Biol Chem. 2007;282:26014–26025. doi: 10.1074/jbc.M702690200. [DOI] [PubMed] [Google Scholar]
- 29.Gilleron M, Quesniaux VF, Puzo G. Acylation state of the phosphatidylinositol hexamannosides from Mycobacterium bovis bacillus Calmette Guérin and Mycobacterium tuberculosis H37Rv and its implication in Toll-like receptor response. J Biol Chem. 2003;278:29880–29889. doi: 10.1074/jbc.M303446200. [DOI] [PubMed] [Google Scholar]
- 30.Nigou J, et al. Mannan chain length controls lipoglycans signaling via and binding to TLR2. J Immunol. 2008;180:6696–6702. doi: 10.4049/jimmunol.180.10.6696. [DOI] [PubMed] [Google Scholar]
- 31.Torrelles JB, et al. Structural differences in lipomannans from pathogenic and nonpathogenic mycobacteria that impact CD1b-restricted T-cell responses. J Biol Chem. 2011 doi: 10.1074/jbc.M111.232587. 10.1074/jbc.M111.232587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schorey JS, Cooper AM. Macrophage signalling upon mycobacterial infection: the MAP kinases lead the way. Cell Microbiol. 2003;5(3):133–142. doi: 10.1046/j.1462-5822.2003.00263.x. [DOI] [PubMed] [Google Scholar]
- 33.Gaestel M. MAPKAP kinases - MKs - two's company, three's a crowd. Nat Rev Mol Cell Biol. 2006;7(2):120–130. doi: 10.1038/nrm1834. [DOI] [PubMed] [Google Scholar]
- 34.Kotlyarov A, et al. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat Cell Biol. 1999;1(2):94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 35.Lehner MD, et al. Mitogen-activated protein kinase-activated protein kinase 2-deficient mice show increased susceptibility to Listeria monocytogenes infection. J Immunol. 2002;168:4667–4673. doi: 10.4049/jimmunol.168.9.4667. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J, et al. MAPK-activated protein kinase 2 differentially regulates plasmodium falciparum glycosylphosphatidylinositol-induced production of tumor necrosis factor-α and interleukin-12 in macrophages. J Biol Chem. 2009;284:15750–15761. doi: 10.1074/jbc.M901111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 38.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 39.Murphy AJ, Guyre PM, Pioli PA. Estradiol suppresses NF-&kappaB activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. J Immunol. 2010;184:5029–5037. doi: 10.4049/jimmunol.0903463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao B, et al. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J Infect Dis. 2009;200:916–925. doi: 10.1086/605443. [DOI] [PubMed] [Google Scholar]
- 41.Wood CD, Thornton TM, Sabio G, Davis RA, Rincon M. Nuclear localization of p38 MAPK in response to DNA damage. Int J Biol Sci. 2009;5:428–437. doi: 10.7150/ijbs.5.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeidan A, Javadov S, Chakrabarti S, Karmazyn M. Leptin-induced cardiomyocyte hypertrophy involves selective caveolae and RhoA/ROCK-dependent p38 MAPK translocation to nuclei. Cardiovasc Res. 2008;77(1):64–72. doi: 10.1093/cvr/cvm020. [DOI] [PubMed] [Google Scholar]
- 43.Gong X, Ming X, Deng P, Jiang Y. Mechanisms regulating the nuclear translocation of p38 MAP kinase. J Cell Biochem. 2010;110:1420–1429. doi: 10.1002/jcb.22675. [DOI] [PubMed] [Google Scholar]
- 44.Rajaram MV, et al. Mycobacterium tuberculosis activates human macrophage peroxisome proliferator-activated receptor γ linking mannose receptor recognition to regulation of immune responses. J Immunol. 2010;185:929–942. doi: 10.4049/jimmunol.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.