Abstract
The suppression of oncogenic levels of MYC is sufficient to induce sustained tumor regression associated with proliferative arrest, differentiation, cellular senescence, and/or apoptosis, a phenomenon known as oncogene addiction. However, after prolonged inactivation of MYC in a conditional transgenic mouse model of Eμ-tTA/tetO-MYC T-cell acute lymphoblastic leukemia, some of the tumors recur, recapitulating what is frequently observed in human tumors in response to targeted therapies. Here we report that these recurring lymphomas express either transgenic or endogenous Myc, albeit in many cases at levels below those in the original tumor, suggesting that tumors continue to be addicted to MYC. Many of the recurring lymphomas (76%) harbored mutations in the tetracycline transactivator, resulting in expression of the MYC transgene even in the presence of doxycycline. Some of the remaining recurring tumors expressed high levels of endogenous Myc, which was associated with a genomic rearrangement of the endogenous Myc locus or activation of Notch1. By gene expression profiling, we confirmed that the primary and recurring tumors have highly similar transcriptomes. Importantly, shRNA-mediated suppression of the high levels of MYC in recurring tumors elicited both suppression of proliferation and increased apoptosis, confirming that these tumors remain oncogene addicted. These results suggest that tumors induced by MYC remain addicted to overexpression of this oncogene.
Keywords: T-cell acute lymphoblastic leukemia, therapeutic resistance, tumor recurrence
The c-MYC proto-oncogene encodes for a basic helix–loop–helix leucine zipper transcription factor that is a key regulator of many cellular processes, including proliferation, growth, apoptosis, metabolism, adhesion, angiogenesis, and DNA repair (1, 2). The deregulated expression of c-MYC (MYC) has been linked to the pathogenesis of a wide variety of human cancers, in particular hematopoietic tumors such as Burkitt's lymphoma and T-cell acute lymphoblastic leukemia (T-ALL), as well as breast, prostate, colon, and liver adenocarcinoma (3, 4). MYC is thought to contribute to neoplasia by deregulating the expression of a large number of genes, resulting in autonomous cellular proliferation and growth, inhibition of differentiation, increased angiogenesis, and genomic destabilization (5).
The essential role of MYC in tumorigenesis suggests that its targeted inactivation may be a highly effective therapy for a large variety of human cancers (6). A number of conditional transgenic mouse models using the tetracycline-regulated system (Tet-system) have been generated to study the role of MYC in both the initiation and maintenance of cancer (7–14). The inactivation of MYC in these tumors is generally, but not always, sufficient to induce sustained tumor regression, although the specific consequences depend upon the tumor type. T-ALLs regress through proliferative arrest, differentiation, senescence, and apoptosis (9, 15). Osteosarcomas regress through proliferative arrest, terminal differentiation, and senescence but not apoptosis (10, 16). Hepatocellular adenocarcinomas undergo proliferative arrest, differentiation, senescence, and apoptosis of many but not all of the cells, leaving a residual population of dormant tumor cells that are able to reacquire a neoplastic phenotype upon MYC reactivation (13). Finally, mammary and lung adenocarcinomas fail to exhibit complete tumor regression upon MYC inactivation (7, 14). Taken together, MYC-induced tumors generally exhibit continued dependence upon this oncogene, a phenomenon known as oncogene addiction (17–19).
Oncogene addiction has been increasingly exploited for the treatment of human cancers. Chronic myeloid leukemia and non–small-cell lung cancer are highly dependent on the activity of the BCR-ABL fusion protein and mutant EGFR, respectively (20, 21). Therapies that target these proteins, such as imatinib against BCR-ABL and erlotinib against EGFR, have proven clinically efficacious (22, 23). However, despite an initial response to treatment, tumors frequently recur, and drug resistance is often associated with mutations in the targeted oncogene product (24, 25). Different mechanisms have been proposed regarding how tumors acquire resistance to targeted therapy, including mutations in the targeted oncogene that impair drug sensitivity and the acquisition of additional genomic abnormalities (7, 26, 27).
We have previously reported that in our conditional transgenic model of MYC-induced T-ALL, MYC inactivation induces sustained tumor regression, but a subset of tumors will recur (9, 26, 28). Recurring lymphomas frequently exhibit unique genomic lesions not present in the primary tumor (28). Lack of p53 is also uniformly associated with tumor recurrence at least in part due to an inability of MYC inactivation to induce TSP-1 and thereby suppress angiogenesis (26). Similarly, it has been shown that mutation of K-Ras is associated with tumor recurrence upon MYC inactivation in mammary adenocarcinomas (7, 29). Here we report that recurring tumors in our model of MYC-induced T-ALL continue to remain dependent upon the oncogene. Our results suggest that although MYC-induced tumors can restore MYC expression through a variety of different mechanisms, they may uniformly remain oncogene addicted.
Results
Recurring T-ALL Tumors Are Phenotypically Identical to Primary Tumors.
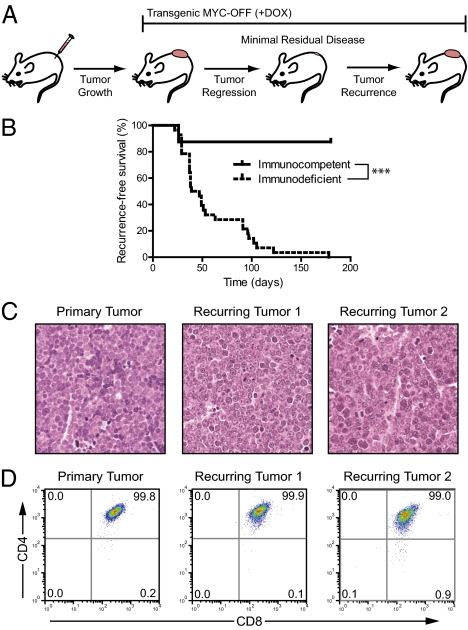
In the Eμ-tTA/tetO-MYC transgenic mouse model, the expression of MYC in hematopoietic cells gives rise to T-ALL (9). Primary tumors, as well as cell lines derived from these tumors, can be transplanted into immunodeficient or immunocompetent syngeneic hosts and undergo rapid tumor regression in response to MYC inactivation upon administration of doxycycline (DOX) to mice (9). However, some of the T-ALLs fail to regress completely and eventually recur (Fig. 1A). In this regard, these tumors mimic what is observed in targeted therapy for human cancer patients.
Fig. 1.
Recurring T-cell lymphomas are phenotypically identical to primary tumors. (A) Schematic of tumor development, regression, and recurrence. (B) Kaplan-Meier survival curve of mice transplanted with lymphoma-derived cell lines and monitored for tumor recurrence (n = 8 for immunocompetent and n = 28 for immunodeficient; ***P < 0.0001 by log–rank test). (C) Representative H&E-stained sections of a primary tumor and two recurring tumors. (D) Cell surface CD4 vs. CD8 expression in a primary tumor and two recurring tumors.
To study the mechanism of recurrence, we generated cohorts of transplanted tumors that were either untreated (referred to as “primary tumors,” n = 8) or treated with DOX to suppress MYC expression (n = 28). Tumors that initially regressed upon DOX treatment until they were no longer detectable by gross examination but then were again palpable after a minimum of 1 wk were defined as “recurring tumors.” Tumors in immunodeficient hosts all recurred (100%), with a median time to recurrence of 43 d, whereas tumor recurrence was infrequent in immunocompetent hosts (12.5%) (Fig. 1B). Primary and recurring tumors were identical histologically (Fig. 1C) and by cell surface phenotype (Fig. 1D). Hence, MYC-induced T-ALLs can recur, especially in immunodeficient hosts, and recurring tumors are phenotypically similar to primary tumors.
Recurring T-ALLs Express MYC.
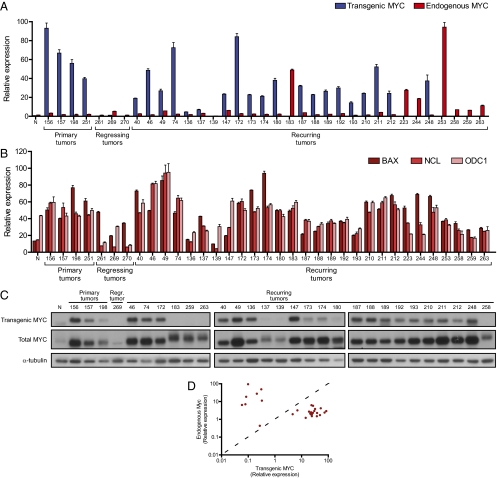
Because primary and recurring tumors were phenotypically identical, we asked whether recurring tumors had become independent of MYC. First we quantified the levels of transgenic human MYC and endogenous mouse Myc in a panel of three primary tumors, three regressing tumors, and 28 recurring T-ALLs (continuous DOX treatment) using RT–quantitative PCR (qPCR), Western blot, and immunohistochemical analysis (Fig. 2 and Fig. S1). As expected, primary T-ALLs expressed high levels of transgenic MYC, and treatment of mice with DOX caused at least a 230-fold decrease in transgenic MYC levels (Fig. 2A). In these tumors, the expression level of endogenous Myc was comparable to a normal thymus control. However, among the recurring tumors, we found that 19 of 28 tumors expressed transgenic MYC at levels similar to those in primary tumors, and 9 tumors instead overexpressed endogenous Myc (47-fold higher than in primary tumors or recurring tumors with transgenic MYC) (Fig. 2A and Table S1). Elevated levels of transgenic and endogenous MYC in recurring tumors were also confirmed at the protein level (Fig. 2C and Fig. S1). Among all of the recurring tumors we examined, we found none that had simultaneous coexpression of transgenic MYC or endogenous mouse Myc (Fig. 2D). Taken together, we conclude that recurring T-ALLs continue to express high levels of transgenic MYC or endogenous Myc.
Fig. 2.
Recurring lymphomas express either transgenic or endogenous Myc. (A) Expression of transgenic and endogenous Myc in transplanted primary, regressing (2 d of DOX), and recurring lymphomas, as determined by RT-qPCR. N, normal thymus control. (B) Transcript levels of the MYC target genes BAX, NCL, and ODC1 as determined by RT-qPCR. (C) Western blot analysis of transgenic and total MYC protein in primary, regressing, and recurring lymphomas. (D) Scatter plot summarizing Myc transcript levels in all analyzed recurring tumors. Each point represents a single sample (n = 28), with levels of transgenic MYC (x axis) vs. endogenous mouse Myc (y axis).
To determine whether the amount of MYC expression in recurring tumors was sufficient to recapitulate the level of MYC target gene activation found in primary tumors, we performed qPCR analysis for the canonical MYC target genes Bax, Ncl, and Odc1 (30–32), in a set of primary, regressing, and recurring tumors. Notably, across all primary and recurring tumors analyzed, MYC target gene levels were closely correlated with the amount of MYC expression (Fig. 2B). Expression of both MYC and MYC target genes in recurring tumors was comparable to that in primary tumors, suggesting that MYC in recurring tumors was not only overexpressed but functionally capable of maintaining target gene expression.
Recurring Tumors Have Frequently Mutated the Tetracycline Transactivator.
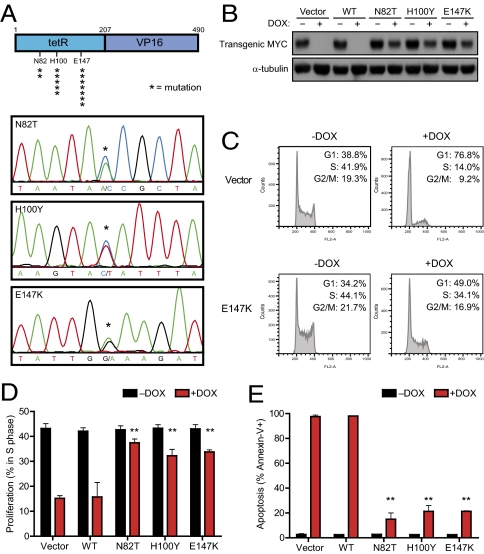
A previous report has suggested that mutations in the Tet-system can explain continued expression of an oncogene in the presence of DOX (33). We sequenced the tetracycline transactivating (tTA) gene in all 28 recurring tumors (Table S1) and found mutations in the majority of recurring tumors that expressed transgenic MYC (74%). We identified three different mutations (N82T, H100Y, and E147K), all corresponding to the DOX-responsive tet-repressor (tetR) domain of tTA (Fig. 3A) (34). To confirm that these mutations altered the sensitivity of tTA to DOX, each mutant tTA was introduced into an Eμ-tTA/tetO-MYC T-ALL cell line. In contrast to wild-type tTA, the activity of the N82T, H100Y, or E147K mutant tTA was no longer inhibited by DOX treatment, and as a consequence, expression of the MYC transgene was not completely suppressed (Fig. 3B). Similarly, lymphoma cells expressing a mutant tTA failed to undergo proliferative arrest or apoptosis upon DOX treatment (Fig. 3 C–E). Thus, mutations in the tTA protein can account for expression of transgenic MYC despite treatment with DOX.
Fig. 3.
Transgenic MYC is reactivated owing to mutations in the tTA protein. (A) Schematic of the tTA protein (asterisks indicate the position and frequency of mutated amino acids) and example chromatograms for the N82T, H100Y, and E147K mutations. (B) Mutant tTAs cloned from recurring lymphomas are insensitive to DOX. A MYC lymphoma line was stably infected with empty vector, wildtype tTA (WT), or mutant tTAs (N82T, H100Y, E147K). Cells were then treated with DOX and analyzed for (B) expression of tTA-dependent transgenic MYC by Western blot (24 h DOX), (C and D) cell cycle status by propidium iodide staining (48 h DOX), or (E) apoptosis rate by Annexin-V staining (5 d DOX). Mean values of a minimum of three independent experiments are shown with SDs where applicable. **P < 0.01, paired t test.
Recurring T-ALLs Overexpress Endogenous Myc Through Multiple Mechanisms.
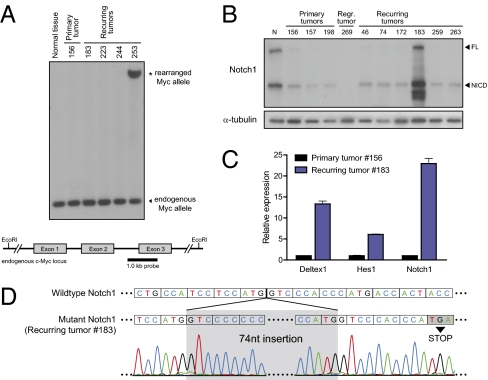
We noted that in the subset of tumors that exhibited high levels of endogenous Myc (9 of 28), there were no tTA mutations. In many human hematopoietic cancers, including Burkitt's lymphoma, overexpression of MYC is mediated by either genomic rearrangement or amplification (35). To determine whether a similar mechanism was responsible in our recurring lymphomas, we examined the endogenous murine c-Myc locus by Southern blotting (Fig. 4A) and identified a rearrangement in one of the recurring lymphomas that had endogenous Myc overexpression (Fig. 4A). Translocations of Myc are also often associated with point mutations that increase the stability of Myc protein (36). However, sequencing of Myc exons 2 and 3 did not reveal mutations in any of the recurring lymphomas with endogenous Myc overexpression. We infer that genomic rearrangement is only one of the possible mechanisms through which recurring tumors restore Myc expression.
Fig. 4.
Up-regulation of endogenous Myc in recurring lymphomas occurs through multiple mechanisms. (A) Southern blot of genomic DNA from normal tissue, a primary tumor, and five recurring tumors, digested with EcoRI and probed with a 1-kb probe specific for the endogenous mouse c-Myc locus. (B) Western blot for Notch1 in a subset of primary and recurring lymphomas. FL, full length; NICD, active intracellular domain. (C) Expression of Notch pathway target genes as determined by RT-qPCR analysis. (D) Schematic depicting the Notch1 gene mutation found in recurring lymphoma #183. Boxes denote codons.
Endogenous Myc can also be up-regulated through activation of signaling pathways that regulate Myc protein stability and/or transcription, such as the Notch, MAPK, and Wnt pathways (37–39). Although we found no consistent difference in activation of the MAPK and Wnt pathways between primary and recurring tumors (Fig. S2), we found that one recurring tumor (#183) significantly overexpressed Notch1 protein (Fig. 4B). In human T-ALL, Notch1 is often activated through mutation of the heterodimerization and/or proline, glutamate, serine, threonine-rich (PEST) degradation domains (40). Sequencing revealed an insertion of 74 nucleotides within the PEST domain in recurring tumor #183, leading to a premature stop codon (Fig. 4D). We also found up-regulation of the Notch target genes Deltex1, Hes1, and Notch1 (41) in recurring tumor #183. We did not detect Notch1 mutations or Notch1 overexpression in any of the other recurring tumors (Fig. S2). Thus, activation of Notch1 is another mechanism by which recurring tumors can gain endogenous Myc overexpression.
Primary and Recurring Tumors Have Similar Gene Expression Programs.
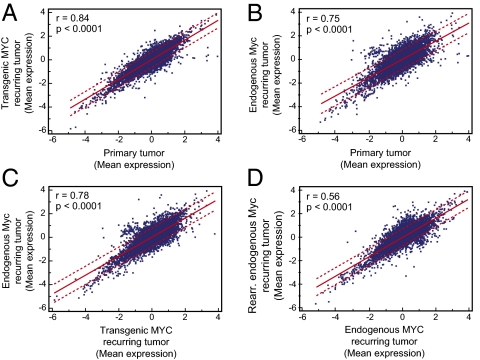
To globally compare the gene expression profiles of primary lymphomas with recurring tumors, we used microarray analysis on a total of 24 different tumor specimens consisting of primary tumors (group I), recurring tumors exhibiting transgenic MYC expression (group II), and recurring tumors in which endogenous Myc was overexpressed (group III). Differentially expressed genes between these groups were identified using the rank product method (42). We compared primary (group I) with recurring tumors exhibiting transgenic MYC expression (group II) and recurring tumors exhibiting endogenous Myc expression (group III) independently. Primary tumors and recurring tumors expressing transgenic MYC were the most similar (Fig. 5A, r = 0.84, P < 0.0001). However, gene expression was also highly similar between primary tumors and recurring tumors that overexpressed endogenous Myc (Fig. 5B, r = 0.75, P < 0.0001). Gene expression was also very similar between recurring tumors expressing transgenic MYC and recurring tumors expressing endogenous Myc (Fig. 5C, r = 0.78, P < 0.0001). The lowest similarity was found between recurring tumors expressing endogenous Myc and recurring tumors expressing endogenous Myc from a rearranged locus (Fig. 5D, r = 0.56, P < 0.0001). Therefore, microarray analysis confirmed that primary and recurring lymphomas exhibited similar gene expression profiles.
Fig. 5.
Microarray comparison of primary and recurring lymphomas. Correlation plots comparing mean expression of genes in (A) primary tumor vs. recurring tumor expressing transgenic MYC, (B) primary tumor vs. recurring tumor expressing endogenous Myc, (C) recurring tumor expressing transgenic MYC vs. recurring tumor expressing endogenous Myc, and (D) recurring tumor expressing endogenous Myc vs. recurring tumor expressing endogenous Myc with genomic rearrangement.
However, not all genes were identical between primary and recurring tumors. We found differential expression of 169 genes (85 up-, 84 down-regulated) between group I and II, and 34 genes (2 up-, 32 down-regulated) between group I and III (Table S2). Between recurring tumors expressing transgenic MYC and recurring tumors expressing endogenous Myc, we found only 32 genes (27 up-, 5 down-regulated) differentially expressed (Table S3). Thus, although the vast majority of genes were expressed similarly, there were still some differences in gene expression between primary and recurrent tumors and between recurring tumors expressing either transgenic or endogenous Myc.
We conclude that the expression of the MYC transgene or overexpression of endogenous Myc was sufficient to completely restore the transcriptome found in primary tumors. Moreover, although there were variations in the absolute level of MYC expression levels, all of the analyzed tumors showed a very similar global gene expression signature, strongly suggesting that MYC acts as a dominating influence on tumor recurrence.
Recurring T-ALLs Remain Addicted to High Levels of MYC.
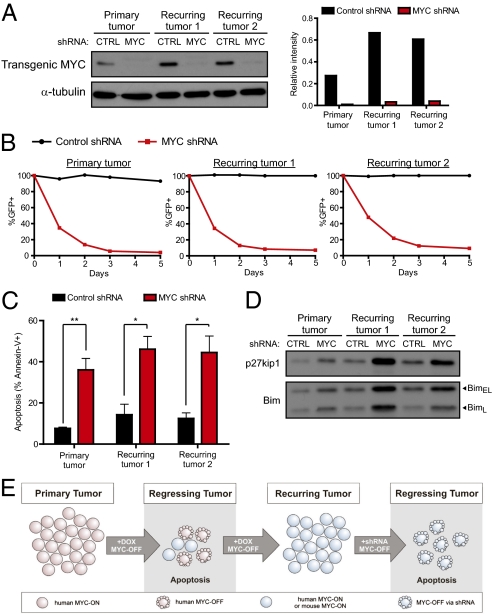
To directly determine whether recurring tumors remained addicted to MYC, we measured the consequences of suppressing MYC using RNAi. In a primary tumor cell line and two lines derived from recurring lymphomas expressing human transgenic MYC (Fig. S3), we introduced either control shRNA or an shRNA against human MYC (Fig. 6A). Cells infected with MYC shRNA failed to proliferate and decreased over time as a percentage of the overall population (Fig. 6B), and MYC knockdown in both the primary and recurring tumor lines led to an increase in the cyclin-dependent kinase inhibitor p27KIP1 (Fig. 6D). Infection with MYC shRNA also resulted in increased apoptosis, as seen by increased Annexin-V staining (Fig. 6C) and increased levels of the proapoptotic protein Bim (Fig. 6D). Hence, primary and recurring lymphomas remain addicted to high levels of MYC for their proliferation and survival.
Fig. 6.
Knockdown of MYC in recurring lymphomas causes cell cycle arrest and increased apoptosis. (A) Levels of MYC protein in parental and recurring tumors lines infected with either control (CTRL) shRNA or MYC shRNA, which specifically targets human MYC. (B) Time course of the percentage of GFP+ shRNA infected populations after MYC knockdown. Values are normalized to the initial percentage of GFP+ cells after infection. (C) Apoptosis in control or MYC shRNA infected cells, as measured by Annexin-V staining. (D) Western blot on shRNA infected cells for expression of p27KIP1 and Bim. Experiments were repeated a minimum of three times, and representative experiments are shown. *P < 0.01, **P < 0.001, paired t test. (E) Model of tumor recurrence and continued dependence of recurring tumors on MYC.
Discussion
We have found in a conditional transgenic mouse model of MYC-induced T-ALL that despite prolonged suppression of the MYC transgene, recurring tumors express and remain addicted to increased levels of MYC. The recurring tumors expressed either transgenic human MYC or endogenous murine Myc. Restoration of transgenic MYC expression was often associated with a variety of mutations in the DOX-responsive tetR domain of tTA (34) that expands upon previously described tTA mutations (33). Importantly, the recurring tumors without expression of transgenic MYC were instead found to express increased levels of endogenous murine Myc, which are higher than the levels found in normal tissue or regressing tumors. In one case this was attributed to genomic rearrangement of the endogenous Myc locus. In another case this was associated with mutation and activation of Notch1. Finally, the suppression of MYC expression in recurring tumors by shRNA resulted in their proliferative arrest and apoptosis, identical to primary tumors. We conclude that T-ALLs that recur remain MYC oncogene addicted (Fig. 6E). Our results have potentially important implications for the development of therapies that target MYC for the treatment of human cancers.
The up-regulation of endogenous Myc can occur through multiple mechanisms, including genomic rearrangement or amplification of the Myc locus, or activation of pathways that regulate Myc transcription or protein stability, including the Notch, RAS/MAPK, and Wnt/β-catenin pathways (37–39). In one recurring tumor, high levels of endogenous Myc expression were attributed to rearrangement of the endogenous locus. Interestingly, we also found a tumor in which Notch1 mutation had occurred. Notch1 is a known transcriptional regulator of Myc (37, 43, 44). The expression of the active intracellular form of Notch1 (ICN1) is sufficient for up-regulation of endogenous Myc upon transgenic MYC inactivation (37). We were unable to find an explanation for the up-regulation of endogenous Myc in the remaining recurring tumors, and therefore we presume that there are additional uncharacterized mechanisms by which Myc can be up-regulated.
Our results at first seem to be in discordance with our previous report that MYC-induced lymphomas can escape oncogene dependence (28). In this study we observed that recurring tumors have many other genomic abnormalities that seem to be associated with a reduced dependence upon MYC. In fact, further analysis has revealed that the majority of recurring tumors find ways to maintain elevated MYC expression. However, we note that most of the recurring tumors seem to express protein levels of transgenic MYC or endogenous Myc that are lower than in the primary tumors (Fig. 2C). Thus, we infer that these additional genetic lesions seem to diminish the need for recurrent tumors to recapitulate the initial high levels but do not obviate the requirement for MYC. We conclude that MYC-induced hematopoietic tumors are addicted to MYC, but the absolute magnitude of the required level of expression can be reduced in recurrent tumors.
Hence, although oncogene addiction may not be escapable, lymphoma cells seem to be heterogeneous in the levels of MYC they require to maintain their tumorigenic state. Correspondingly, any circumstances that facilitate the ability of tumors to reduce their requirement for MYC would increase tumor recurrence. We and others have described that the loss of tumor suppressor genes, desensitization to inhibitory cytokines, or absence of specific host immune effectors can greatly accelerate the ability of tumors to recur despite initial oncogene suppression. Mutant Ras or the loss of p19ARF have been shown to prevent oncogene inactivation from inducing sustained regression of breast cancers (8, 45). The absence of p53 facilitates tumor recurrence by preventing the induction of the antiangiogenic factor TSP-1 and the consequential shutdown of angiogenesis after MYC inactivation (26). The loss of p53, p16, or Rb expression, or the inability to respond to an autocrine TGF-β signal, can also increase tumor recurrence by impairing the ability of MYC inactivation to induce cellular senescence (15, 16). Finally, the absence of an intact immune system also abrogates MYC inactivation from inducing senescence and suppressing angiogenesis, leading to frequent tumor recurrence (46). Therefore, a variety of both tumor cell-intrinsic and extrinsic factors can each play a pivotal role in facilitating the ability of rare tumor cells that maintain MYC expression to contribute to tumor recurrence.
Our findings provide further optimism that a therapy directly targeting MYC will have sustained clinical activity against human tumors, and lymphoma in particular. However, our results also point to the fact that tumors will incessantly attempt to reacquire high levels of MYC expression. Additionally, a therapeutic agent against MYC has not yet been identified. We note that currently available targeted therapies are often unable to induce sustained tumor regression. In the case of B-Raf inhibition in melanoma or EGFR inhibition in lung cancer, acquired resistance to targeted therapy can occur through the activation of parallel pathways or alternative signaling molecules (47–49). Our results suggest that tumors addicted to MYC may be less likely to achieve resistance through a similar mechanism. MYC may hold a unique position as a convergent downstream target of multiple signaling pathways that are critical to maintenance of a tumorigenic state.
Finally, our results suggest that in the case of MYC-driven cancers, tumor recurrence may not be generally due to an escape of oncogene addiction but rather to the ability to accommodate reduced MYC expression. We speculate that MYC oncogene addiction may be an intransient feature of at least some cancers. Therefore, the targeted inhibition of MYC offers great potential as a therapeutic strategy.
Materials and Methods
Tumorigenicity Assays.
Tumor cell lines were derived from Eμ-tTA/tetO-MYC mice as previously described (9). Cell lines were injected s.c. into recipient mice (7.5–10 × 10E6 cells per mouse). Once tumors had grown to 2 cm3, DOX was administered to the mice in the drinking water (100 μg/mL). Animals were housed in the Stanford vivarium as per animal protocols approved by Stanford University.
Analysis of tTA Sequence and Function.
tTA was amplified by PCR from genomic DNA and directly sequenced (primers described in Table S4). Wild-type and mutant tTAs were cloned into pMSCV-puro and infected into an MYC lymphoma line. Cells were treated with 20 ng/mL of DOX.
Detection of Genomic Rearrangements by Southern Blotting.
Genomic DNA was digested, separated by gel electrophoresis, and transferred to a nitrocellulose membrane using standard protocols. Probes against mouse c-Myc were generated by PCR. Probe labeling, hybridization, and detection were done using the AlkPhos Direct Labeling and Detection System (Amersham).
Sequencing of Myc and Notch1.
Myc exons 2 and 3 and Notch1 exons 26, 27, and 34 were PCR amplified (primers in Table S4) and directly sequenced. Notch1 exon 34 (sample #183) was cloned into pBluescript KS+ to obtain allele-specific sequences.
shRNA-Mediated Knockdown.
Cell lines derived from primary and recurring tumors were infected with the pLKO.1 lentiviral vector containing either control or MYC-specific shRNA (Sigma TRCTRCN0000039642) and GFP.
Supplementary Material
Acknowledgments
We thank members of the D.W.F. laboratory for helpful discussion and suggestions. This work was funded by the Burroughs Welcome Fund, the Damon Runyon Foundation, National Institutes of Health (NIH) RO1 Grant CA 089305, 105102, National Cancer Institute's In-vivo Cellular and Molecular Imaging Center Grant CA 114747, Integrative Cancer Biology Program Grant CA 112973, NIH/National Cancer Institute PO1 Grant CA034233, and Leukaemia and Lymphoma Society Translational Research Grant R6223-07 (to D.W.F.). P.S.C. was funded by NIH Training Grant 5 T32 AI07290, J.v.R. was supported by the Lymphoma Research Foundation, and S.J.A. was supported by National Cancer Institute Public Health Service Grant T32CA09151.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE32341).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107303108/-/DCSupplemental.
References
- 1.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang CV, et al. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 5.Pelengaris S, Khan M. The many faces of c-MYC. Arch Biochem Biophys. 2003;416:129–136. doi: 10.1016/s0003-9861(03)00294-7. [DOI] [PubMed] [Google Scholar]
- 6.Felsher DW. Cancer revoked: Oncogenes as therapeutic targets. Nat Rev Cancer. 2003;3:375–380. doi: 10.1038/nrc1070. [DOI] [PubMed] [Google Scholar]
- 7.Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 8.D'Cruz CM, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 9.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 10.Jain M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science (New York, N.Y.) 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 11.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 12.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 13.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 14.Tran PT, et al. Combined Inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PLoS ONE. 2008;3:e2125. doi: 10.1371/journal.pone.0002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Riggelen J, et al. The interaction between Myc and Miz1 is required to antagonize TGFbeta-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev. 2010;24:1281–1294. doi: 10.1101/gad.585710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CH, et al. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci USA. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsher DW. Oncogene addiction versus oncogene amnesia: Perhaps more than just a bad habit? Cancer Res. 2008;68:3081–3086. doi: 10.1158/0008-5472.CAN-07-5832. discussion 3086. [DOI] [PubMed] [Google Scholar]
- 18.Singh A, Settleman J. Oncogenic K-ras “addiction” and synthetic lethality. Cell Cycle. 2009;8:2676–2677. doi: 10.4161/cc.8.17.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 20.Sawyers CL. Chronic myeloid leukemia. N Engl J Med. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida T, Zhang G, Haura EB. Targeting epidermal growth factor receptor: Central signaling kinase in lung cancer. Biochem Pharmacol. 2010;80:613–623. doi: 10.1016/j.bcp.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 23.Shepherd FA, et al. National Cancer Institute of Canada Clinical Trials Group Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 24.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah NP, Sawyers CL. Mechanisms of resistance to STI571 in Philadelphia chromosome-associated leukemias. Oncogene. 2003;22:7389–7395. doi: 10.1038/sj.onc.1206942. [DOI] [PubMed] [Google Scholar]
- 26.Giuriato S, et al. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc Natl Acad Sci USA. 2006;103:16266–16271. doi: 10.1073/pnas.0608017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah NP, et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science (New York, N.Y.) 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 28.Karlsson A, et al. Genomically complex lymphomas undergo sustained tumor regression upon MYC inactivation unless they acquire novel chromosomal translocations. Blood. 2003;101:2797–2803. doi: 10.1182/blood-2002-10-3091. [DOI] [PubMed] [Google Scholar]
- 29.Jang JW, Boxer RB, Chodosh LA. Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol Cell Biol. 2006;26:8109–8121. doi: 10.1128/MCB.00404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell KO, et al. Bax is a transcriptional target and mediator of c-myc-induced apoptosis. Cancer Res. 2000;60:6318–6325. [PubMed] [Google Scholar]
- 32.Greasley PJ, Bonnard C, Amati B. Myc induces the nucleolin and BN51 genes: Possible implications in ribosome biogenesis. Nucleic Acids Res. 2000;28:446–453. doi: 10.1093/nar/28.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci USA. 2008;105:5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 36.Bahram F, von der Lehr N, Cetinkaya C, Larsson LG. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood. 2000;95:2104–2110. [PubMed] [Google Scholar]
- 37.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 39.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 40.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 41.Yashiro-Ohtani Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 43.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma VM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debies MT, et al. Tumor escape in a Wnt1-dependent mouse breast cancer model is enabled by p19Arf/p53 pathway lesions but not p16 Ink4a loss. J Clin Invest. 2008;118:51–63. doi: 10.1172/JCI33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rakhra K, et al. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell. 2010;18:485–498. doi: 10.1016/j.ccr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science (New York, N.Y.) 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 48.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.