Abstract
Staphylococcal γ-hemolysin is a bicomponent pore-forming toxin composed of LukF and Hlg2. These proteins are expressed as water-soluble monomers and then assemble into the oligomeric pore form on the target cell. Here, we report the crystal structure of the octameric pore form of γ-hemolysin at 2.5 Å resolution, which is the first high-resolution structure of a β-barrel transmembrane protein composed of two proteins reported to date. The octameric assembly consists of four molecules of LukF and Hlg2 located alternately in a circular pattern, which explains the biochemical data accumulated over the past two decades. The structure, in combination with the monomeric forms, demonstrates the elaborate molecular machinery involved in pore formation by two different molecules, in which interprotomer electrostatic interactions using loops connecting β2 and β3 (loop A: Asp43-Lys48 of LukF and Lys37-Lys43 of Hlg2) play pivotal roles as the structural determinants for assembly through unwinding of the N-terminal β-strands (amino-latch) of the adjacent protomer, releasing the transmembrane stem domain folded into a β-sheet in the monomer (prestem), and interaction with the adjacent protomer.
Pathogenic bacteria secrete various virulence factors to attack host cells. The pore-forming toxins (PFTs) are among the most sophisticated virulence factors, and are expressed as water-soluble monomeric proteins that assemble on the membranes of the target cells to form bilayer-spanning pores (1). With the appearance of the pore on the membrane, the cells are killed through leakage. It is interesting to note that PFTs are expressed not only by bacteria but also by eukaryotes, such as the immune proteins perforin and complement C9, suggesting the universality of these molecules in a wide range of organisms (2). PFTs can be classified into two families according to the secondary structure of the transmembrane region in the pore structure; i.e., α-helical PFT (α-PFT) and β-barrel PFT (β-PFTs) (3, 4).
Staphylococcus aureus, a ubiquitous and pernicious human pathogen, secretes several β-PFTs including αHL, γ-hemolysin (γHL), leukocidin (LUK), and Panton–Valentine leukocidin (PVL) (5). The αHL consists of a single polypeptide, whereas the others are bicomponent β-PFTs that require the synergistic association of a class F component and a class S component. The γHL, LUK, and PVL are composed of LukF and Hlg2, LukF and LukS, and LukF-PV and LukS-PV, as class F and S components, respectively. The components of bicomponent β-PFTs are similar to each other and to αHL in amino acid sequence: Within a class, S and F proteins are approximately 70% identical, whereas between classes the identity drops to approximately 30%. Class F proteins are more closely related to αHL (approximately 30%) than class S proteins (approximately 20%) (5, 6). Extensive experiments have been carried out for more than two decades, and have suggested that the pore formation mechanism of bicomponent toxins is as follows (7, 8). The soluble forms of F and S components bind sequentially to the target cells and form a heterodimer (9, 10). Each heterodimer assembles into an oligomer on the target cell to form a ring-shaped particle called a prepore, in which the β-barrel pore is not yet formed (11–14). After forming a stable prepore, the β-barrel pore is formed. Pore formation requires the binding of phosphatidylcholine (PC) head groups to a cleft in the LukF component surrounded by Trp177 and Arg198 (Trp176 and Arg197 of LukF-PV) (13, 15, 16). The crystal structures of the monomeric forms of bicomponent β-PFTs [i.e., LukF (15), LukF-PV (17), and LukS-PV (18)], have been determined. However, the structures of the pore forms have not been reported at atomic resolution, which has hindered detailed discussion of the complicated molecular mechanism of action of bicomponent pore-forming toxins. Although bicomponent PFTs are found in several species, such as the edible mushroom Pleurotus ostreatus (19), the structures of these pores have not been reported.
One of the most important issues for staphylococcal bicomponent PFTs is the stoichiometry of the class F and S components. Electron microscopy and cross-linking experiments of purified γHL pores on human erythrocyte membranes demonstrated the existence of a heptamer with a 3∶4 or 4∶3 molar ratio of F to S components (20, 21), whereas biochemical analyses of pores of engineered covalent γHL heterodimers on erythrocytes and leukocytes suggested octameric stoichiometry (22). Several reports using LUK pores formed on rabbit erythrocytes also demonstrated the existence of an octamer consisting of 4-plus-4 subunits (23–25). However, hexamer was also proposed based on structure modeling using monomeric structures of PVL (17) and electron microscopy of LUK pores on leukocytes and erythrocytes (26). It is also important to determine the significance of using two components. Although a role was proposed for the F component in initiating the pore formation process (27), that of the S component remains unclear.
In the present study, we determined the crystal structure of the pore form of bicomponent β-PFT, γHL. This is a unique report of the crystal structure of a heterocomponent β-barrel- type transmembrane protein. This is also a unique bicomponent β-PFT of which both monomer- and pore-form structures have been determined by X-ray crystallography, which allowed us to discuss the pore formation mechanism based on their real structures at atomic resolution. Based on the structural differences between pore and monomer forms in combination with biological data accumulated over the past two decades, we propose a mechanism of pore formation by β-PFTs along with the roles of each component. The electrochemical properties of the pore are also discussed from a structural viewpoint.
Results
Molecular Architecture of γHL Pore.
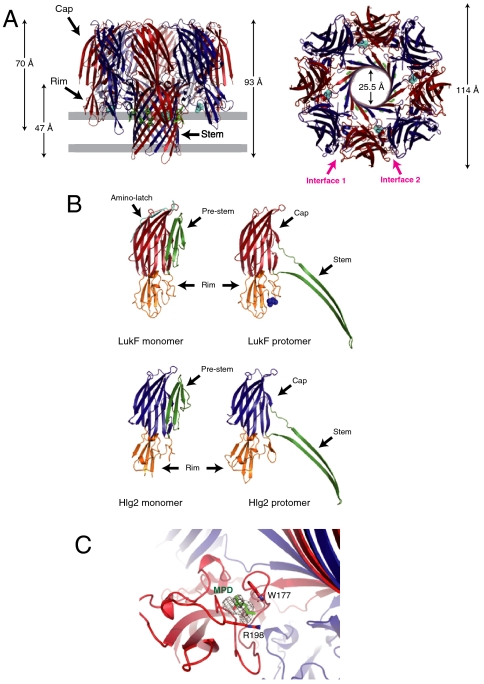
Recently, we found that monomeric αHL in the presence of 2-methyl-2,4-pentanediol (MPD) can spontaneously form a pore structure identical to that in the biological membrane (28). We applied this method to prepare crystals of γHL pores. Purified monomeric soluble LukF and Hlg2 formed SDS-resistant oligomers in the presence of 20% MPD, suggesting that MPD can induce pore formation of γHL (Fig. S1). Fortunately, crystals were grown from a buffer containing 50% MPD, and the structure was determined by the molecular replacement method at a resolution of 2.5 Å. The revealed structure was an octameric pore form consisting of four molecules of each of LukF and Hlg2 (Fig. 1A). Each component was located alternately in a circular pattern along a noncrystallographic fourfold axis. The height and diameter of the octameric structure were 93 and 114 Å, respectively. In analogy to the heptameric structure of αHL (29, 30), the protomer was composed of cap, rim, and stem domains (Fig. 1B). The cap domain interacts with those of adjacent protomers, and the rim domain is located beneath the cap domain. The height of the ellipsoidal aqueous domain composed of cap and rim domains was approximately 70 Å. The protruding stem domains formed a transmembrane β-barrel 25 Å in diameter and 47 Å in height (measured from Cα positions), composed of 16 antiparallel β-strands from eight protomers (four protomers from each of LukF and LukS). The N-terminal amino-latches, which participate in the interprotomer interaction in the heptameric pore structure of αHL (28–30), were disordered. MPD molecules were bound at the base of the rim domain of four LukF protomers, and recognized by Trp177 and Arg198, which participate in recognition of the lipid head group in the structure of monomeric LukF (15) (Fig. 1C). In heptameric αHL, the lipid head group and/or MPD were captured by their corresponding residues (28–30). In contrast, no significant electron density was observed at the same region of Hlg2. An aromatic residue layer, which is commonly observed in transmembrane β-barrel proteins near the lipid/solvent interface (31, 32), was formed by Tyr117, Phe119, and Phe139 of LukF and Tyr111, Phe129, and Tyr131 of Hlg2 (Fig. 1A). MPD molecules were located close to the aromatic layer, which defines the position of the surface of membrane bound to the toxin pore.
Fig. 1.
Overall octameric pore structure of γHL. (A) Side and top views of the heptamer. LukF and Hlg2 are shown in red and blue, respectively. MPD molecules bound with LukF are shown as cyan spheres. The aromatic side chains located around the putative membrane surface are shown as green sticks. The putative membrane region is also shown in gray. (B) Structures of the protomers of LukF (Upper) and Hlg2 (Lower). Monomeric structures of each molecule are also shown. Red, cap of LukF; blue, cap of Hlg2; green, stem (prestem in monomers); orange, rim; cyan, amino-latch. Blue spheres represent MPD bound with LukF protomer. (C) Close-up view of the MPD binding site. The bound MPD, Trp177, and Arg198 are shown as sticks. The Fo-Fc map (contoured at 1.5σ) around the MPD is also shown.
The diameter and height of the octameric pore γHL are 14 Å larger and 4 Å shorter than those of heptameric αHL (100 Å in diameter and 97 Å in height), respectively, and 21 Å and 47 Å shorter, respectively, compared to those of the cytolysin from Vibrio cholerae (VCC) (135 Å in diameter and 140 Å in height) (32), which is a heptameric β-PFT (Fig. S2). The shape of the inner hole of γHL flared out into both sides, and did not have deep constriction at the top of the β-barrel as observed for αHL and VCC, although Lys108 and Gln118 of Hlg2 formed a shallow constriction at the middle (Fig. S2). The vestibule formed by the cap domains is also wider than in other molecules, which is due to the absence of the amino-latch and the abundance of protomers.
Interprotomer Interactions.
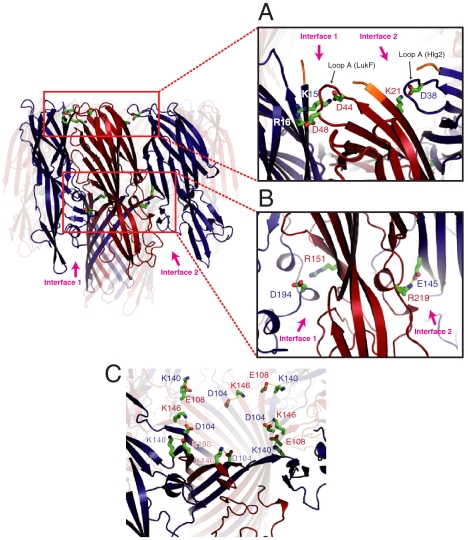
There are two types of interface between protomers in the γHL octamer (Fig. 1A), [i.e., the interface between the left side of LukF and the right side of Hlg2 (hereafter, interface 1)], and that between the right side of LukF and the left side of Hlg2 (interface 2). The buried surface areas at interfaces 1 and 2 were calculated as approximately 1148 Å2 and 963 Å2, respectively. Interactions between the cap domains made predominant contributions in both interfaces, in which more than 160 atoms from each protomer were involved (Table S1). In interface 1, an interprotomer electrostatic interaction cluster, was observed between a loop connecting β2 and β3 of LukF (Asp43–Lys48, referred to as loop A below) and β1 of Hlg2, to which Asp44 and Asp48 of LukF and Lys15 and Arg16 of Hlg2 contributed (Fig. 2A). In a similar region of interface 2, Asp38 located in loop A of Hlg2 (Lys37–Lys43) formed an electrostatic interaction with Lys21 in β1 of LukF (Fig. 2A), suggesting that the electrostatic interaction using loop A is conserved in both interfaces. Moreover, there was an additional ionic interaction between Arg219 of LukF and Glu145 of Hlg2 at interface 2 (Fig. 2B). Around the corresponding region of interface 1, Arg151 of LukF and Asp194 of Hlg2 came into close apposition (distance between Cα atoms: 10.4 Å), although they did not form electrostatic interactions (Fig. 2B).
Fig. 2.
Electrostatic interactions at the interprotomer interface. (A) Interactions between loop A and β-strand 1. The N-terminal residues that are not necessary for pore formation are shown in orange. (B) Electrostatic interaction between Arg219 (LukF) and Glu145 (Hlg2) located at interface 2. Structurally corresponding residues at interface 1 [Asp194 (Hlg2) and Arg151 (LukF)] are also shown. (C) Ion pairs stabilizing the upper end of the β-barrel.
The stem region was composed of an interprotomer antiparallel β-barrel, in which 34 interprotomer hydrogen bonds were formed (Fig. 3C). The upper ends of the interprotomer β-sheets were stabilized by two ion pairs between Glu108 (LukF) and Lys140 (Hlg2), and Lys146 (LukF) and Asp104 (Hlg2) (Fig. 2C). As Lys140 and Asp140 of Hlg2 are substituted in Gln and Asn in LukS and LukS-PV, these interprotomer ion pairs would be a characteristic of γHL.
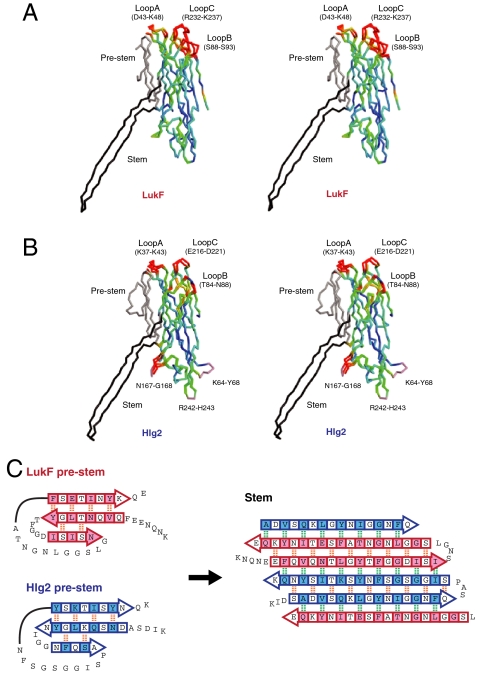
Fig. 3.
Structure comparison between monomer and protomer. (A and B) Stereo representations of the superposed monomer and protomer LukF (A) and Hlg2 (B). The Cα trace of each molecule is colored according to the distance between Cα atoms of corresponding residues in monomer and protomer from blue (0 Å) to red (1.5 Å). Residues with Cα distances greater than 1.5 Å are shown in red. The prestem and stem are shown in gray and black, respectively. Loops disordered in the monomer are also shown in pink. (C) Schematic representation of the structural change between prestem (Left) and stem. Arrows represent β-strands. Intra- and interprotomer backbone hydrogen bonds are shown as orange and green dashed lines, respectively. Residues the side chains of which face the hydrophobic environment (interface between prestem and cap domain in monomers or outside of the β-barrel in octamer) are shown in pink (LukF) and/or blue (Hlg2) shaded boxes.
Structure Comparison of Protomers in Pore Form with Monomeric Components.
In both LukF and Hlg2, the stem domain, which was folded into a three-stranded antiparallel β-sheet as the prestem in the monomer (15, 33), protruded outward to form a transmembrane β-barrel in the octamer (Figs. 1B and 3C). Two β-strands and their connecting loop of the prestem transformed into a β-strand heading toward the bottom of the β-barrel, and a β-strand and the following long loop (which was disordered in LukF monomer) changed into β-strands heading back toward the rim domain (Fig. 3C). Through this process, all 12 hydrogen bonds formed between β-strands in the prestem were disrupted, and instead 18 and 34 new hydrogen bonds were formed within and between protomers, respectively. Interestingly, the carbonyl and amino groups that formed hydrogen bonds between the first and third β-strands of the prestem faced outside of the stem in the octamer, and formed hydrogen bonds with the adjacent protomers. Although the first and second β-strands formed an antiparallel β-sheet in the prestem, they turned toward the same direction and formed one β-strand after transformation. These observations suggested that the secondary structure of the stem region is transiently completely disrupted, and reoriented as a β-barrel with formation of interprotomer interactions.
In addition to the stem domain, three loops located at the top of the cap domain—hereafter, loops A [Asp43–Lys48 (LukF) and Lys37-Lys43 (Hlg2)], B [Ser88–Ser93 (LukF) and Thr84–Asn88 (Hlg2)], and C [Arg232–Lys237 (LukF) and Glu216–Asp221 (Hlg2)], respectively—showed significant conformational changes in both LukF and Hlg2 (Fig. 3 A and B). Loop A formed interprotomer electrostatic interactions with the β-1 strand of the adjacent molecule in both interfaces of the octamer (Fig. 2A), whereas no direct interprotomer interactions by loops B and C were observed. In the monomer, loop A captures the folded prestem (Figs. 1B and 3 A and B). The conformational change of loop A upon oligomer formation is likely to induce structural perturbation of other loops.
The core of the cap and rim domains was well superposed between monomer and protomer [rmsd 1.13 Å and 1.18 Å for LukF (220 Cα atoms) and Hlg2 (243 Cα atoms), respectively], suggesting that these two domains are rigid and that their relative orientation does not change upon octamer assembly. The loops in the rim domain disordered in the Hlg2 monomer (33) (i.e., Asn167–Gly168, Arg242–His243, and Lys64–Tyr68), were clearly observed in the octamer. Asn167–Gly168 was stabilized by interaction with the β-barrel, whereas the other two showed no significant interactions (Fig. 3B).
Discussion
Assembly Mechanism.
Pore formation is initiated by the binding of LukF onto the cell surface, followed by heterodimer formation (10, 34). LukF has two possible interfaces, and it was unclear which interface has priority in heterodimer formation with Hlg2. Our previous study indicated that a heterodimer with shorter distance between Ser45 (LukF) and Lys222 (Hlg2) acts as a structural unit for oligomerization (34). The distance between these residues through interface 1 was approximately 21 Å, whereas that with interface 2 was 43 Å (Fig. S3A), indicating that the heterodimer interacting with interface 1 must be formed first, followed by assembly into an octamer. This was supported by the difference in the buried surface area at interfaces 1 and 2 (1,148 Å2 and 963 Å2, respectively). This is also consistent with the report by Joubert et al. that artificial heterodimers covalently linked by genetically introduced cysteines at Thr28 (Hlg2) and Asn157 (LukF), and/or Thr21 (Hlg2) and Thr158 (LukF), efficiently formed pores (22). In the revealed octameric structure, residues in each pair are located close to each other at interface 1 (Cβ atoms are within 11 Å) (Fig. S3B).
Tyr72, Trp257, Phe260, and Tyr261 of LukF are necessary for stable binding to human erythrocytes (13, 27, 35). All of these residues are located at the bottom of the rim domain in the octameric pore structure. Moreover, the MPD binding residues, Trp177 and Arg198, are located above these residues (Fig. S3C). Although these residues are thought to make contact with the cell surface, the vertical interval between Cα atoms of the bottommost and topmost residues (Phe260 and Trp177, respectively) was 14 Å. This suggested that monomeric LukF binds to the cell surface at an angle with respect to the molecular axis bringing the side chains into contact with the cell surface. Assuming this binding, interface 1 is exposed to the solvent, and consequently, Hlg2 would bind preferentially to LukF with this interface [Fig. 4 (1)]. For the tetrameric assembly, the inclined orientation of the LukF–Hlg2 heterodimer must change to expose interface 2 for the interaction with other dimer molecules. The interaction of Hlg2 with its ligand(s) located at the surface of the membrane, reported to be proteinaceous components (36), would induce reorientation of the heterodimer.
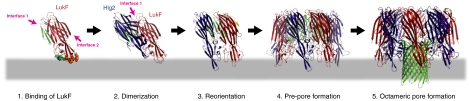
Fig. 4.
Mechanism of assembly for staphylococcal γ-HL. (1) Binding of LukF. LukF binds to the erythrocyte surface via Tyr72, Trp257, Phe260, and Tyr 261 (shown as orange spheres) inclined with respect to its molecular axis. Trp177 and Arg198 (shown as green spheres) capture lipid head groups. As a consequence of the orientation, interface 1 is exposed to the surface, whereas interface 2 is hindered. The membrane is shown as a gray bar. (2) Dimerization. Hlg2 binds to LukF through the surface-exposed interface 1. Upon binding, the amino-latch of Hlg2 is released from the β-sheet due to the structural clash with loop A (LukF). At this step, the prestem is not released completely from the cap domain. (3) Reorientation. The heterodimer changes the orientation to expose interface 2. Binding of Hlg2 with the proteinaceous component(s) may induce this motion. (4) Prepore formation. The heterodimer assembles into a prepore. Upon octameric assembly, the long amino-latch of LukF [yellow in (3)] is released from the cap domain. The electrostatic interactions between loop A and β-1 are formed completely in both interfaces, which induces a conformation change of the prestem into the partially unfolded state and release of the prestem from the cap domain (green loops). (5) Octameric pore formation. The prestem completely unfolds and all hydrogen bonds are disrupted. Unfolded prepore penetrates into the membrane and forms a transmembrane β-barrel (green cartoon).
Based on these observations, the following processes were proposed to occur at the initial step of pore formation: (i) LukF first binds at the cell surface with exposure of interface 1; (ii) LukF is bound by Hlg2 through the exposed interface 1; (iii) the binding of Hlg2 with its receptor then induces reorientation of the heterodimer to expose interface 2; followed by (iv) assembling into tetramer, hexamer, and octamer (Fig. 4).
Function of the N-Terminal Region and Loop A in Interprotomer Interaction.
In the octameric pore structure, the amino-latches of both LukF and Hlg2 were disordered, although that of αHL contributed strongly to interprotomer interactions in the heptameric pore structure, implying that the amino-latches do not contribute to stabilization of octameric assembly. This is consistent with our previous report and that of Miles et al. (37) indicating that the amino-latches of bicomponent β-PFTs are not essential for their hemolytic activity (38, 37). Surprisingly, the essential residues coincided with those that formed interactions with loop A (Fig. 2A). In both LukF and Hlg2, loop A was located alongside the essential N-terminal region of the adjacent protomer, and made electrostatic interactions. These observations indicated a pivotal role of loop A in the interprotomer interaction. In the monomeric state, loop A made interactions with the prestem region, whereas it interacted with the adjacent protomer in the pore structure. These observations indicated that two events (i.e., release of the prestem and formation of the interaction with the adjacent protomer), occur on loop A upon oligomerization. Loop A would act as a clamp for the folded prestem in the monomer, but act as an adhesive material between protomers after releasing the prestem. A similar function was recently also proposed for the corresponding loop in VCC (32).
A Model of the Prepore State.
The heterodimer is known to assemble into a ring-shaped nonlytic oligomeric intermediate called a prepore before formation of the pore (11–13). Formation of the prepore was first identified for staphylococcal αHL (39–42), and is now recognized as the general mechanism adopted by a wide variety of PFTs (43). A structure model of the prepore state oligomer was constructed by superposing monomeric LukF and LukS-PV onto the octamer. The prestems were packed inside the ring composed of the cap domains, and the amino-latches were located at the interface of protomers (Fig. S4A). Several steric constraints were observed at the molecular interfaces, which were mainly due to the prestem and amino-latch regions (Fig. S4B).
Around the top of interface 1, significant steric restraints were observed among loop A and the prestem of LukF, and the amino-latch and cap domain of LukS–PV. Similar steric restraints were also observed at interface 2. Considering the rigidity of the cap domains, the conformational change in the amino-latch and prestem must occur upon oligomer formation. The amino-latch may regulate assembly by steric restraint in a similar manner to the prodomain of VCC, that hinders the interprotomer surface and proteolytic removal of the domain initiates pore formation (32).
A conformational change in the amino-latch was reported to precede those in the prestem and prepore formation (11). Therefore, the release of the amino-latch from the cap domain would occur first in protomer assembly. As the amino-latch showed steric conflict with loop A in both interfaces, loop A is likely to cause the conformation change of the amino-latch. We demonstrated previously that the conformation change of the prestem from a rigid to a flexible structure is essential for prepore formation (11). The transition of the prestem would be coupled with prepore formation, to which interprotomer electrostatic interaction of loop A makes a strong contribution.
Taken together, we propose the following dynamics upon oligomerization: (i) the conformation change of the amino-latch is induced with the aid of loop A upon heterodimer formation; (ii) then each heterodimer assembles into a prepore with formation of electrostatic interactions between loop A and β1; (iii) this causes a conformation change and releases the prestem (Fig. 4).
A Model of Transition from Prepore to Pore.
The MPD molecule added to induce pore formation was bound between Trp177 and Arg198 of LukF, which is known to be a lipid head group binding site. We demonstrated previously that binding of the lipid head group with these residues governs β-barrel formation (13), which is strongly supported by the lack of an MPD molecule in Hlg2 in the octameric structure. In contrast, the crystal structure of monomeric LukF showed that LukF exists as a monomer even if the lipid head group is captured at that site (15). These observations indicate that both oligomerization and capture of the lipid head group are necessary for β-barrel formation.
Structure comparison of stem regions between protomer and monomer clearly showed that its secondary structure is transiently disrupted upon β-barrel formation, strongly supporting the sliding model proposed by Viero et al. in which β-strands of the prestem are proposed to open as a zip and slide downward (44). As described above, it is plausible that the prestem is partially unfolded when the prepore is formed. The W177T/R198T mutant can form the prepore but cannot penetrate the partially unfolded stem into the membrane to form a pore (13). Binding of the lipid head group may accelerate sliding of the unfolded prestem into the membrane using a mechanism similar to an allosteric effect.
Relationship to the Reported Hexameric and Heptameric Structure.
With the aid of MPD, octameric pores were formed spontaneously, indicating that the octamer is the most stable oligomeric form of γHL. This agreed well with several previous reports showing that the pore of bicomponent β-PFTs prepared on cells consisted of four molecules of each of the F and S components (22–25). In contrast, several reports indicated that γHL pores formed on the erythrocyte membrane are mainly composed of six (17, 26) or seven protomers (12, 20, 34). However, there is considerable steric constraint in the heptameric γHL model constructed by superposing individual protomers onto heptameric αHL. Moreover, electrostatic repulsion by the residues forming interprotomer ion pairs in the octamer (Fig. 2B) was observed in the homodimer interface formed when heptameric assembly was constructed by two components. These observations indicated that each protomer does not make a tight heptameric assembly as observed for αHL. Based on the results of imaging analysis, we recently reported that seven protomers in heptameric γHL lay asymmetrically (21). The observed heptameric or hexameric pore of γHL may have been a pore in which one or two protomer(s) were absent from the stable octamer.
Structural Explanation of the Characteristics of the Pore.
The pore of γHL has a flared structure in comparison to αHL and VCC (Fig. S2), which is consistent with the conductance of γHL (115 ± 15 pS) higher than that of αHL (92 ± 12 pS) (45). The wider vestibule space may also contribute to the higher conductance. Substitution of Lys108 of Hlg2, which formed shallow constriction at the middle of the transmembrane β-barrel, by Thr was reported to cause a significant increase in the conductance (up to 170 ± 12 pS) and more cationic selectivity (45). This may indicate that constriction with an electric charge significantly influenced both conductance and ion selectivity, even if the constriction was shallow.
Recently, a pore of αHL was developed as a nano device for applications such as DNA sequencing using its electrochemical properties (46, 47). Bicomponent pores have significant benefits for application as tools in biotechnology, as the two components can be engineered independently. The crystal structure of the octameric pore of γHL will facilitate future developments in both bioengineering and fundamental scientific research.
Methods
Preparation of LukF and Hlg2.
LukF and Hlg2 were overexpressed in the pET Escherichia coli expression system, and purified as a monomeric protein from the soluble fraction by passage through a Ni sepharose 6 Fast Flow column (GE Healthcare Biosciences AB) and HiLoad 26/60 Superdex 200-pg column (GE Healthcare Biosciences AB). Full details of the experimental procedures are presented in SI Text.
Crystallization and X-ray Diffraction Data Collection.
Crystals of octameric pore γ-hemolysin were grown from a solution containing 0.1 M sodium acetate (pH 4.6), 0.5 M ammonium acetate, and 50% (v/v) MPD. X-ray diffraction experiments were performed on the beamline BL41XU at SPring-8. The structure of γ-hemolysin was determined by the molecular replacement method. The refinement statistics are summarized in Table S2. The details of the structure determination procedures are presented in SI Text.
Supplementary Material
Acknowledgments.
This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan (Y. T., M. Y, and I. T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3B07).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110402108/-/DCSupplemental.
References
- 1.Iacovache I, van der Goot FG, Pernot L. Pore formation: An ancient yet complex form of attack. Biochim Biophys Acta. 2008;1778:1611–1623. doi: 10.1016/j.bbamem.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 2.Peitsch MC, Tschopp J. Assembly of macromolecular pores by immune defense systems. Curr Opin Cell Biol. 1991;3:710–716. doi: 10.1016/0955-0674(91)90045-z. [DOI] [PubMed] [Google Scholar]
- 3.Gouaux E. Channel-forming toxins: tales of transformation. Curr Opin Struct Biol. 1997;7:566–573. doi: 10.1016/s0959-440x(97)80123-6. [DOI] [PubMed] [Google Scholar]
- 4.Parker MW, Feil SC. Pore-forming protein toxins: From structure to function. Prog Biophys Mol Biol. 2005;88:91–142. doi: 10.1016/j.pbiomolbio.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Menestrina G, Serra MD, Prevost G. Mode of action of beta-barrel pore-forming toxins of the staphylococcal alpha-hemolysin family. Toxicon. 2001;39:1661–1672. doi: 10.1016/s0041-0101(01)00153-2. [DOI] [PubMed] [Google Scholar]
- 6.Montoya M, Gouaux E. Beta-barrel membrane protein folding and structure viewed through the lens of alpha-hemolysin. Biochim Biophys Acta. 2003;1609:19–27. doi: 10.1016/s0005-2736(02)00663-6. [DOI] [PubMed] [Google Scholar]
- 7.Kaneko J, Kamio Y. Bacterial two-component and hetero-heptameric pore-forming cytolytic toxins: Structures, pore-forming mechanism, and organization of the genes. Biosci Biotechnol Biochem. 2004;68:981–1003. doi: 10.1271/bbb.68.981. [DOI] [PubMed] [Google Scholar]
- 8.Prevost G, et al. In: The Comprehensive Sourcebook of Bacterial Protein Toxins. Alouf JE, Popoff MR, editors. Amsterdam: Academic; 2006. pp. 560–607. [Google Scholar]
- 9.Meyer F, Girardot R, Piemont Y, Prevost G, Colin DA. Analysis of the specificity of Panton–Valentine leucocidin and gamma-hemolysin F component binding. Infect Immun. 2009;77:266–273. doi: 10.1128/IAI.00402-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozawa T, Kaneko J, Kamio Y. Essential binding of LukF of staphylococcal gamma-hemolysin followed by the binding of H gamma II for the hemolysis of human erythrocytes. Biosci Biotechnol Biochem. 1995;59:1181–1183. doi: 10.1271/bbb.59.1181. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen VT, Higuchi H, Kamio Y. Controlling pore assembly of staphylococcal gamma-haemolysin by low temperature and by disulphide bond formation in double-cysteine LukF mutants. Mol Microbiol. 2002;45:1485–1498. doi: 10.1046/j.1365-2958.2002.03125.x. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen AH, Nguyen VT, Kamio Y, Higuchi H. Single-molecule visualization of environment-sensitive fluorophores inserted into cell membranes by staphylococcal gamma-hemolysin. Biochemistry. 2006;45:2570–2576. doi: 10.1021/bi0514156. [DOI] [PubMed] [Google Scholar]
- 13.Monma N, Nguyen VT, Kaneko J, Higuchi H, Kamio Y. Essential residues, W177 and R198, of LukF for phosphatidylcholine-binding and pore-formation by staphylococcal gamma-hemolysin on human erythrocyte membranes. J Biochem. 2004;136:427–431. doi: 10.1093/jb/mvh140. [DOI] [PubMed] [Google Scholar]
- 14.Werner S, et al. Retrieving biological activity from LukF-PV mutants combined with different S components implies compatibility between the stem domains of these staphylococcal bicomponent leucotoxins. Infect Immun. 2002;70:1310–1318. doi: 10.1128/IAI.70.3.1310-1318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson R, Nariya H, Yokota K, Kamio Y, Gouaux E. Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel. Nat Struct Biol. 1999;6:134–140. doi: 10.1038/5821. [DOI] [PubMed] [Google Scholar]
- 16.Potrich C, et al. The influence of membrane lipids in Staphylococcus aureus gamma-hemolysins pore formation. J Membr Biol. 2009;227:13–24. doi: 10.1007/s00232-008-9140-6. [DOI] [PubMed] [Google Scholar]
- 17.Pedelacq JD, et al. The structure of a Staphylococcus aureus leucocidin component (LukF-PV) reveals the fold of the water-soluble species of a family of transmembrane pore-forming toxins. Structure. 1999;7:277–287. doi: 10.1016/s0969-2126(99)80038-0. [DOI] [PubMed] [Google Scholar]
- 18.Guillet V, et al. Crystal structure of leucotoxin S component: new insight into the Staphylococcal beta-barrel pore-forming toxins. J Biol Chem. 2004;279:41028–41037. doi: 10.1074/jbc.M406904200. [DOI] [PubMed] [Google Scholar]
- 19.Tomita T, et al. Pleurotolysin, a novel sphingomyelin-specific two-component cytolysin from the edible mushroom Pleurotus ostreatus, assembles into a transmembrane pore complex. J Biol Chem. 2004;279:26975–26982. doi: 10.1074/jbc.M402676200. [DOI] [PubMed] [Google Scholar]
- 20.Sugawara-Tomita N, Tomita T, Kamio Y. Stochastic assembly of two-component Staphylococcal–Hemolysin into heteroheptameric transmembrane pores with alternate subunit arrangements in ratios of 3∶4 and 4∶3. J Bacteriol. 2002;184:4747–4756. doi: 10.1128/JB.184.17.4747-4756.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomita T, Abe K, Ohta M. Quantitative analysis of subunit mismatch arrangement in Staphylococcal gamma-hemolysin heteroheptameric transmembrane pore; Proceedings of ASME 2011 International Mechanical Engineering Congress and Exposition; 2011. IMECE2011-63645. [Google Scholar]
- 22.Joubert O, et al. Engineered covalent leucotoxin heterodimers form functional pores: Insights into S–F interactions. Biochem J. 2006;396:381–389. doi: 10.1042/BJ20051878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miles G, Movileanu L, Bayley H. Subunit composition of a bicomponent toxin: Staphylococcal leukocidin forms an octameric transmembrane pore. Protein Sci. 2002;11:894–902. doi: 10.1110/ps.4360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayasinghe L, Bayley H. The leukocidin pore: Evidence for an octamer with four LukF subunits and four LukS subunits alternating around a central axis. Protein Sci. 2005;14:2550–2561. doi: 10.1110/ps.051648505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das SK, Darshi M, Cheley S, Wallace MI, Bayley H. Membrane protein stoichiometry determined from the step-wise photobleaching of dye-labelled subunits. ChemBioChem. 2007;8:994–999. doi: 10.1002/cbic.200600474. [DOI] [PubMed] [Google Scholar]
- 26.Sugawara N, Tomita T, Sato T, Kamio Y. Assembly of Staphylococcus aureus leukocidin into a pore-forming ring-shaped oligomer on human polymorphonuclear leukocytes and rabbit erythrocytes. Biosci Biotechnol Biochem. 1999;63:884–891. doi: 10.1271/bbb.63.884. [DOI] [PubMed] [Google Scholar]
- 27.Yokota K, Kamio Y. Tyrosine72 residue at the bottom of rim domain in LukF crucial for the sequential binding of the staphylococcal gamma-hemolysin to human erythrocytes. Biosci Biotechnol Biochem. 2000;64:2744–2747. doi: 10.1271/bbb.64.2744. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka Y, et al. 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal α-hemolysin into heptameric pore structure. Protein Sci. 2011;20:448–456. doi: 10.1002/pro.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song L, et al. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 30.Galdiero S, Gouaux E. High resolution crystallographic studies of α-hemolysin-phospholipid complexes define heptamer–lipid head group interactions: Implication for understanding protein–lipid interactions. Protein Sci. 2004;13:1503–1511. doi: 10.1110/ps.03561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouaux E. Roll out the barrel. Nat Struct Biol. 1998;5:931–932. doi: 10.1038/2804. [DOI] [PubMed] [Google Scholar]
- 32.De S, Olson R. Crystal structure of the Vibrio cholerae cytolysin heptamer reveals common features among disparate pore-forming toxins. Proc Natl Acad Sci USA. 2011;108:7385–7390. doi: 10.1073/pnas.1017442108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roblin P, et al. A covalent S-F heterodimer of leucotoxin reveals molecular plasticity of β-barrel pore-forming toxins. Proteins. 2008;71:485–496. doi: 10.1002/prot.21900. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen VT, Kamio Y, Higuchi H. Single-molecule imaging of cooperative assembly of gamma-hemolysin on erythrocyte membranes. EMBO J. 2003;22:4968–4979. doi: 10.1093/emboj/cdg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen VT, Kamio Y. Cooperative assembly of beta-barrel pore-forming toxins. J Biochem. 2004;136:563–567. doi: 10.1093/jb/mvh160. [DOI] [PubMed] [Google Scholar]
- 36.Kaneko J, Ozawa T, Tomita T, Kamio Y. Sequential binding of Staphylococcal gamma-hemolysin to human erythrocytes and complex formation of the hemolysin on the cell surface. Biosci Biotechnol Biochem. 1997;61:846–851. doi: 10.1271/bbb.61.846. [DOI] [PubMed] [Google Scholar]
- 37.Miles G, Jayasinghe L, Bayley H. Assembly of the bi-component leukocidin pore examined by truncation mutagenesis. J Biol Chem. 2006;281:2205–2214. doi: 10.1074/jbc.M510842200. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko J, Mascarenas AL, Huda MN, Tomita T, Kamio Y. An N-terminal region of LukF of staphylococcal leukocidin/gamma-hemolysin crucial for the biological activity of the toxin. Biosci Biotechnol Biochem. 1998;62:1465–1467. doi: 10.1271/bbb.62.1465. [DOI] [PubMed] [Google Scholar]
- 39.Walker B, Krishnasastry M, Zorn L, Bayley H. Assembly of the oligomeric membrane pore formed by Staphylococcal alpha-hemolysin examined by truncation mutagenesis. J Biol Chem. 1992;267:21782–21786. [PubMed] [Google Scholar]
- 40.Panchal RG, Bayley H. Interactions between residues in staphylococcal alpha-hemolysin revealed by reversion mutagenesis. J Biol Chem. 1995;270:23072–23076. doi: 10.1074/jbc.270.39.23072. [DOI] [PubMed] [Google Scholar]
- 41.Walker B, Bayley H. Key residues for membrane binding, oligomerization, and pore forming activity of staphylococcal alpha-hemolysin identified by cysteine scanning mutagenesis and targeted chemical modification. J Biol Chem. 1995;270:23065–23071. doi: 10.1074/jbc.270.39.23065. [DOI] [PubMed] [Google Scholar]
- 42.Walker B, Braha O, Cheley S, Bayley H. An intermediate in the assembly of a pore-forming protein trapped with a genetically-engineered switch. Chem Biol. 1995;2:99–105. doi: 10.1016/1074-5521(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 43.Bayley H. Membrane-protein structure: Piercing insights. Nature. 2009;459:651–652. doi: 10.1038/459651a. [DOI] [PubMed] [Google Scholar]
- 44.Viero G, et al. A molecular pin to study the dynamics of beta-barrel formation in pore-forming toxins on erythrocytes: A sliding model. Cell Mol Life Sci. 2008;65:312–323. doi: 10.1007/s00018-007-7491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comai M, et al. Protein engineering modulates the transport properties and ion selectivity of the pores formed by staphylococcal gamma-haemolysins in lipid membranes. Mol Microbiol. 2002;44:1251–1267. doi: 10.1046/j.1365-2958.2002.02943.x. [DOI] [PubMed] [Google Scholar]
- 46.Astier Y, Braha O, Bayley H. Toward single molecule DNA sequencing: direct identification of ribonucleoside and deoxyribonucleoside 5′-monophosphates by using an engineered protein nanopore equipped with a molecular adapter. J Am Chem Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 47.Asandei A, Apetrei A, Luchian T. Uni-molecular detection and quantification of selected beta-lactam antibiotics with a hybrid alpha-hemolysin protein pore. J Mol Recognit. 2011;24:199–207. doi: 10.1002/jmr.1038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.