Abstract
RIG-I–like receptors (RLRs) activate host innate immune responses against virus infection through recruiting the mitochondrial adaptor protein MAVS (also known as IPS1, VISA, or CARDIF). Here we show that MAVS also plays a pivotal role in maintaining intestinal homeostasis. We found that MAVS knockout mice developed more severe mortality and morbidity than WT animals in an experimental model of colitis. Bone marrow transplantation experiments revealed that MAVS in cells of nonhematopoietic origin plays a dominant role in the protection against colitis. Importantly, RNA species derived from intestinal commensal bacteria activate the RIG-I–MAVS pathway to induce the production of multiple cytokines and antimicrobial peptides, including IFN-β and RegIIIγ. These results unveil a previously unexplored role of MAVS in monitoring intestinal commensal bacteria and maintaining tissue homeostasis.
The mammalian intestine is colonized by trillions of microorganisms, including bacteria, fungi, and viruses (1, 2). Commensal bacteria constitute the dominant microbial population and contribute to host physiology and health in many ways, including enhancing digestive efficiency, sculpting the gut-associated lymphoid system, and blocking pathogen invasion. However, abnormal interactions between the intestinal mucosal system and luminal microbiota may lead to sustained inflammation, an important mechanism for human inflammatory bowel disease (3, 4).
In recent years, accumulating evidence has suggested a central role of innate immunity in the homeostasis of the gastrointestinal tract and the pathogenesis of inflammatory bowel disease. The mammalian innate immune system employs pattern-recognition receptors (PRRs), including Toll-like receptors (TLRs), NOD-like receptors (NLRs) and retinoic acid inducible gene-1 (RIG-I)–like receptors (RLRs), for the detection of pathogens (5, 6). TLRs are localized on the cell surface and endosomal membranes and NLRs and RLRs reside in the cytoplasm. TLRs recognize microbial ligands, such as LPS, nucleic acids, and peptidoglycans. Ligand binding triggers the association of these receptors with adaptor molecules, such as myeloid differentiation primary response gene 88 (MyD88) and TIR domain-containing adaptor-inducing IFN-β (TRIF), which subsequently activate a number of transcriptional factors, including NF-κB and IRF3. Some NLRs detect bacterial peptidoglycans and activate NF-κB, whereas others activate the inflammasome, which leads to the production of cytokines, such as IL-1 and IL-18. The RIG-I pathway senses cytoplasmic viral RNAs to elicit the production of type I interferons (IFN-α/β). RIG-I recruits MAVS, a mitochondria-localized antiviral protein (also known as IPS1, VISA, and CARDIF), which acts as an essential adaptor to activate the downstream transcription factors IRF3 and NF-κB (7–11). Both 5′-triphosphate and double-stranded structures of RNAs have been identified as the structural elements important for recognition by RIG-I (12–15). Using RIG-I, MDA5 (a homolog of RIG-I), and MAVS knockout mice, genetic studies have clearly demonstrated the essential function of the RIG-I–MAVS pathway in antiviral immune defense (16–18).
Besides their role in antimicrobial defense, TLRs and NLRs are crucial for the homeostasis of gastrointestinal epithelium. Using MyD88 and TLR knockout mice, it has been demonstrated that the TLR pathway is essential for mouse survival during dextran sulfate sodium salt (DSS)-induced colitis (19, 20). These important findings show that TLR-mediated signaling is not only involved in microbial sensing and inflammatory response, but also controls intestinal epithelial homeostasis and prevents tissue injury. The role of NLRs in controlling intestinal homeostasis is best illustrated by the mutations of NOD2 that are closely linked to Crohn disease (21, 22). Recent studies also provide evidence that loss-of-function mutations of the inflammasome sensitize mice to DSS-induced colitis (23, 24).
Emerging evidence has also suggested a role of the RIG-I pathway in the gut. RIG-I is expressed at the apical surface of intestinal epithelium in biopsies from human colonic tissues (25). Moreover, RIG-I knockout mice have increased sensitivity to chemical-induced colitis and are defective in antibacterial defense (26, 27). However, the mechanism by which RIG-I regulates intestinal inflammation remains poorly understood. In particular, the relationship between the RIG-I pathway and intestinal commensal bacteria is not clear.
In this article, we show that Mavs-deficient mice are highly susceptible to DSS-induced colitis and that MAVS in cells of nonhematopoietic origin mediates protection against intestinal injury. Furthermore, we demonstrate that RNA species derived from commensal bacteria in the gastrointestinal tract act as a ligand for RIG-I and induce an immune response through MAVS. These results demonstrate that the RIG-I–MAVS pathway is not only essential for immune defense against viral infection, but also controls tissue homeostasis by monitoring commensal bacteria.
Results
Mavs−/− Mice are Highly Susceptible to DSS-Induced Colitis.
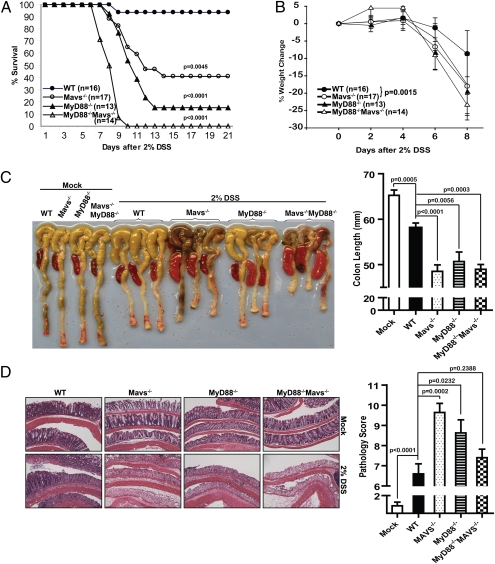
To investigate the potential role of MAVS in tissue homeostasis, we used a colitis model induced by DSS that works primarily through disruption of colonic epithelium (28). In pilot experiments, we found that male Mavs−/− mice were more sensitive to DSS-induced mortality than female mice (Fig. S1B). Therefore, in all subsequent experiments, 6- to 10-wk-old male mice were used. Four groups of mice, WT, Mavs−/−, MyD88−/−, and MyD88−/− Mavs−/−, were administered 2% DSS in drinking water for 7 d followed by normal drinking water for 14 d. Mortality was monitored up to the 21st day. Mavs−/− mice showed dramatically increased mortality (10 of 17, or 59%, of mice died) (Fig. 1A) and severe weight loss starting on the sixth day (Fig. 1B). Under the same condition, almost all WT mice (15 of 16 mice) survived and the weight loss was more modest compared with the mutant mice. Consistent with a previous report (19), 11 of 13 (or 85%) MyD88−/− mice died from complications of colitis. Interestingly, MyD88 and MAVS seemed to cooperate to protect mice from DSS-induced injuries because none of the MyD88−/− Mavs−/− mice survived after day 10. To analyze the morphology and histology of the colons and spleens, we harvested these organs after the mice were treated with 2% DSS for 7 d followed by 1 d with normal drinking water. As shown in Fig. 1C, the colon lengths were similar among mock-treated animals, but DSS treatment led to a significant reduction in length, which was more evident in all of the mutant mice. In addition, enlarged spleens were observed in DSS-treated mutant animals but not in mock-treated mice or DSS-treated WT mice. We further analyzed pathological changes of the colons by histology. H&E staining of the colons showed that after DSS-treatment, WT mice had more leukocyte infiltration and more remaining epithelium than all of the mutant animals (Fig. 1D). Naive mice, regardless of genotype, showed no signs of spontaneous colitis clinically and histopathologically (data not shown). Mavs−/−, MyD88−/−, and MyD88−/−Mavs−/− mice were also more susceptible than WT mice to lower concentration of DSS (1%) (Fig. S1A).
Fig. 1.
Mavs−/− mice are highly susceptible to DSS-induced colitis. Four groups of mice—WT, Mavs−/−, MyD88−/−, and MyD88−/−Mavs−/−—were given 2% DSS in drinking water for 7 d, followed by normal drinking water for another 14 d. (A) Increased mortality in Mavs-deficient mice. Statistical analysis was performed using Mantel-Cox test. Compared with WT: Mavs−/− P = 0.0045, MyD88−/− P < 0.0001, MyD88−/−Mavs−/− P < 0.0001. Compared with MyD88−/−: MyD88−/−Mavs−/− P = 0.0002. (B) Weight change following DSS administration. Error bars represent SEMs. Statistics of the weight changes on day 8 was determined using the Student's test, Mavs−/− P = 0.0015, MyD88−/− P = 0.0022, MyD88−/−Mavs−/− P < 0.0001. (C) Gross morphological changes of colon and spleen. On day 8 after 7 d of 2% DSS and 1 d drinking water, colons and spleens were excised and photographed. Representative photos of colon and spleen are shown on the left. Colon lengths are shown on the right. The numbers of mice used in the experiments are as follows: WT: n = 10; Mavs−/−: n = 11; MyD88−/−: n = 5; MyD88−/−Mavs−/−: n = 5. (D) Colons were cut longitudinally, fixed, and then stained with H&E. Representative histological images are shown on the left and pathological scores on the right. WT: n = 10; Mavs−/−: n = 11; MyD88−/−: n = 8; MyD88−/−Mavs−/−: n = 10.
To determine the role of commensal bacteria in the pathogenesis, we treated animals with antibiotics (ampicillin, metronidazol, neomycin, and vancomycin) in the drinking water for 4 wk, followed by 2% DSS for 7 d. Consistent with previous reports (19), depletion of commensal bacteria by antibiotics in WT mice led to dramatically increased mortality (Fig. S2A), indicating that commensal bacteria protect the mice from DSS-induced colitis. Furthermore, germ-free mice had increased susceptibility to DSS treatment, which might be partly because of decreased proliferation potential of intestinal epithelial cells (Fig. S2 B and C) (see also ref. 19). The antibiotic treatment did not rescue the DSS-induced mortality of MAVS knockout mice, suggesting that the death of the animals was not simply because of bacterial overgrowth. Rather, the commensal bacteria might have a protective role, and the Mavs-deficiency might result in an inability to repair DSS-induced tissue damage, which caused the death.
MAVS in Nonhematopoietic Cells Is Critical for Protection Against Colonic Epithelial Damage.
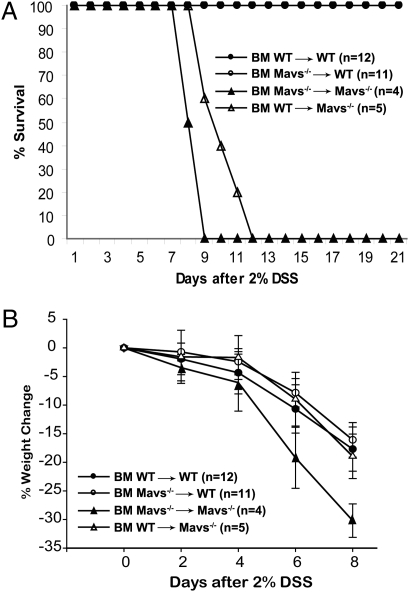
The interaction between epithelial cells and their surrounding stromal cells is an important mechanism for tissue regenerative responses. To examine the relative contribution of cells of hematopoietic versus epithelial origin for protection from DSS-induced colitis, we generated bone marrow chimeric mice expressing MAVS in either of the two compartments. All mice used in bone marrow transplantation experiments were C57BL/6J or had been crossed to C57BL/6J background for more than 10 generations. Control experiments using congenic surface markers CD45.1 and CD45.2 showed that the chimeric mice contained 80% to 90% of the donor cells (Table S1). Transplantation of Mavs−/− bone marrow to irradiated WT mice did not cause these mice to become more sensitive to DSS-induced mortality (Fig. 2A). In reciprocal transplantation, WT bone marrow provided only limited protection to MAVS knockout mice (Fig. 2B). Similar results were also observed in WT animals reconstituted with bone marrow from double-knockout (MyD88−/−Mavs−/−) mice. The efficiency of chimerism in spleen and intestine was around 95% and 85%, respectively (Fig. S3). These findings indicate that MAVS expression in nonhematopoietic cells plays a dominant role in the resistance of gut epithelium to insults. However, we cannot rule out the possibility that residual radiation-resistant hematopoietic cells such as lamina propria macrophages may contribute to the prevention of DSS-induced colitis.
Fig. 2.
MAVS in cells of nonhematopoietic origin plays a dominant role in preventing DSS-induced colitis. Bone marrow transplantation experiments were carried out and the efficiency of transplantation was verified by FACS analysis of leukocyte surface markers, as described in Methods. (A) All animals were treated with 2% DSS in drinking water before switching back to normal water, and mortality was monitored until the 21st day. Mavs−/− mice reconstituted with WT bone marrow show slightly better survival than those reconstituted with Mavs−/− bone marrow (P = 0.0448). (B) Weight loss of the mice after DSS treatment was monitored, as described in Fig. 1B. The results are representative of three independent experiments.
RNA Species from Intestinal Commensal Bacteria Stimulate the RIG-I–MAVS Pathway.
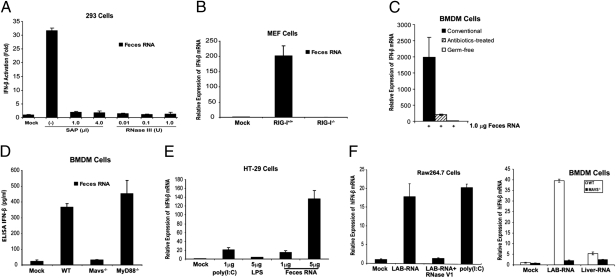
Although it is well known that viral RNA activates the RIG-I pathway, it is not clear whether bacterial RNA could also function as a RIG-I ligand. Like viral RNA, bacterial RNA is expected to contain 5′-triphosphate and some secondary structures that are hallmarks of a RIG-I ligand (29). However, RNA isolated from some Escherichia coli strains does not induce IFN-β, although it is capable of inducing inflammatory cytokines, including TNF-α and IL-1β (30). Because mice deficient in type-I IFN receptor (IFN-α/βR−/−) are also highly sensitive to DSS-induced colitis (31), we examined whether commensal bacterial RNA could induce IFN-β through the RIG-I–MAVS pathway. Total RNA from feces collected from conventionally reared mice were transfected into a HEK293T cell line stably integrating a luciferase reporter driven by the IFN-β promoter (32). As shown in Fig. 3A, the feces RNA strongly induced the IFN-β reporter, and this activity was abolished by treatment with shrimp alkaline phosphatase (SAP), which removes 5′-triphosphate, or with RNase III, which digests double-stranded RNA. Transfection of feces RNA into WT, but not RIG-I–deficient, mouse embryonic fibroblast cells led to a robust induction of IFN-β mRNA (Fig. 3B). Control experiments showed that RNAs from mouse liver and several E. coli strains (e.g., DH5α, TOP10) did not induce IFN-β (Fig. S4 A and B). Furthermore, transfection is required for IFN-β–inducing activity of feces RNA or RNA isolated from Listeria monocytogenes (Fig. S4C). These results suggest that the feces RNA contain 5′-triphosphate and dsRNA segments that activate RIG-I. Transfection of feces RNA isolated from conventionally raised mice into bone marrow-derived macrophages (BMDM) also strongly induced IFN-β (Fig. 3C). To determine if commensal bacterial RNA was responsible for inducing interferons, we isolated feces RNA from antibiotic-treated mice as well as germ-free mice. We noted that the amounts of RNA isolated from the feces of antibiotic-treated or germ-free mice were much reduced (>100-fold less) compared with those from conventionally raised mice (Table S2). However, even if we used the same amount (1 μg) of RNA, those from bacteria-depleted mice were devoid of IFN-inducing activity (Fig. 3C), indicating that commensal bacterial RNA was responsible for IFN induction. ELISA experiments showed that the bacterial RNA induced the production of IFN-β protein in BMDM in a manner that depends on MAVS, but not MyD88 or TRIF (Fig. 3D and Fig. S4D). Transfection of feces RNA into the intestinal epithelial cell line HT-29 also led to a marked induction of IFN-β (Fig. 3E). To determine the types of bacteria that could generate IFN-inducing RNA, the fecal bacteria from WT mice were cultured in vitro under aerobic and anaerobic conditions and then RNA was extracted. Similar to RNA isolated from the intracellular bacterium L. monocytogenes, RNA from the fecal bacteria grown under both conditions was able to induce IFN-β in BMDM (Fig. S5). We also isolated RNA from Lactobacillus salivarius (LAB), a Gram-positive facultative anaerobic bacterium commonly found in the gastrointestinal tract. Strikingly, LAB RNA strongly induced IFN-β in the mouse macrophage cell line Raw264.7, and this activity was abolished by -RNase V1, which cleaves RNA containing double-stranded regions (Fig. 3F, Left). Transfection of LAB RNA into BMDM from WT and Mavs−/− mice showed that IFN-β induction by the RNA was dependent on MAVS (Fig. 3F, Right). Taken together, these results strongly suggest that RNA species from commensal bacteria function as a bona fide RIG-I ligand and induce IFN-β through MAVS.
Fig. 3.
RNA derived from mouse feces induces IFN-β through the RIG-I–MAVS pathway. (A) Feces RNA was treated with or without SAP or RNase III, then transfected into HEK293T IFN-β-luciferase reporter cells. Luciferase activity was measured 24 h after transfection. (B) RIG-I–dependent induction of IFN-β mRNA by feces RNA. Feces RNA was transfected into WT or RIG-I–deficient MEF cells, then IFN-β mRNA was measured by qRT-PCR 8 h after transfection. (C) One-microgram of RNA from the feces of conventional, antibiotics-treated, or germ-free mice was transfected into BMDM. IFN-β mRNA was measured by qRT-PCR 8 h after transfection. (D) Feces RNA from conventionally raised mice was transfected into BMDM from mice of the indicated genotypes, and the secretion of IFN-β protein in the media was measured by ELISA 24 h after transfection. (E) Feces RNA or poly(I:C) was transfected into HT-29 cells, then IFN-β RNA was measured by qRT-PCR 8 h after transfection. LPS was added to the culture media without transfection reagent. (F) RNA isolated from L. salivarius (LAB) was treated with RNase V1 or not treated before transfection into Raw264.7 cells (Left) and the induction of IFN-β RNA was measured by qRT-PCR. (Right) LAB or liver RNA was transfected into BMDM from WT or Mavs−/− mice, followed by measurement of IFN-β RNA. Error bars represent the variation range of duplicate experiments (A) or SD of triplicate experiments (B–F).
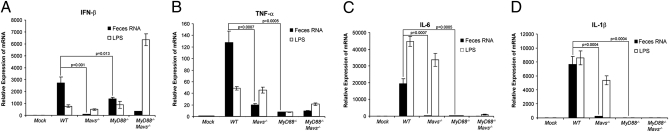
To determine if and how feces RNA induces proinflammatory cytokines, BMDM from WT or mutant mice lacking either Mavs or MyD88, or both, were transfected with feces RNA or incubated in the presence of LPS, and then the production of different cytokines was measured by quantitative RT-PCR (qRT-PCR). The induction of all cytokines, including IFN-β, TNF-α, IL-6, and IL-1β by the feces RNA was dependent on MAVS (Fig. 4). Interestingly, although IFN-β induction by the feces RNA was partially reduced in MyD88−/− mice (Fig. 4A), the induction of TNF-α, IL-6, and IL-1β was abolished in the absence of MyD88 (Fig. 4 B–D). Thus, both MyD88 and MAVS regulate the induction of inflammatory cytokines in response to commensal bacterial RNA. In contrast to feces RNA, LPS induced TNF-α, IL-6, and IL-1β through a mechanism dependent on MyD88, but not MAVS. The induction of IFN-β by LPS was further enhanced in the absence of MyD88 and MAVS, suggesting the possibility of a compensatory up-regulation of the TRIF pathway, which can induce IFN-β (5).
Fig. 4.
(A–D) Feces RNA induces multiple cytokines through MAVS and MyD88. Feces RNA was transfected into BMDM cells from WT, Mavs−/−, MyD88−/−, and MyD88−/−Mavs−/− mice, and the indicated cytokines were measured by qRT-PCR 8 h after transfection. LPS was added to the culture media without transfection reagent.
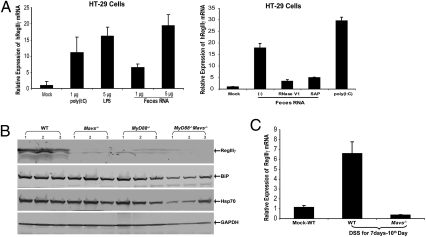
RegIIIγ is a secreted C-type lectin exhibiting potent antimicrobial activities (33). Colonization by commensal bacteria in the gut has been shown to strongly induce RegIIIγ through the MyD88-mediated pathway, with the highest expression in the distal region (ileum) of the intestine (33–35). To determine if commensal bacterial RNA could induce RegIIIγ, HT-29 cells were transfected with feces RNA and the expression of RegIIIγ measured by qRT-PCR. Similar to stimulation with LPS and the dsRNA analog poly(I:C), the feces RNA induced RegIIIγ expression (Fig. 5A). Treatment of the feces RNA with SAP or RNaseIII abolished its ability to induce RegIIIγ (Fig. 5A, Right), suggesting that 5′-triphosphate and dsRNA regions of feces RNA are required for RegIIIγ induction. Immunoblotting of protein extracts from the ileum of conventionally raised mice showed that RegIIIγ was expressed in WT mice, but this expression was markedly reduced in mice lacking Mavs, MyD88, or both (Fig. 5B). We also observed that chaperons, such as BiP and HSP70, were reduced significantly only in MyD88−/− Mavs−/− mice (Fig. 5B). Following DSS damage, the expression of RegIIIγ was also much higher in WT than in Mavs−/− mice (Fig. 5C). Similarly, colon culture experiments indicated that the level of IL-6 protein was lower in Mavs−/− mice than in WT mice (Fig. S6). Taken together, these results show that MAVS is critical for the expression of RegIIIγ as well as cyto-protective cytokines and proteins in mouse intestine.
Fig. 5.
MAVS-dependent expression of RegIIIγ in gut mucosa. (A) Feces RNA and poly(I:C) were transfected into HT-29 cells and RegIIIγ was measured by qRT-PCR 8 h after transfection. LPS was added to the culture media without transfection reagent. (Right) Feces RNA was treated with RNase V1 or SAP before transfection into HT-29 cells. (B) Tissue samples were prepared from ileums of WT and mutant mice, as described in Methods, and the protein extracts were analyzed by immunoblotting with antibodies against RegIIIγ, chaperone proteins BiP and Hsp70, and GAPDH. (C) RegIIIγ mRNA in the small intestines from DSS-treated mice was measured by qRT-PCR. The result represents two independent experiments.
Discussion
In this article, we show that, similar to MyD88, MAVS is essential for protecting mice from DSS-induced colitis and death. Furthermore, we demonstrate that RNA species derived from intestinal commensal bacteria act through the RIG-I–MAVS pathway to produce effector molecules, including interferons, cytokines, and antimicrobial peptides. Thus, RIG-I is capable of detecting both viral and bacterial RNA ligands. Our results suggest that the RIG-I pathway is not only involved in innate immunity against viral and bacterial infections, but also plays a key role in monitoring commensal bacteria and maintaining intestinal homeostasis.
Although the role of IFNs, cytokines, and antimicrobial peptides in immune defense against microbial pathogens is well established, how these molecules maintain tissue homeostasis is just beginning to be unraveled (36, 37). Recent studies have demonstrated a critical role of the JAK-STAT pathway in stem cell proliferation in the gut and intestinal epithelia homeostasis in mice, as well as in Drosophila (37). IFNs have also been shown to activate dormant stem cells (38). Moreover, IFN receptor-deficient (IFN-α/βR−/−) mice are highly susceptible to DSS-induced colitis (31). These findings suggest that defective production of type-I IFNs in Mavs-deficient mice is an important factor that contributes to colitis. We tried to rescue Mavs−/− mice from DSS-induced colitis by administration of IFN-β or TLR ligands, such as LPS and poly(I:C). However, these treatments proved to be insufficient to reduce the mortality from DSS-induced colitis (Fig. S7). Thus, other factors regulated by MAVS, such as immune cytokines and antimicrobial peptides, are also important in preventing experimental colitis. It is likely that the RIG-I–MAVS pathway controls the production of multiple effector molecules that function together to maintain tissue homeostasis and facilitate the regeneration of the intestinal mucosa. Given the critical role of MAVS in immune response to commensal bacteria, it is possible that the compositions of microbiota in MAVS-deficient mice may differ from those of the WT mice and such difference may contribute to the colitis phenotypes in MAVS-deficient mice. A systemic analysis of microbiome in these mice could yield interesting results.
Recent studies have suggested that TLRs and NLRs are involved in the detection of intestinal commensal bacteria, which are critical in shaping the host immune system (6). Our study provides another mechanism of bacterial detection in the gut through the RIG-I pathway. We show that commensal bacterial RNA is a functional ligand of RIG-I that strongly induces the production of IFNs and cytokines through MAVS. Although prokaryotic RNA is expected to contain 5′-triphosphate and duplex regions in the secondary structure, certain bacterial RNAs lack the ability to induce IFN-β (30). Currently it is not understood why some bacterial RNAs induce IFN-β, whereas others do not. One possibility is that some bacterial RNAs contain modifications that prevent them from functioning as a RIG-I ligand. In any case, it is unlikely that the IFN-inducing activity we observed resulted from some animal viruses in the gut, because RNA from antibiotic-treated mice did not induce IFNs (Fig. 3C). Although we cannot completely rule out the possibility that some intestinal bacteriophages may engage the RIG-I pathway, we found that RNA isolated from an E. coli strain (K91) harboring a λ- (M15) phage failed to induce IFN-β in BMDM. It is also possible that some bacterial DNA could be converted by cellular RNA polymerase III to 5′-triphosphorylated RNA, which in turn activates the RIG-I–MAVS pathway (32, 39). However, in many cell types, cytosolic DNA could activate a MAVS-independent pathway that involves the endoplasmic reticulum adaptor protein STING (also known as MITA) (40). It will be interesting to determine whether the interaction between commensal bacterial DNA and the host innate-immune system (i.e., MAVS and STING) could play a role in maintaining intestinal tissue homeostasis.
Our bone-marrow chimera experiments show that MAVS in nonhematopoietic, rather than hematopoietic cells, play a dominant role in suppressing DSS-induced colitis. Similarly, a recent report showed that MyD88 in nonhematopoietic cells is important for protection against DSS-induced colitis (34). These results suggest that intestinal epithelial cells, which are constantly exposed to the microbial community, are involved in detecting commensal bacteria to activate MAVS. Despite their intracellular location, RLRs and NLRs apparently have the ability to detect bacterial ligands, including RNA and peptidoglycans. How bacteria deliver these materials to the cytosol of intestinal epithelial cells is not well understood. It is possible that tissue injury, such as that caused by DSS, facilitates the uptake of intestinal bacteria by intestinal epithelial cells, which then deploy PRRs, including RLRs, NLRs, and TLRs, to orchestrate an immune and stress response that facilitates wound healing and suppress bacteria overgrowth. It is remarkable to note that the signaling pathways triggered by different PRRs are apparently not redundant, because mutations of RLRs, NLRs, or TLRs all exacerbate experimental colitis. Thus, different PRRs likely activate distinct signaling pathways that together maintain intestinal homeostasis.
Methods
Mice.
WT, Mavs−/−, MyD88−/−, and MyD88−/−Mavs−/− mice were bred and maintained under specific pathogen-free conditions in the animal care facility of University of Texas Southwestern Medical Center at Dallas. Germ-free C57BL/6 mice were maintained in plastic gnotobiotic isolators, as previously described (41). These strains were maintained as C57BL/6J and 129SvEv mixed background. The generation of Mavs−/− mice has been previously described (18). Mavs−/− mice have been backcrossed into C57BL/6J background for more than 10 generations. MyD88−/− mice on C57BL/6 background were kindly provided by Shizuo Akira (Osaka University, Osaka, Japan). Unless otherwise indicated, the mice used in the experiments were 6- to 10-wk-old male mice on mixed background. All mice were engineered, housed and used according to the experimental protocols approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center.
Induction of Colitis with DSS.
Sex- and age-matched animals received 2% DSS (molecular weight 36,000–50,000; MP Biomedicals) in drinking water for 7 d. The amount of DSS water consumed per animal was noted; there was no marked difference between experimental groups. For survival studies, mice were monitored every day up to 21 d postinitiation of DSS-treatment. For weight-change studies, the percentage of weight loss of animals was determined through the following equation: weight loss (%) = 100 × (weight at the xth day – weight on the 0th day)/(weight on the 0th day). Animals were also monitored for rectal bleeding, diarrhea, and general signs of morbidity, including hunched posture and failure to groom.
Statistics.
The statistical analysis was done using software GraphPad Prism 5. P value <0.05 was considered statistically significant. Additional methods and supplementary figures can be found in the SI.
Supplementary Material
Acknowledgments
We thank Xiang Chen and Yilun Sun for assistance with the maintenance of mouse colonies, Katalin Tus and John D. Schatzle for assistance with bone marrow transplantation experiments, Angela Mobley for help with flow cytometry, and John Shelton for histology. This work was supported by grants from the National Institutes of Health and by The Howard Hughes Medical Institute (to Z.J.C. and L.V.H).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107114108/-/DCSupplemental.
References
- 1.Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514–521. doi: 10.1172/JCI30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Lavelle EC, Murphy C, O'Neill LA, Creagh EM. The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 2010;3:17–28. doi: 10.1038/mi.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 8.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 9.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 10.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 13.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 14.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 17.Kumar H, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Q, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Vijay-Kumar M, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 22.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 23.Zaki MH, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen IC, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi S, et al. Retinoic acid-inducible gene-I is constitutively expressed and involved in IFN-gamma-stimulated CXCL9-11 production in intestinal epithelial cells. Immunol Lett. 2009;123:9–13. doi: 10.1016/j.imlet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Kong L, et al. An essential role for RIG-I in toll-like receptor-stimulated phagocytosis. Cell Host Microbe. 2009;6:150–161. doi: 10.1016/j.chom.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, et al. Rig-I−/− mice develop colitis associated with downregulation of G alpha i2. Cell Res. 2007;17:858–868. doi: 10.1038/cr.2007.81. [DOI] [PubMed] [Google Scholar]
- 28.Okayasu I, et al. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 29.Bieger CD, Nierlich DP. Distribution of 5′-triphosphate termini on the mRNA of Escherichia coli. J Bacteriol. 1989;171:141–147. doi: 10.1128/jb.171.1.141-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eberle F, Sirin M, Binder M, Dalpke AH. Bacterial RNA is recognized by different sets of immunoreceptors. Eur J Immunol. 2009;39:2537–2547. doi: 10.1002/eji.200838978. [DOI] [PubMed] [Google Scholar]
- 31.Katakura K, et al. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J Clin Invest. 2005;115:695–702. doi: 10.1172/JCI22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brandl K, et al. MyD88 signaling in nonhematopoietic cells protects mice against induced colitis by regulating specific EGF receptor ligands. Proc Natl Acad Sci USA. 2010;107:19967–19972. doi: 10.1073/pnas.1014669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: Implications for inflammatory diseases. Nat Rev Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 37.Pitsouli C, Apidianakis Y, Perrimon N. Homeostasis in infected epithelia: Stem cells take the lead. Cell Host Microbe. 2009;6:301–307. doi: 10.1016/j.chom.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 39.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 42.Xiao H, et al. The Toll-interleukin-1 receptor member SIGIRR regulates colonic epithelial homeostasis, inflammation, and tumorigenesis. Immunity. 2007;26:461–475. doi: 10.1016/j.immuni.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 43.Turer EE, et al. Homeostatic MyD88-dependent signals cause lethal inflamMation in the absence of A20. J Exp Med. 2008;205:451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.