Abstract
The worldwide spread of antibiotic-resistant bacteria has lent urgency to the search for antibiotics with new modes of action that are devoid of preexisting cross-resistances. We previously described a unique class of acyldepsipeptides (ADEPs) that exerts prominent antibacterial activity against Gram-positive pathogens including streptococci, enterococci, as well as multidrug-resistant Staphylococcus aureus. Here, we report that ADEP prevents cell division in Gram-positive bacteria and induces strong filamentation of rod-shaped Bacillus subtilis and swelling of coccoid S. aureus and Streptococcus pneumoniae. It emerged that ADEP treatment inhibits septum formation at the stage of Z-ring assembly, and that central cell division proteins delocalize from midcell positions. Using in vivo and in vitro studies, we show that the inhibition of Z-ring formation is a consequence of the proteolytic degradation of the essential cell division protein FtsZ. ADEP switches the bacterial ClpP peptidase from a regulated to an uncontrolled protease, and it turned out that FtsZ is particularly prone to degradation by the ADEP–ClpP complex. By preventing cell division, ADEP inhibits a vital cellular process of bacteria that is not targeted by any therapeutically applied antibiotic so far. Their unique multifaceted mechanism of action and antibacterial potency makes them promising lead structures for future antibiotic development.
Keywords: proteolysis, divisome, tubulin, multidrug-resistant Staphylococcus aureus
Bacterial resistance to important antibiotic classes has become a major public health problem concerning the treatment of nosocomial and community-acquired infections. During the last decades, there has been a significant spread of bacterial resistance, along with a constantly declining number of antibiotic drug approvals (1, 2). For instance, the proportion of healthcare-related staphylococcal infections that are caused by multidrug-resistant Staphylococcus aureus (MRSA) has dramatically increased from 2% in 1974 to 64% in 2004 in intensive care units of the United States (3). These pathogens are now spreading into the community (4), representing a rising hygienic and economical threat to the public healthcare system, and clinical MRSA strains that have gained additional resistances to so-called “last resort antibiotics” including vancomycin, daptomycin, and linezolid have already been described (5–8). Due to the strong intrinsic capacity of bacteria to respond to any antibiotic treatment by genetic as well as physiological adaptations, it cannot be expected that the resistance situation will substantially relax in the future (9, 10). Therefore, there is an urgent and constant need to evaluate new antimicrobial agents that provide alternatives to commonly applied antibiotics.
In this context, a new class of acyldepsipeptide antibiotics, designated ADEPs, has recently attracted considerable attention. Distinct from all clinically applied antibiotics so far, which predominantly target DNA, RNA, protein, folic acid, or cell wall biosynthesis pathways, ADEPs act via an unprecedented mechanism by dysregulating ClpP (11, 12), the proteolytic core of the bacterial ATP-dependent caseinolytic protease. The Clp protease is a crucial factor in maintaining vital cellular functions in protein quality control and protein homeostasis as well as in controlling developmental processes like cell motility, genetic competence, cell differentiation, and sporulation (13, 14). ClpP is a tightly regulated protein, which is unable to degrade proteins on its own, and strictly depends on Clp ATPases and accessory proteins for proteolytic activation (15). The Gram-positive model organism Bacillus subtilis possesses three Clp ATPases, namely ClpX, ClpC, and ClpE, which are indispensable for recognizing, unfolding, and feeding the protein substrates through the tiny entrance pores into the proteolytic chamber of ClpP. ADEPs overcome these strict control mechanisms by turning ClpP into an uncontrolled protease that now degrades flexible proteins like casein in the absence of Clp ATPases (11). Biochemical studies demonstrated that ADEPs trigger oligomerization of ClpP monomers and activate the resulting tetradecamer to bind and degrade unfolded, nascent polypeptides and flexible proteins independently. Additionally, ADEPs abrogate the interaction of ClpP with cooperating Clp ATPases, thus preventing degradation of its physiological substrates and all natural functions of ClpP (16). Crystal structure determinations and electron microscopic images of ClpP from B. subtilis and Escherichia coli in its free form, and in complex with ADEPs, recently provided a rationale for these biochemical observations. The ADEPs increase subunit interaction between ClpP monomers, compete with the Clp ATPases for the same binding site, and trigger a closed- to open-gate structural transition of the substrate entrance pore, which is otherwise tightly closed (17, 18). Although this explained the activity of the ADEPs on the molecular level of its target, the specific series of events that finally leads to bacterial cell death remained unknown. In preliminary morphological studies, we observed that ADEP treatment induces filamentation of B. subtilis cells, implying a more specific cellular damage than overall degradation of unfolded proteins. Because understanding the mechanism of a novel antibiotic is a prerequisite for successful antibacterial drug discovery, we analyzed this phenomenon further. It turned out that ADEPs primarily inhibit bacterial cell division in various Gram-positive species, and fluorescence microscopy studies indicated that this inhibition is related to the delocalization of essential cell division mediators.
Results
ADEP Induces Filamentation of B. subtilis and Swelling of S. aureus and Streptococcus pneumoniae.
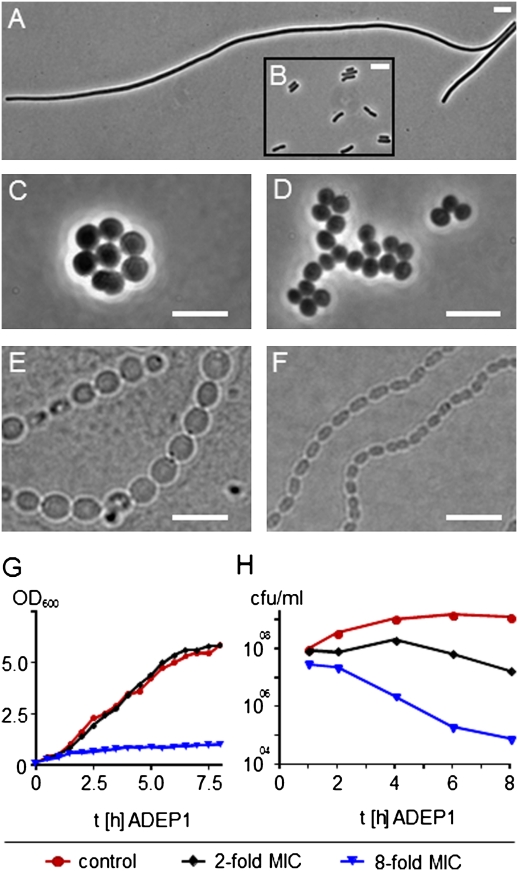
To identify the particular events that destine the bacteria to death upon ADEP treatment, we first analyzed the impact of ADEPs on the synthesis of essential biopolymers in B. subtilis 168, by measuring the incorporation of radioactively labeled precursors (Fig. S1). Even ADEP levels representing fourfold the minimal inhibitory concentration (MIC) did not inhibit the syntheses of DNA, RNA, protein, and cell wall, which are typically targeted by classical antibiotics. However, at the same ADEP concentrations, the rod-shaped cells of B. subtilis 168 grew into very long filaments, which reached 60- to 100-fold the length of untreated cells, and coccoid cells of S. aureus HG001 and S. pneumoniae G9A swelled to more than 3-fold the volume of wild-type cells (Fig. 1 A–F). Accordingly, when the optical density of an ADEP-treated B. subtilis culture was measured together with the number of colony forming units (cfu), the cell mass increased along with the untreated control cells at twice the MIC, whereas the cell numbers stagnated (Fig. 1 G and H). Only at eight times the MIC, ADEP exhibited bactericidal effects against B. subtilis, indicated by cell growth inhibition and the concomitant drop in cfu. Thus, in the presence of lower inhibitory ADEP concentrations, cells maintain their capacity to produce biomass, but they are no longer able to properly divide. These observations suggested that one of the early effects of ADEP treatment is the perturbation of bacterial cell division. Next, we set out to identify the link between ADEP-mediated ClpP dysregulation and the phenotypic consequence of cell division inhibition.
Fig. 1.
Effects of ADEPs on the growth of Gram-positive bacteria. ADEP treatment results in strong filamentation of B. subtilis 168 (A) as well as swelling of S. aureus HG001 (C) and S. pneumoniae G9A (E) compared with untreated control cells (B, D, and F, respectively), indicating cell division inhibition. Cells were treated for 4–5 h with inhibitory ADEP concentrations (A, 0.25 μg/mL ADEP2; C, 1 μg/mL ADEP2; and E, 0.8 μg/mL ADEP1). (Scale bars, 5 μm.) Accordingly, growth of ADEP-treated B. subtilis 168 (G) at 0.39 μg/mL ADEP1 (twice the MIC) increases along with the control, whereas the colony forming units (cfu) stagnate (H). At higher concentrations (1.56 μg/mL ADEP1, eight times the MIC), ADEP1 exhibits bactericidal effects, indicated by cell growth inhibition and the concomitant drop in cfu. Growth curves were confirmed by three independent experiments and values of a representative experiment are depicted. MIC, minimal inhibitory concentration.
ADEP Inhibits Septum Formation in a ClpP-Dependent Manner.
The filamentation phenotype was induced by all antibacterially active derivatives of the ADEP class that we tested. ADEP2 was selected for the following detailed studies on cell division in B. subtilis and S. aureus. ADEP2 is a well-characterized synthetic congener with improved activity against Gram-positive bacteria compared with the natural product ADEP1, and ADEP2 had already served as a model for the ADEP mechanism in our previous studies (Fig. S2).
Cell division in Gram-positive bacteria is executed by a macromolecular complex known as the divisome, which is highly dynamic and characterized by a time-dependent assembly of specific cell division proteins (19, 20). Divisome formation is driven by the GTP-dependent polymerization of FtsZ (21), a structural homolog of eukaryotic tubulin (22), into a ring-like structure at the prospective division site. The resulting FtsZ ring or “Z-ring” then functions as a scaffold for the recruitment of other proteins necessary for cell division, which, e.g., tether FtsZ to the membrane (FtsA), promote Z-ring assembly (ZapA), contribute to the dynamic nature of the Z-ring (EzrA), carry out cell wall synthesis at the septum (penicillin-binding proteins, PBP1 and PBP2B), and regulate the positioning of future Z-ring (DivIVA) (19, 20).
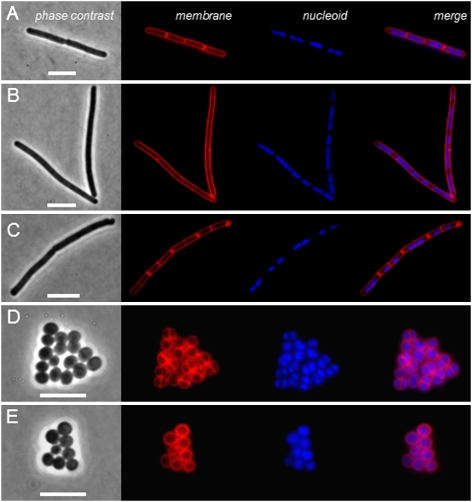
As a first approach to identify the reasons for ADEP-induced inhibition of cell division, we monitored septum formation and nucleoid segregation in ADEP-treated B. subtilis and S. aureus cells (Fig. 2). To this end, exponentially growing cells were treated with inhibitory concentrations of ADEP for 60 min, and were subsequently costained with FM5-95 membrane dye and DAPI nucleoid dye for analysis with fluorescence microscopy. In the elongated filaments of ADEP-treated B. subtilis, septum formation was clearly inhibited (Fig. 2 A and B), and equivalent results were obtained with S. aureus (Fig. 2 D and E). Inhibition of septum formation was confirmed by electron microscopy (Fig. S3). To find out whether the inhibition of septum formation was caused by ADEP-induced dysregulation of ClpP (11), we investigated septum formation also in a clpP deletion mutant of B. subtilis. When B. subtilis ΔclpP (23) was treated with similar ADEP concentrations as the wild-type strain 168, we observed no filamentation and cells divided normally, demonstrating the crucial role of ClpP for ADEP activity (Fig. 2C). Inhibition of septum formation can either be caused by direct interference with the cell division apparatus or by an imperfect segregation of nucleoids. To evaluate impaired chromosome segregation as a potential cause of cell division inhibition, we analyzed the localization pattern of GFP-tagged Spo0J in B. subtilis. Together with Soj, the chromosomal protein Spo0J is required for proper chromosome partitioning, separation of sister origins, and synchronous DNA replication, thus representing an indicator for correct nucleoid segregation (24, 25). Spo0J usually binds to eight known parS sites located in the origin region of the chromosome, and a delocalization of Spo0J would indicate perturbation of nucleoid segregation. However, our results showed that the packaging of the DAPI-stained nucleoids as well as Spo0J localization at the replication origins remained undisturbed upon ADEP treatment (Fig. 2 and Fig. S4). Therefore, imperfect nucleoid segregation can be excluded as a trigger for the inhibition of septum formation.
Fig. 2.
ADEP inhibits septum formation in B. subtilis and S. aureus. Fluorescence images show B. subtilis 168 and S. aureus HG001 cells costained with FM5-95 membrane dye and DAPI nucleoid dye after ADEP treatment. (A) B. subtilis 168, control; (B) B. subtilis 168, plus 0.25 μg/mL ADEP2; (C) B. subtilis 168 ΔclpP (strain QB4916), plus 0.25 μg/mL ADEP2; (D) S. aureus HG001, control; and (E) S. aureus HG001, plus 1.0 μg/mL ADEP2. After 60 min of ADEP treatment, septum formation is strictly inhibited in both wild-type species, whereas septa were normally formed in the ADEP-treated clpP deletion background, demonstrating an essential role of ClpP for ADEP activity. (Scale bars, 5 μm.)
ADEP Causes Delocalization of FtsZ and Inhibition of Z-Ring Assembly.
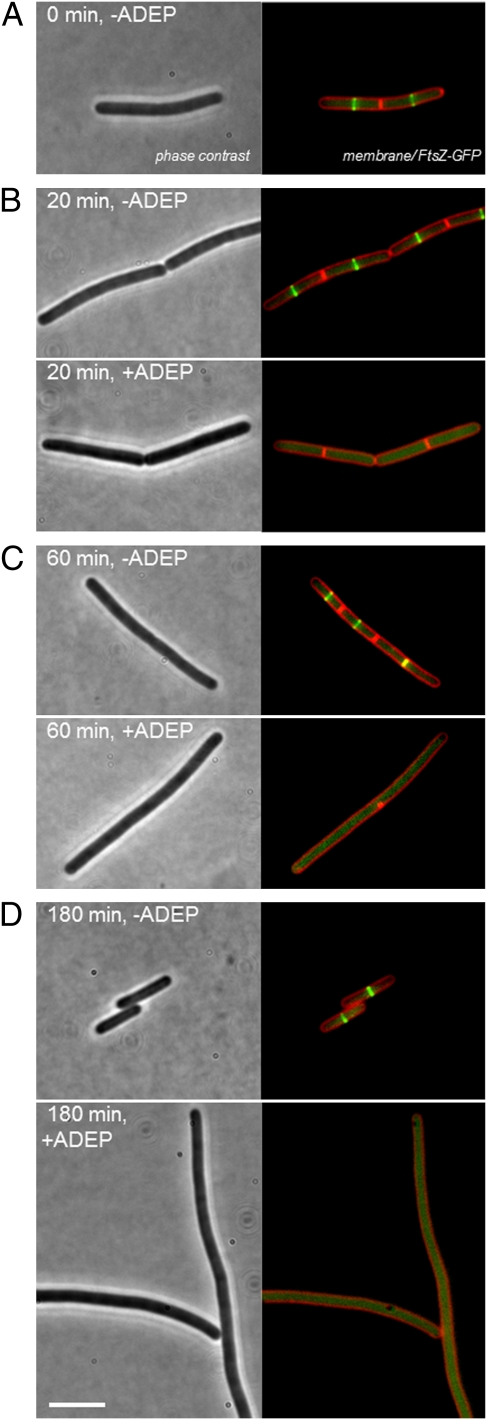
Because inhibition of septum formation was not due to disturbed nucleoid segregation (Fig. 2 and Fig. S4), we assumed that the ADEP–ClpP complex might directly affect the divisome. To test whether the ADEP–ClpP complex interferes with specific components of the divisome, we performed localization studies using GFP-labeled cell division proteins, including the first and major cell division protein FtsZ (Fig. 3A). When we treated exponentially growing B. subtilis cells with ADEP, we observed that the cells were characterized by the uniform loss of Z-rings after 20 min of antibiotic exposure, indicated by the absence of the typical GFP signal at midcell (Fig. 3B). This phenomenon became even more apparent after 60 min of treatment (Fig. 3C), when the bacteria became filamentous. Finally, after 180 min of exposure, the cells had grown into very long filaments with no apparent Z-rings (Fig. 3D). Accordingly, further divisome components including the late-phase cell division protein DivIVA were no longer recruited to the prospective cell division sites, indicating the perturbation of the whole cell division apparatus (Fig. S5).
Fig. 3.
ADEP inhibits Z-ring formation in B. subtilis. Fluorescence images show the localization of GFP-tagged FtsZ of B. subtilis 168 strain 2020 during exponential growth in the absence or presence of 0.25 μg/mL ADEP2. The fluorescence images shown are overlays of GFP (green) and FM5-95 (red) channels. (A) In untreated cells, FtsZ condenses at midcell to form the Z-ring. (B) After 20 min of ADEP treatment, cells uniformly showed delocalization of GFP–FtsZ along with a significant loss of Z-ring formation. (C and D) Lack of Z-rings and resulting filamentous growth after prolonged ADEP treatment. (Scale bars, 5 μm.)
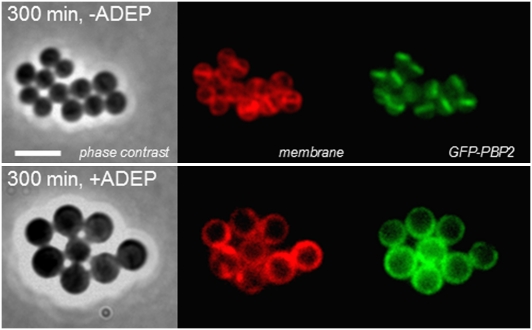
To test whether these findings also apply to S. aureus, we followed the localization of GFP-tagged penicillin-binding protein 2 (PBP2) in the presence of ADEP (Fig. 4). PBP2 is involved in the late stages of cell division and it depends on FtsZ for its localization at midcell, where it contributes to septal cell wall synthesis by catalyzing transpeptidation and transglycosylation reactions (26). We observed that GFP–PBP2 of ADEP-treated S. aureus cells was completely delocalized instead of being present at the division site as seen in the control cells. Considering the results obtained with FtsZ from B. subtilis (Fig. 3), the delocalization of PBP2 in S. aureus is most likely the direct consequence of the inhibition of Z-ring assembly in this organism. Our results clearly indicate that the ADEP–ClpP complex perturbs normal localization of FtsZ.
Fig. 4.
ADEP triggers the delocalization of PBP2 in S. aureus. In correspondence to the results shown for B. subtilis (Fig. 3), the fluorescence images of GFP-tagged PBP2 of S. aureus strain RNpPBP2-31 show the delocalization of GFP–PBP2 upon ADEP treatment (1 μg/mL), which is most probably because of the inhibition of septum formation at the stage of Z-ring assembly. (Scale bars, 2.5 μm.)
Effect of ADEP Is Independent of Clp ATPases.
In previous reports it was shown that the Clp ATPase ClpX plays an important role in balancing the ratio of assembled versus unassembled FtsZ and overexpression of ClpX inhibits Z-ring assembly in B. subtilis cells (27, 28). Because ADEP displaces the ClpP ATPases from ClpP (16), it is possible that the presence of ADEP results in an increased concentration of free ClpX, which then prevents FtsZ polymerization. To test this possibility, we determined the impact of ADEP in a ΔclpX mutant, as well as in deletion backgrounds of the other Clp ATPases ClpC or ClpE. In all three mutants, cell division remained affected by ADEP treatment (Fig. S6), demonstrating that the ADEP-mediated inhibition of Z-ring assembly occurs independently of Clp ATPases.
ADEP Treatment Results in Decreased Abundance of FtsZ.
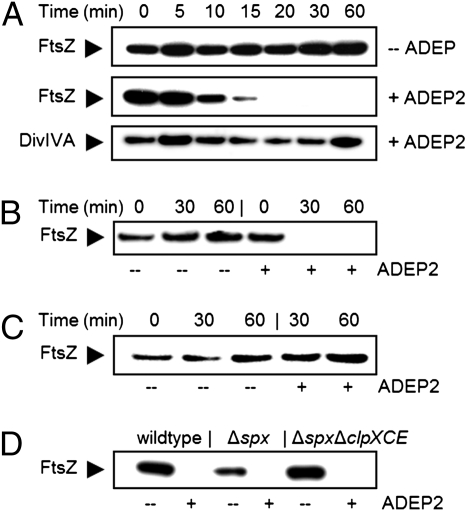
Next, we measured the amount of FtsZ in untreated and treated B. subtilis cells using immunoblotting. Interestingly, the FtsZ signal disappeared after 10–20 min of ADEP treatment (Fig. 5A). This phenomenon was not restricted to B. subtilis and similar results were obtained with S. aureus (Fig. 5B). To investigate whether ADEP-activated ClpP was responsible for the degradation of FtsZ, we repeated the experiment with ΔclpP mutants, and indeed no decrease of the FtsZ signal was observed in such background (Fig. 5C and Fig. S7). Apart from a mutant carrying a clpP deletion in the wild-type background, we also used a ΔclpP strain carrying an additional deletion in spx, encoding a global transcriptional regulator. Both B. subtilis clpP and clpX null mutants suffer from severe defects in growth and competence (29), whereas the additional deletion of the spx gene relieves most of the adverse effects of clpP and clpX deletions (30). Noteworthy, it has been previously reported that spx deletions do not significantly alter intracellular FtsZ levels (27). We also tested a Δspx ΔclpXCE triple Clp-ATPase mutant and again the FtsZ signal disappeared after incubation with ADEP (Fig. 5D). Clearly none of the Clp ATPases are involved in the ADEP-dependent degradation of FtsZ.
Fig. 5.
ADEP induces the ClpP-dependent degradation of FtsZ in bacterial cells. ADEP treatment of exponentially growing WT cells of (A) B. subtilis 168 and (B) S. aureus HG001 resulted in a decreased abundance of FtsZ over time, compared with the untreated control. Immunodetection of FtsZ or DivIVA was performed using specific anti-FtsZ or anti-DivIVA antibodies, respectively, as indicated in the margin. DivIVA served as a housekeeping protein control. (C) Immunodetection of FtsZ in untreated or ADEP-treated B. subtilis strain QB4916 (ΔclpP). (D) ADEP-induced degradation of FtsZ after 60 min in B. subtilis strain BJK474 (Δspx ΔclpXCE) and the respective control strains B. subtilis strain BJK424 (Δspx) and B. subtilis PY79 (WT).
ADEP Induces ClpP-Dependent Degradation of FtsZ in Vitro.
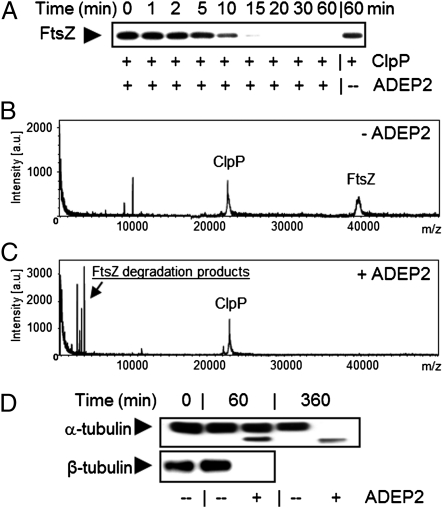
To confirm that the ADEP–ClpP complex is directly responsible for FtsZ degradation, we set up an in vitro test system using purified ClpP and FtsZ from B. subtilis 168. When we incubated FtsZ with a ClpP reaction mixture in the presence of ADEP, we observed that the FtsZ signal disappeared after 10–15 min, and this was clearly ADEP dependent (Fig. 6A). To determine the nature of the degradation products, we performed MS of the samples. Although ClpP (∼22.8 kDa) and FtsZ (∼39.8 kDa) could be easily detected in the untreated controls (Fig. 6B), the ADEP-treated samples were characterized by the complete loss of the FtsZ peak and the concomitant appearance of various signals in the low-molecular range (<4 kDa), most probably representing FtsZ degradation products (Fig. 6C). Because other folded proteins resisted degradation in vitro (Fig. S8), the ADEP–ClpP complex appears to select for especially sensitive target proteins, one of which is FtsZ.
Fig. 6.
ADEP–ClpP degrades FtsZ and αβ-tubulin in vitro. (A) Time-dependent degradation of purified FtsZ by the ADEP–ClpP complex in vitro. (B and C) Mass spectra of in vitro FtsZ degradation assays containing purified, native FtsZ protein (∼39.8 kDa) and ClpP–6HIS protein (∼22.8 kDa) after incubation for 60 min at 37 °C (B) in the absence or (C) in the presence of 10 μg/mL ADEP2. Compared with the control assay, the mass spectrum of the ADEP2-supplemented degradation assay was characterized by the loss of the FtsZ peak and the concomitant appearance of several peaks in the low-molecular range (<4 kDa). (D) Time-dependent degradation of purified αβ-tubulin by the ADEP–ClpP complex in vitro. Immunodetection of αβ-tubulin was performed using anti–α-tubulin or anti–β-tubulin antibodies as indicated. Both α- and β-tubulin are targets for ADEP–ClpP-dependent degradation; however, β-tubulin is degraded faster than α-tubulin for yet unknown reasons.
FtsZ is a close structural homolog of tubulin but is rather dissimilar in its amino acid sequence (22). To examine whether the degradation by ClpP was specific for FtsZ, we decided to test bovine αβ-tubulin in our degradation assay. Interestingly, the concentration of α- and β-tubulin clearly decreased in the presence of ADEP (Fig. 6D). Apparently, it is the topology of FtsZ, and not so much a specific amino acid sequence, that makes it vulnerable for proteolytic attack by the ADEP–ClpP complex.
Discussion
The mechanism of antibacterial action of the ADEPs presents itself as a unique and multifaceted process. At the molecular level, ADEPs target ClpP and trigger a variety of distinct events. By binding to the interphase of two adjacent ClpP monomers, the ADEPs support the oligomerization process that is a prerequisite of forming a functional proteolytic core (17). They sterically block an important contact site for the Clp ATPases at ClpP, thereby abrogating the interaction between the two partners and the normal functions of ClpP (17). Most importantly, the compounds trigger a conformational change in ClpP that widens the gated pore and makes particular groups of proteins prone to untimely degradation (17, 18).
Dissecting the cascade of events that leads to bacterial death, we noticed two distinct phenotypic reactions of ADEP-treated bacteria. At antibiotic concentrations close to the MIC, cell size increased, indicating considerable remaining biosynthetic capacity, and especially rod-shaped B. subtilis cells grew into very long filaments. In contrast, at several times the MIC, biomass increase ceased early, resulting only in small filaments or spheres. The latter phenotype can be readily explained by previously identified general effects of the ADEPs. The widened entrance pore of ClpP is large enough to allow access of a small bundle of protein strands (17), and we reported earlier that ADEP-activated ClpP can degrade the flexible model substrate protein casein or nascent protein chains in the course of translation (11, 16). Imagining such a broad destructive capacity, it is highly plausible that essential proteins are depleted in various vital processes of the bacterial metabolism, resulting in the cessation of biomass production and growth.
In the current report, we now present the reason for the filamentation phenotype at lower ADEP concentrations. Our results show that ADEP treatment prevents the formation of FtsZ rings, and thereby cell division, due to the ClpP-dependent degradation of the major cell division protein FtsZ. FtsZ appears to be a preferred target for the ADEP–ClpP complex at low inhibitory concentrations, because cells treated this way are not impaired in their general metabolic activity, yet incapable of septum formation. In our in vitro assays, FtsZ was almost completely degraded into short peptides, whereas even the flexible casein was digested only into discrete protein fragments under the same conditions (11, 16). Why FtsZ is so particularly sensitive for ADEP–ClpP is not apparent. We could show that αβ-tubulin, a structural homolog of FtsZ, is also prone to ADEP–ClpP-dependent proteolysis, whereas different proteins resisted degradation. Therefore, it appears that the efficient degradation of FtsZ is most probably the result of a particular structural feature that is shared with tubulin.
Previous studies have shown that ClpX and ClpP regulate the availability of FtsZ in E. coli (31, 32). In B. subtilis, ClpX is also involved in the regulation of FtsZ; however, ClpP was not involved in this activity and it was clearly shown that FtsZ was not degraded by ClpXP in B. subtilis (27, 28). ADEP renders the activity of ClpP independent of Clp ATPases including ClpX, and we have shown that the ADEP activity does not require ClpX; therefore, the mechanism by which ADEP-activated ClpP degrades FtsZ seems unrelated to the natural Clp controls, at least in B. subtilis.
FtsZ is a particularly sensitive target of ADEP-dysregulated ClpP and consequently cell division is the primarily targeted pathway of the ADEPs from a physiological point of view. Bacterial cell division emerged in recent years as a new pathway for antibiotic attack and benzamide inhibitors have been described that interfere with FtsZ depolymerization dynamics by inhibiting GTPase function (33–35). These compounds showed promising MIC values and moderate in vivo activity against staphylococcal infections, but other Gram-positive pathogens were not affected. The ADEPs now demonstrate a unique and highly effective way of preventing FtsZ function. Their strong antibacterial potency in the ng/mL range against multiresistant staphylococci, streptococci, and enterococci, and their considerable efficacy in animal models of infection (11), prove the value of cell division inhibition as a pathway for antibiotic intervention and make ADEPs a promising model for the development of new and effective antibacterial agents.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains and plasmids used in this study are listed in Table S1. Strains were grown in lysogeny broth (LB) at 37 °C supplemented with appropriate antibiotics or inducing compounds when required. For filamentation experiments in vivo, B. subtilis cells were treated with 0.25 μg/mL ADEP2 (eight times the MIC), and S. aureus cells with 1.0 μg/mL ADEP2 (twice the MIC) unless otherwise stated. MICs were determined according to the guidelines of the Clinical Laboratory Standards Institute except for using the corresponding test medium for the assay of interest.
Incorporation of Radioactive Metabolites.
The incorporation of radioactive metabolites was performed as previously described (36) and as detailed in Fig. S1.
Fluorescence and Electron Microscopy.
For fluorescence microscopy, cells were grown to midexponential phase at 37 °C and analyzed by phase contrast or fluorescence microscopy on microscope slides covered with a thin film of 1% agarose in PBS. When ADEP2 was used, the antibiotic was usually added to the cultures in advance at an OD600 of 0.1 and samples were then taken at distinct time points as indicated. DNA was visualized within the cells by staining with 4′,6-diamidino-1-phenylindole (DAPI; 0.25 μg/mL; Sigma-Aldrich) and membranes by staining with FM5-95 membrane dye (1 μg/mL; Molecular Probes). Fluorescence microscopy was carried out using a Zeiss Axiovert 200M microscope equipped with a Photometrics CoolSnap HQ CCD camera (Roper Scientific). Image acquisition and analysis were performed with Metamorph 6 (Molecular Devices) and ImageJ v1.43 software (National Institutes of Health).
EM was performed as previously described (37) and as detailed in Fig. S3.
Purification of ClpP and FtsZ.
C-terminally 6His-tagged ClpP of B. subtilis 168 was expressed in E. coli BL21(DE3) harboring the pClpP11 plasmid (38) and purified as described before (39). Native FtsZ of B. subtilis was purified as described by Wang and Lutkenhaus (40) using E. coli strain W3110 (pBS58) (pCXZ) with the following modifications. Cells were resuspended and lysed in buffer A (50 mM Tris/HCl, 50 mM KCl, 1 mM EDTA, 10% glycerol, pH 8). Cell debris was pelleted and the supernatant was filtered using 0.45-μm membrane filters (Whatman Schleicher & Schuell) and applied to an AEX Source Q 30-mL column (GE Healthcare). Proteins were eluted by applying a gradient of 0–50% of buffer B (50 mM Tris/HCl, 1 M KCl, 1 mM EDTA, 10% glycerol, pH 8). Quality and quantity of the protein eluates were determined via SDS/PAGE analyses and measured by using the Bradford assay (Bio-Rad) and the Nanodrop spectrophotometer (Nanodrop Technologies).
In Vitro FtsZ Degradation Assay.
For in vitro degradation of FtsZ or αβ-tubulin, target proteins (4 μM of B. subtilis FtsZ, or 5 μM of bovine αβ-tubulin; Cytoskeleton) were incubated with B. subtilis 168 ClpP (3 μM) in ClpP activity buffer (50 mM Tris/HCl pH 8, 25 mM MgCl2, 100 mM KCl, 2 mM DTT) at 37 °C in the absence or presence of 10 μg/mL ADEP2. Samples were taken at distinct time points and analyzed via standard SDS/PAGE and immunodetection techniques.
MS and Data Analyses.
Mass spectra analyses of in vitro FtsZ degradation assays were performed using the Bruker Biflex III MALDI-TOF mass spectrometer (Bruker Daltonics). Samples of 400 μL of the respective in vitro degradation assay reaction were dialyzed against ultrapure H2O using Slide-A-Lyzer dialysis cassettes (2K MWCO, 0.1–0.5 mL; ThermoFisherScientific) for 16–20 h. Aliquots of 1 μL were mixed with 2 μL of matrix, consisting of a saturated solution of 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid) in 33% acetonitrile-0.1% trifluoroacetic acid (1:2), and were subsequently spotted onto a ground steel MALDI target plate and air dried at room temperature. Mass spectra were measured in the linear positive ion mode within a mass range of 1,000–50,000 Da and further analyzed using FlexAnalysis (version 2.0) software (Bruker Daltonics). The spectra were externally calibrated by peptide calibration standards.
Supplementary Material
Acknowledgments
We thank U. Gerth, R. Losick, T. Msadek, M. Pinho, K. Turgay, and P. Zuber for the kind gift of strains and plasmids. We also thank S. Raddatz and H. Paulsen for the synthesis of ADEP derivatives; A. Popp and H.-P. Kroll for conducting the electron microscopic study; and A. Mogk, A. Hoffmann, J. Kirstein, and K. Turgay for performing the protein degradation assays depicted in Fig. S8. We acknowledge the technical expertise of A. Berscheid, U. Sauer, and H. Strahl. The authors appreciate financial support from the German Research Foundation (FOR 854) and the research fund of the Medical Faculty, University of Bonn (BONFOR).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110385108/-/DCSupplemental.
References
- 1.Infectious Diseases Society of America The 10 x ’20 Initiative: Pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis. 2010;50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 2.Souli M, Galani I, Giamarellou H. Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Euro Surveill. 2008;13:pii:19045. [PubMed] [Google Scholar]
- 3.Klevens RM, et al. National Nosocomial Infections Surveillance System Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. Clin Infect Dis. 2006;42:389–391. doi: 10.1086/499367. [DOI] [PubMed] [Google Scholar]
- 4.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46(Suppl 5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiramatsu K, et al. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 6.Tenover FC, et al. Characterisation of a Staphylococcus aureus strain with progressive loss of susceptibility to vancomycin and daptomycin during therapy. Int J Antimicrob Agents. 2009;33:564–568. doi: 10.1016/j.ijantimicag.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsiodras S, et al. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet. 2001;358:207–208. doi: 10.1016/S0140-6736(01)05410-1. [DOI] [PubMed] [Google Scholar]
- 8.Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2010;23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brötz-Oesterhelt H, Sass P. Postgenomic strategies in antibacterial drug discovery. Future Microbiol. 2010;5:1553–1579. doi: 10.2217/fmb.10.119. [DOI] [PubMed] [Google Scholar]
- 10.Sass P, Brötz-Oesterhelt H. In: Stress Responses in Foodborne Microorganisms. Wong HC, editor. Hauppauge, NY: Nova Science; 2011. [Google Scholar]
- 11.Brötz-Oesterhelt H, et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 12.Hinzen B, et al. Medicinal chemistry optimization of acyldepsipeptides of the enopeptin class antibiotics. ChemMedChem. 2006;1:689–693. doi: 10.1002/cmdc.200600055. [DOI] [PubMed] [Google Scholar]
- 13.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Frees D, Savijoki K, Varmanen P, Ingmer H. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol Microbiol. 2007;63:1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x. [DOI] [PubMed] [Google Scholar]
- 15.Kirstein J, Molière N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- 16.Kirstein J, et al. The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol Med. 2009;1:37–49. doi: 10.1002/emmm.200900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee BG, et al. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat Struct Mol Biol. 2010;17:471–478. doi: 10.1038/nsmb.1787. [DOI] [PubMed] [Google Scholar]
- 18.Li DH, et al. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chem Biol. 2010;17:959–969. doi: 10.1016/j.chembiol.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams DW, Errington J. Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 21.Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 22.Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 23.Msadek T, et al. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol Microbiol. 1998;27:899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee PS, Grossman AD. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol Microbiol. 2006;60:853–869. doi: 10.1111/j.1365-2958.2006.05140.x. [DOI] [PubMed] [Google Scholar]
- 25.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 26.Pinho MG, Errington J. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol Microbiol. 2005;55:799–807. doi: 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- 27.Weart RB, Nakano S, Lane BE, Zuber P, Levin PA. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol Microbiol. 2005;57:238–249. doi: 10.1111/j.1365-2958.2005.04673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haeusser DP, Lee AH, Weart RB, Levin PA. ClpX inhibits FtsZ assembly in a manner that does not require its ATP hydrolysis-dependent chaperone activity. J Bacteriol. 2009;191:1986–1991. doi: 10.1128/JB.01606-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerth U, Krüger E, Derré I, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- 30.Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol Microbiol. 2001;42:383–394. doi: 10.1046/j.1365-2958.2001.02639.x. [DOI] [PubMed] [Google Scholar]
- 31.Camberg JL, Hoskins JR, Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci USA. 2009;106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camberg JL, Hoskins JR, Wickner S. The interplay of ClpXP with the cell division machinery in Escherichia coli. J Bacteriol. 2011;193:1911–1918. doi: 10.1128/JB.01317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haydon DJ, et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science. 2008;321:1673–1675. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 34.Andreu JM, et al. The antibacterial cell division inhibitor PC190723 is an FtsZ polymer-stabilizing agent that induces filament assembly and condensation. J Biol Chem. 2010;285:14239–14246. doi: 10.1074/jbc.M109.094722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams DW, Wu LJ, Czaplewski LG, Errington J. Multiple effects of benzamide antibiotics on FtsZ function. Mol Microbiol. 2011;80:68–84. doi: 10.1111/j.1365-2958.2011.07559.x. [DOI] [PubMed] [Google Scholar]
- 36.Freiberg C, et al. Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors with antibacterial activity. J Biol Chem. 2004;279:26066–26073. doi: 10.1074/jbc.M402989200. [DOI] [PubMed] [Google Scholar]
- 37.Pauluhn J, Emura M, Mohr U, Popp A, Rosenbruch M. Two-week inhalation toxicity of polymeric diphenylmethane-4, 4′-diisocyanate (PMDI) in rats: Analysis of biochemical and morphological markers of early pulmonary response. Inhal Toxicol. 1999;11:1143–1163. doi: 10.1080/089583799196637. [DOI] [PubMed] [Google Scholar]
- 38.Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sass P, Bierbaum G. Lytic activity of recombinant bacteriophage phi11 and phi12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl Environ Microbiol. 2007;73:347–352. doi: 10.1128/AEM.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.