Abstract
Members of the Myb oncoprotein and E2F-Rb tumor suppressor protein families are present within the same highly conserved multiprotein transcriptional repressor complex, named either as Myb and synthetic multivuval class B (Myb-MuvB) or as Drosophila Rb E2F and Myb-interacting proteins (dREAM). We now report that the animal-specific C terminus of Drosophila Myb but not the more highly conserved N-terminal DNA-binding domain is necessary and sufficient for (i) adult viability, (ii) proper localization to chromosomes in vivo, (iii) regulation of gene expression in vivo, and (iv) interaction with the highly conserved core of the MuvB/dREAM transcriptional repressor complex. In addition, we have identified a conserved peptide motif that is required for this interaction. Our results imply that an ancient function of Myb in regulating G2/M genes in both plants and animals appears to have been transferred from the DNA-binding domain to the animal-specific C-terminal domain. Increased expression of B-MYB/MYBL2, the human ortholog of Drosophila Myb, correlates with poor prognosis in human patients with breast cancer. Therefore, our results imply that the specific interaction of the C terminus of Myb with the MuvB/dREAM core complex may provide an attractive target for the development of cancer therapeutics.
Keywords: oncogene, transcription, repression, evolution
The Myb oncogene family was discovered because of the retroviral transduction of the chicken cellular myb protooncogene that generated the avian myeloblastosis virus (1, 2). Both the cellular Myb and viral Myb proteins are present within the nucleus of the cell, bind to specific DNA sequences, and can regulate gene expression (3–9). The Myb DNA-binding domain is present in members of all major classes of eukaryotic organisms, including plants, animals, fungi, cellular slime molds, and protists (10, 11). Simpler plants have only one or a few Myb transcription factors, whereas all flowering plant species have hundreds of related Myb transcription factors that arose via massive gene duplication and divergence (12). The picture in animals is simpler. All vertebrates have three closely related Myb transcription factors, whereas most invertebrates have only one Myb transcription factor. In addition to the presence of an aminoterminal Myb DNA-binding domain, most of these Myb transcription factors of animals share a conserved C-terminal domain that appears to be animal-specific. We have recently reported that, rather surprisingly, this C-terminal Myb domain is sufficient to rescue the lethality of a Myb null mutation in Drosophila when expressed via a heterologous promoter (13). We have now used a combination of genetic, cell biological, and biochemical approaches to explore the function of this animal-specific C-terminal Myb domain. We find that this domain is both necessary and sufficient for interaction with the synthetic multivulval class B (MuvB) core, a highly conserved transcriptional repressor complex that also interacts with the E2F-Rb tumor suppressor proteins (14–16).
Results
Evolutionarily Guided Alanine Substitution Mutants of Drosophila Myb.
Several temperature-sensitive and null alleles of Drosophila Myb have previously been described (17–20). However, attempts to isolate additional mutant alleles via traditional chemical mutagenesis were unsuccessful despite the screening of large numbers of mutagenized animals. We surmised that new mutants might be difficult to obtain for two reasons. First, many amino acid substitutions might be phenotypically silent, and would therefore not be isolated in traditional screens for altered morphology, sterility, or lethality. Second, nonsense and frameshift mutants might result in dominant loss-of-function (antimorphic) or gain-of-function (neomorphic) alleles that cause lethality and would be unrecoverable. We therefore adapted a previously described strategy in which individual amino acids or small patches of adjacent amino acids are replaced by alanines using site-directed mutagenesis (21). Rather than targeting patches of highly charged residues that are likely to be exposed on the hydrophilic surface of a properly folded protein, we used multiple protein sequence alignments as a guide for identifying both charged and uncharged regions subject to evolutionarily guided alanine substitution. Our rationale was that because Drosophila Myb is present within the conserved Myb-MuvB or Drosophila Rb E2F and Myb-interacting (dREAM) multiprotein complex, some residues critical for protein function might be required for hydrophobic protein-protein interactions, protein-DNA interactions, or the creation of structural motifs or might be targets for posttranslational modifications (14–16).
We constructed a set of 17 alanine mutations that targeted (i) residues within the highly conserved DNA-binding domain that had previously been shown to be critical for DNA binding or for transcriptional regulation, (ii) residues within the highly conserved animal-specific C-terminal domain, and (iii) an acidic patch N-terminal to the DNA-binding domain that is present in all closely related Myb proteins of animals and that we have previously shown can regulate DNA binding in vitro and in vivo (5, 22–30) (Fig. 1). To facilitate detection in vivo, the WT Myb and each mutant protein were fused to a fluorescent GFP domain. To avoid the potentially unrecoverable nature of dominant lethal mutations, the mutant cDNAs were initially expressed under control of the binary GAL4-upstream activation sequence (UAS) transcriptional regulatory system in transgenic Drosophila (31). We also expressed either the N-terminal Myb DNA-binding domain or the animal-specific C-terminal Myb domain using the GAL4-UAS system. All the alanine mutant proteins were expressed at similar levels when tested by transient transfection in Drosophila S2 cells as determined by immunofluorescence and immunoblotting.
Fig. 1.
Mutants of Drosophila Myb. (A) (Top) Cartoon of the WT protein. The cyan boxes indicate the three conserved Myb repeats that constitute the paneukaryotic N-terminal DNA-binding domain. The red box indicates the conserved animal-specific C-terminal domain. The lines below the cartoon indicate the location of 17 previously undescribed alanine substitution mutants. Also shown are cartoons of the N-terminal (Middle; N-term) mutant (residues 1–316) and C-terminal (Bottom; C-term) mutant (residues 247–657). (B) Alignment of a conserved animal-specific C-terminal region that includes a previously described heat-sensitive hypomorphic Myb1 mutant was made using MACAW software (NCBI) (34, 51). The three Myb genes of vertebrates arose via duplications that occurred before the divergence of all species examined thus far and are represented here by human MYB/C-MYB, MYBL1/A-MYB, and MYBL2/B-MYB (11).
Animal-Specific C Terminus of Myb Is Necessary and Sufficient for Adult Viability.
As previously reported, we found that the WT and C-terminal Myb proteins were capable of rescuing Myb-null mutant animals to adult viability (Fig. 2 and Fig. S1). In contrast, the N-terminal Myb DNA-binding domain functioned as a dominant lethal. An analysis of chromosome condensation by antibody staining for histone H3 phosphorylated on serine 10 (PH3) in imaginal wing disks revealed an increased number of PH3-positive cells in the posterior compartment expressing the N-terminal Myb protein (Fig. S2). The majority of these PH3-positive cells were delayed or arrested in prophase. These results are similar to those previously reported in Myb-null and hypomorphic mutants, consistent with a dominant loss-of-function phenotype for the N terminus (19, 32, 33).
Fig. 2.
Rescue of Myb-null Drosophila by Myb WT and mutant proteins. (Upper) Rescue of adult viability of the otherwise lethal MybMH107-null mutant via the GAL4-UAS binary system at 25 °C. (Lower) Rescue of adult viability of the otherwise lethal MybMH107-null mutant via the native Myb promoter. Details of the Drosophila genetics are provided in Figs. S1 and S4. ND, indicated mutant not tested for rescue at the indicated temperature.
Of the 17 alanine substitution mutants tested, most rescued the lethality of a Myb-null mutant at 25 °C using the GAL4-UAS system driven by the ubiquitously expressed daughterless promoter (Fig. 2 and Fig. S1). Three mutants rescued poorly at 25 °C and not at all at 29 °C (TPT445AAA, KW606AA, and DQ617AA). Another five mutants rescued reasonably well at 25 °C but only weakly at 29 °C (DKD36AAA, PP374AA, FLK393AAA, EDL475AAA, and RK523AA). Interestingly, the C terminus itself displayed a similar heat-sensitive rescue phenotype in the GAL4-UAS system. Because the GAL4-UAS system itself is cold-sensitive in Drosophila, with higher expression levels as temperatures increase in the range of 18 °C to 29 °C, we reasoned that the observed heat sensitivity of a Myb mutant might be attributable either to classic heat-sensitive behavior via protein instability or to a dominant gain-of-function phenotype at higher levels of expression. We therefore tested these 8 alanine substitution mutants as well as the C terminus for dominant gain-of-function phenotype in a Myb WT genetic background. No mutant phenotypes were detected when the 8 alanine substitution mutants were expressed using the GAL4-UAS system driven by the ubiquitously expressed daughterless promoter at 29 °C. However, the C terminus did cause a nonlethal mutant phenotype under similar conditions with an apparent failure of abdominal cuticle differentiation in the dorsal midline (Fig. S3). Interestingly, a similar phenotype has been reported in Myb1-mutant animals raised at the permissive temperature (18). These results imply that the C terminus can function as a dominant loss-of-function allele at high levels of expression.
To study the function of the mutant proteins at physiological levels, we therefore constructed transgenes in which the eight alanine substitution mutants of interest, the C terminus, or the WT Myb protein was expressed under control of the native Myb regulatory sequences embedded within their normal context within a 14-kb segment of genomic DNA (Fig. 2 and Fig. S4). We had previously shown that when the WT Myb protein was expressed in this context, it was capable of rescuing a Myb-null mutant to full viability and fertility (19). Five of the alanine substitution mutants were capable of rescuing a Myb-null mutant to viability at either 25 °C or 29 °C when expressed via native Myb regulatory sequences. In contrast, the three alanine substitution mutants that rescued only weakly via the GAL4-UAS system failed to rescue at either 25 °C or 29 °C when expressed via native Myb regulatory sequences. Two of these mutants (KW606AA and DQ617AA) were capable of rescuing weakly at 18 °C when expressed via the native Myb promoter, implying that these mutant proteins themselves are heat-sensitive. Interestingly, the heat-sensitive hypomorphic Myb1 mutant has a single amino acid substitution in the same highly conserved animal-specific region of the protein (G613S) (34) (Fig. 1). Unexpectedly, the C terminus exhibited a similar heat-sensitive rescue when expressed via the native Myb regulatory sequences, whereas a third alanine substitution mutant (TPT445AAA) rescued only very rarely and only at 18 °C. In summary, these results imply that the animal-specific C terminus is both necessary and sufficient for adult viability.
Animal-Specific C Terminus of Myb Is Necessary and Sufficient for Transcriptional Regulation.
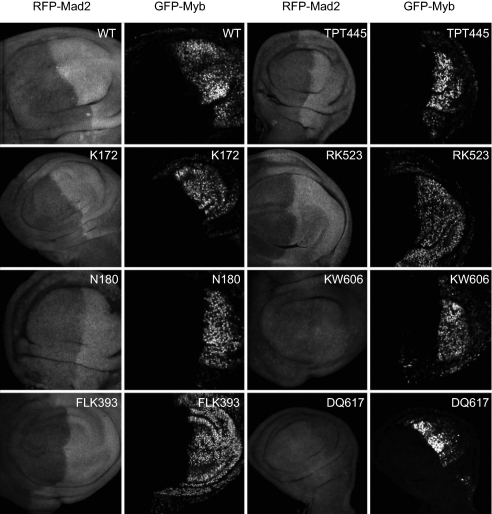
Several critical regulators of G2/M are themselves transcriptionally regulated by Drosophila Myb in cultured cell lines and in vivo (13, 35). We wished to examine the correlation between transcriptional regulation in vivo and rescue of adult viability for a series of Myb mutants. For this purpose, we used the GAL4-UAS system driven by the engrailed promoter to express either WT or mutant GFP-Myb fusion protein in the posterior compartment of the imaginal wing disks of Myb-null third-instar larvae. As an assay of transcriptional regulation, we used a red fluorescent protein (RFP)-Mad2 fusion protein expressed under control of the native Mad2 promoter. WT Myb and all the mutants tested, with the notable exception of KW606AA and DQ617AA, were able to induce expression of RFP-Mad2 in vivo (Fig. 3). We had previously reported that the Myb C terminus alone was also capable of inducing expression of RFP-Mad2 in vivo (13). Together, these results imply that the animal-specific Myb C terminus is both necessary (KW606AA and DQ617AA) and sufficient for transcriptional regulation by Myb in vivo.
Fig. 3.
Regulation of Mad2 expression by Myb WT and mutant proteins. The indicated Myb proteins were expressed via engrailed-GAL4 in the posterior compartments of third-instar MybMH107-null mutant larvae. Imaginal disks were dissected, fixed, stained with anti-RFP antibodies, and visualized by laser scanning confocal microscopy at 400× magnification. Each row displays RFP-Mad2 (columns 1 and 3) and GFP-Myb (columns 2 and 4) in two different imaginal wing disks.
Animal-Specific C Terminus of Myb Is Necessary and Sufficient for Proper Chromosomal Localization.
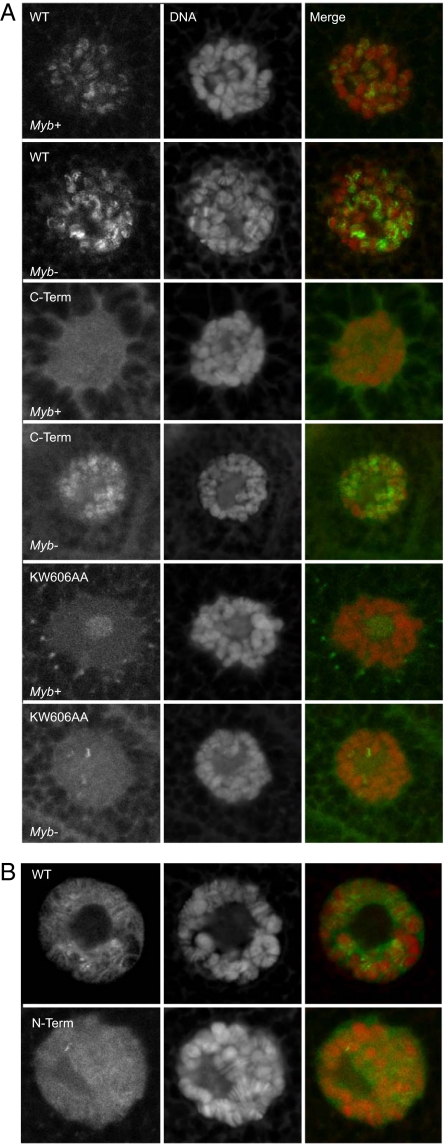
To investigate the function of the Myb C terminus further, we examined the intracellular localization of WT and mutant GFP-fusion proteins. The salivary glands of third-instar Drosophila larvae contain giant polytene chromosomes that permit the visualization of condensed chromosomal bands and relatively decondensed interbands (36). As we have previously reported, the WT GFP-Myb fusion protein expressed under control of the native Myb regulatory sequences decorates a large number of euchromatic interband regions but neither the highly condensed pericentric heterochromatin nor the central nucleolar region (33) (Fig. 4). The GFP-C terminus displayed a diffuse nuclear pattern in a Myb WT background with only some hints of an underlying banding pattern. In contrast, in a Myb-null background, the same GFP-C terminus clearly decorated chromosomal interbands in a pattern similar to that of WT GFP-Myb, although perhaps a bit more diffuse. These results imply that the animal-specific Myb C terminus is capable of localizing to the cell nucleus and interacting specifically with euchromatic regions of the chromosomes in the complete absence of the highly conserved Myb DNA-binding domain. However, the C terminus does not compete efficiently with the WT protein for what appears to be a limiting number of binding sites within the chromosomes.
Fig. 4.
Chromosomal localization of Myb WT and mutant proteins. (A) Indicated GFP-Myb fusion proteins were expressed via the native Myb promoter in either Myb WT (Myb+) or Myb-null (Myb−) larvae. Salivary glands were dissected, fixed, and visualized by laser scanning confocal microscopy at 600× magnification. Each row displays GFP-Myb (Left), DNA staining (Center), and merged images (Right) of the same salivary gland nucleus. The central area of lightly stained DNA visible in some nuclei is the nucleolus. (B) Full-length WT or N-terminal GFP-Myb protein was expressed via daughterless-GAL4.
The GFP-Myb-KW606AA substitution mutant within the C-terminal region of the full-length Myb protein also displayed a diffuse nuclear localization in the presence of the WT Myb protein, including a filling of the nucleolar region not observed with either the GFP-Myb WT or C-terminal protein. The nearby GFP-Myb-DQ617AA substitution mutant behaved similar to the GFP-Myb-KW606AA mutant protein. In a Myb-null background, the GFP-Myb-KW606AA mutant protein was able to decorate only a few bands within the polytene chromosomes, with the remainder of the protein being present diffusely throughout the nucleus. These results imply that although the KW606AA and DQ617AA mutants have an intact DNA-binding domain, they are incapable of localizing efficiently to all but a few chromosomal regions, even in the absence of any competition with WT Myb protein.
To determine the role of the highly conserved Myb DNA-binding domain itself in chromosomal localization, either the full-length Myb protein or the dominant lethal N terminus was expressed in third-instar larval salivary glands as a GFP fusion via the GAL-UAS system using the daughterless promoter. Both proteins appeared to be expressed at somewhat higher levels than was the full-length Myb protein when driven by the native Myb promoter. Nevertheless the full-length protein clearly localized to numerous interband regions within the polytene chromosomes, whereas the localization of the N terminus was much more diffuse throughout the chromosomes.
Animal-Specific C Terminus of Myb Is Necessary and Sufficient for Interaction with MuvB Complex.
We reasoned that the animal-specific C terminus might function via specific interactions with other members of the Myb-MuvB/dREAM complex to rescue Myb-null mutant lethality, the regulation of gene expression in vivo, and proper chromosomal localization. To test this hypothesis, we expressed FLAG-tagged WT or mutant Myb protein in Drosophila S2 cells in which RNAi had been used to knock down expression of the endogenous Myb protein to undetectable levels. Following immunoprecipitation of cell lysates with anti-FLAG antibodies, samples were analyzed by immunoblotting for the coprecipitation of the other members of the MuvB core complex (Mip130/LIN-9, Mip120/LIN-54, Mip40/LIN-37, and p55 CAF1/LIN-53) (14, 37). The WT Myb and C terminus coprecipitated the entire MuvB core complex, whereas the highly conserved N-terminal DNA-binding domain did not (Fig. 5, Left). Furthermore, the KW606AA and DQ617AA mutants failed to coprecipitate any components of the MuvB core complex (Fig. 5, Right). These results imply that the animal-specific Myb C terminus is both necessary and sufficient for interaction with the MuvB core complex (Fig. 6).
Fig. 5.
Coprecipitation of the MuvB core complex by Myb WT and mutant proteins. FLAG-tagged Myb WT or mutant protein was expressed in Drosophila S2 cells by transient DNA transfection. RNAi directed against the 5′-UTR was used to knock down endogenous Myb expression. Immunoblots of total extracts (input) or of immunoprecipitated complexes (FLAG-IP) were analyzed using the indicated antibodies. (Left) Analysis of interaction with the MuvB core complex by full-length (WT), N-terminal, or C-terminal Myb. The anti-Myb antibody recognizes the N-terminal repeats of the Myb DNA-binding domain; therefore, it cannot detect the C-terminal mutant (19). A 50-kDa background band detected by anti-FLAG in all FLAG-IP samples is mouse Ig-heavy chain that is released from the anti-FLAG beads used for immunoprecipitation. (Right) Analysis of interaction with the MuvB core complex by WT or the indicated alanine substitution mutants of Myb.
Fig. 6.
Function and evolutionary conservation of Myb and MuvB core proteins. (Left) Model for the function of WT and mutant Myb proteins in gene regulation. The MuvB core and associated proteins repress gene expression. This repression is relieved by contact with the animal-specific C terminus of Myb. The N-terminal mutant of Myb acts as a dominant loss-of-function allele, presumably by blocking access by the WT Myb protein. (Right) Evolutionary conservation of Myb and MuvB core proteins. MYB-N indicates a Myb DNA-binding domain closely related to that of animal Myb proteins. MYB-C indicates the animal-specific C-terminal domain. The core MuvB components are indicated by their names in Drosophila and in C. elegans. Homologous protein sequences were identified by searching the NCBI databases using TBlastn. Black squares indicate the presence of a homologous protein. Light gray squares indicate the absence of a homologous protein in the current NCBI database.
Discussion
The Myb DNA-binding domain is not encoded within any prokaryotic genomes sequenced thus far. On the other hand, it is present in members of all major groups of eukaryotes, including fungi, cellular slime molds, protists, plants, and animals (10). The other members of the core MuvB complex are variably conserved among all the major groups of eukaryotes (37) (Fig. 6). CAF1/LIN-53 is the only MuvB core protein with a significant homology in prokaryotes, with homologs being present in some Archaebacteria. This is perhaps consistent with the function of this protein as a histone chaperone in that Archaebacteria are the only known group of prokaryotes that encode homologs of the eukaryotic histones (38). In contrast, both the Myb DNA-binding domain and the other MuvB core proteins appear to have arisen during the evolution of eukaryotes. Early during the evolution of animals, the animal-specific C-terminal Myb domain appears to have been selected specifically to provide a protein-protein bridge between the Myb DNA-binding domain and the core members of the MuvB complex.
An interesting exception occurs in Caenorhabditis elegans, the nematode worm in which the synthetic multivulval class B genes were first described (39, 40). Nematodes lack an animal-like Myb protein but do contain all other members of the larger Myb-MuvB/dREAM complex, including homologs of the vertebrate and Drosophila Rb and E2F proteins (37, 40). These observations suggest that during the evolution of nematodes, there was a selection for a decoupling of Myb and the remaining members of the Myb-MuvB/dREAM complex. We have previously shown that vertebrate B-Myb can complement cell cycle defects in a Drosophila Myb-null mutant (41). Consistent with this observation, others have shown that vertebrate B-Myb is present in a complex together with the homologs of the MuvB core complex (42–44). Furthermore, we have shown that Drosophila Myb acts in vivo to inhibit transcriptional repression by the remainder of this complex (13). Together, these observations imply that the highly stereotypical and largely postmitotic development of the nematode might not require, and perhaps would be adversely affected by, inhibition of this repressive activity by Myb.
In flowering plants, “three-repeat” Myb proteins with a DNA-binding domain similar to that of the animal Myb proteins regulate the transcription of key regulators of the G2/M phases of the cell cycle (45, 46). Drosophila Myb and its vertebrate ortholog, B-MYB/MYBL2, regulate many of the same genes that are critical for progression through G2/M. However, as we have shown, the animal-specific C terminus rather than the more widely conserved Myb DNA-binding domain is required for this regulatory circuit in Drosophila. These results suggest that an ancient function of Myb, regulation of G2/M genes, has been conserved among plants and animals but that the critical molecular mechanism underlying this function has been transferred from the DNA-binding domain to an animal-specific C-terminal domain.
Members of the Myb oncogene family have been strongly implicated in different types of human cancer. Elevated levels of expression of B-MYB/MYBL2, the ortholog of Drosophila Myb, correlate with poor prognosis in patients who have breast cancer (47, 48). Duplications and chromosomal translocations of the C-MYB/MYB gene appear likely to be causal in human leukemias and adenoid cystic carcinomas, respectively (49, 50). Our results therefore suggest that the specific interaction of a conserved domain within the C terminus of Myb proteins with the MuvB/dREAM core complex may provide an attractive target for the development of cancer therapeutics.
Materials and Methods
Genetics.
The Myb mutant Df(1)MH107 has been previously described (19). The engrailed-GAL4 stock was obtained from Jeff Axelrod (Stanford University, Palo Alto, CA). The RFP-MAD2 stock was obtained from Roger Karess (Centre National de la Recherche Scientifique, Gif sur Yvette, France). Other GAL4 stocks and balancer chromosome stocks were obtained from the Bloomington Stock Center. All new transgenic stocks were prepared by injection of w1118 embryos (BestGene) and then balanced. The absence of an FM7 Actin-GFP balancer chromosome was used to identify MybMH107/Y male larvae as needed. Stocks and crosses were maintained on standard cornmeal-molasses-agar food at 22 °C unless otherwise indicated. Rescue test crosses were scored for the number of round-eyed (Bar+) Myb-null males vs. the number of bar-eyed (FM7, Bar1) Myb-WT males (Figs. S1and S4).
Molecular Biology.
All alanine substitution mutants of Drosophila Myb were constructed using QuikChange site-directed mutagenesis (Stratagene) in the previously described pSP72-GFP-Myb plasmid (13). For expression under control of GAL4, the BglII-BglII restriction fragment containing the GFP-Myb ORF was inserted into the BglII site of the pUAST plasmid (31). All constructs were first tested for expression by transient cotransfection, together with a tubulin promoter-GAL4 driver plasmid in Drosophila S2 cells. Transfected cells were examined for GFP fluorescence and for protein expression by immunoblotting using anti-GFP and anti-Myb antibodies. For expression under control of the native Myb promoter, the same BglII-BglII restriction fragment was inserted into the previously described pSP73-MluI-Casper intermediate plasmid (33). The MluI-MluI restriction fragment was then inserted into the previously described pCASPER-Myb plasmid that contains a 14.2-kb insert of genomic DNA surrounding the Myb gene (19). This resulted in intronless GFP-Myb cDNA being expressed under control of the native Myb promoter. A control WT GFP-Myb cDNA construct was prepared in a similar fashion. For the DQ617AA mutant that lies outside of the MluI-MluI fragment, a different strategy was used. A pSP73 plasmid containing a 4-kb BamHI fragment of genomic DNA, including the 3′ ORF of Myb was digested with SpeI, blunt-ended with Klenow fragment and dNTPs, and then digested with MluI. This plasmid backbone was ligated to the MluI-PsiI fragment of pSP72-GFP-Myb containing the DQ617AA mutant. The BamHI-BamHI fragment containing the 3′ ORF of Myb was then ligated to BamHI-digested pCASPER-Myb to create pCASPER-Myb-DQ617AA.
The N-terminal 316 amino acids of Myb were fused in-frame to GFP and then expressed under control of UASGAL4 by first cloning the BglII-BamHI fragment of pSP73-Myb into pEGFP-N3 (Clontech). The BglII-NotI fragment encoding the Myb (amino acids 1–316)-GFP fusion protein was then inserted into pUAST. The expression of the C terminus of Myb (amino acids 247–647) under control of UASGAL4 has been previously described (13).
Immunofluorescence.
Larval tissues were dissected, fixed, and mounted for laser scanning confocal microscopy using a Nikon PCM 2000 as previously described (33). The RFP-Mad2 fusion protein was detected in third-instar larval imaginal wing disks by immunostaining with anti-RFP antiserum as previously described (13). Where indicated, TO-PRO-3 (Invitrogen) was used to stain DNA without any RNase treatment to preserve nuclear structure.
Immunoprecipitation and RNAi Treatment.
Immunoprecipitation and immunoblotting of Drosophila S2 cells treated with RNAi directed against Myb were performed as previously described (13).
Supplementary Material
Acknowledgments
We thank Lajja Mani and Klara Fekete for assistance with Drosophila food and stock maintenance and Patty Winningham for administrative support. This work was supported by US Public Health Service Grant R01CA128836 (to J.S.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111855108/-/DCSupplemental.
References
- 1.Lipsick JS, Wang DM. Transformation by v-Myb. Oncogene. 1999;18:3047–3055. doi: 10.1038/sj.onc.1202745. [DOI] [PubMed] [Google Scholar]
- 2.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 3.Klempnauer KH, Symonds G, Evan GI, Bishop JM. Subcellular localization of proteins encoded by oncogenes of avian myeloblastosis virus and avian leukemia virus E26 and by chicken c-myb gene. Cell. 1984;37:537–547. doi: 10.1016/0092-8674(84)90384-2. [DOI] [PubMed] [Google Scholar]
- 4.Boyle WJ, Lampert MA, Lipsick JS, Baluda MA. Avian myeloblastosis virus and E26 virus oncogene products are nuclear proteins. Proc Natl Acad Sci USA. 1984;81:4265–4269. doi: 10.1073/pnas.81.14.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biedenkapp H, Borgmeyer U, Sippel AE, Klempnauer KH. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- 6.Nishina Y, Nakagoshi H, Imamoto F, Gonda TJ, Ishii S. Trans-activation by the c-myb proto-oncogene. Nucleic Acids Res. 1989;17(1):107–117. doi: 10.1093/nar/17.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weston K, Bishop JM. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell. 1989;58(1):85–93. doi: 10.1016/0092-8674(89)90405-4. [DOI] [PubMed] [Google Scholar]
- 8.Ness SA, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 9.Ibanez CE, Lipsick JS. trans activation of gene expression by v-myb. Mol Cell Biol. 1990;10:2285–2293. doi: 10.1128/mcb.10.5.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 11.Davidson C, Ray E, Lipsick J. In: Myb Transcription Factors: Their Role in Growth, Differentiation and Disease. Frampton J, editor. Boston: Kluwer Academic; 2004. pp. 1–33. [Google Scholar]
- 12.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66(1):94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 13.Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 2008;22:601–614. doi: 10.1101/gad.1626308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beall EL, et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- 15.Lewis PW, et al. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korenjak M, et al. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119(2):181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 17.Katzen AL, Kornberg TB, Bishop JM. Isolation of the proto-oncogene c-myb from D. melanogaster. Cell. 1985;41:449–456. doi: 10.1016/s0092-8674(85)80018-0. [DOI] [PubMed] [Google Scholar]
- 18.Katzen AL, Bishop JM. myb provides an essential function during Drosophila development. Proc Natl Acad Sci USA. 1996;93:13955–13960. doi: 10.1073/pnas.93.24.13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manak JR, Mitiku N, Lipsick JS. Mutation of the Drosophila homologue of the Myb protooncogene causes genomic instability. Proc Natl Acad Sci USA. 2002;99:7438–7443. doi: 10.1073/pnas.122231599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada M, Akimaru H, Hou DX, Takahashi T, Ishii S. Myb controls G(2)/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J. 2002;21:675–684. doi: 10.1093/emboj/21.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wertman KF, Drubin DG, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane T, Ibanez C, Garcia A, Graf T, Lipsick J. Transformation by v-myb correlates with trans-activation of gene expression. Mol Cell Biol. 1990;10:2591–2598. doi: 10.1128/mcb.10.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frampton J, Gibson TJ, Ness SA, Döderlein G, Graf T. Proposed structure for the DNA-binding domain of the Myb oncoprotein based on model building and mutational analysis. Protein Eng. 1991;4:891–901. doi: 10.1093/protein/4.8.891. [DOI] [PubMed] [Google Scholar]
- 24.Gabrielsen OS, Sentenac A, Fromageot P. Specific DNA binding by c-Myb: Evidence for a double helix-turn-helix-related motif. Science. 1991;253:1140–1143. doi: 10.1126/science.1887237. [DOI] [PubMed] [Google Scholar]
- 25.Garcia A, LaMontagne K, Reavis D, Stober-Grässer U, Lipsick JS. Determinants of sequence-specific DNA-binding by p48v-myb. Oncogene. 1991;6:265–273. [PubMed] [Google Scholar]
- 26.Grässer FA, LaMontagne K, Whittaker L, Stohr S, Lipsick JS. A highly conserved cysteine in the v-Myb DNA-binding domain is essential for transformation and transcriptional trans-activation. Oncogene. 1992;7:1005–1009. [PubMed] [Google Scholar]
- 27.Dini PW, Lipsick JS. Oncogenic truncation of the first repeat of c-Myb decreases DNA binding in vitro and in vivo. Mol Cell Biol. 1993;13:7334–7348. doi: 10.1128/mcb.13.12.7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myrset AH, et al. DNA and redox state induced conformational changes in the DNA-binding domain of the Myb oncoprotein. EMBO J. 1993;12:4625–4633. doi: 10.1002/j.1460-2075.1993.tb06151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogata K, et al. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 30.Ko ER, Ko D, Chen C, Lipsick JS. A conserved acidic patch in the Myb domain is required for activation of an endogenous target gene and for chromatin binding. Mol Cancer. 2008;7:77. doi: 10.1186/1476-4598-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 32.Fung SM, Ramsay G, Katzen AL. Mutations in Drosophila myb lead to centrosome amplification and genomic instability. Development. 2002;129:347–359. doi: 10.1242/dev.129.2.347. [DOI] [PubMed] [Google Scholar]
- 33.Manak JR, Wen H, Van T, Andrejka L, Lipsick JS. Loss of Drosophila Myb interrupts the progression of chromosome condensation. Nat Cell Biol. 2007;9:581–587. doi: 10.1038/ncb1580. [DOI] [PubMed] [Google Scholar]
- 34.Katzen AL, et al. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev. 1998;12:831–843. doi: 10.1101/gad.12.6.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georlette D, et al. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev. 2007;21:2880–2896. doi: 10.1101/gad.1600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M. Drosophila. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Lipsick JS. synMuv verite—Myb comes into focus. Genes Dev. 2004;18:2837–2844. doi: 10.1101/gad.1274804. [DOI] [PubMed] [Google Scholar]
- 38.Sandman K, Reeve JN. Archaeal histones and the origin of the histone fold. Curr Opin Microbiol. 2006;9:520–525. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Ferguson EL, Horvitz HR. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123(1):109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison MM, Ceol CJ, Lu X, Horvitz HR. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci USA. 2006;103:16782–16787. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davidson CJ, Tirouvanziam R, Herzenberg LA, Lipsick JS. Functional evolution of the vertebrate Myb gene family: B-Myb, but neither A-Myb nor c-Myb, complements Drosophila Myb in hemocytes. Genetics. 2005;169:215–229. doi: 10.1534/genetics.104.034132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Litovchick L, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 43.Pilkinton M, Sandoval R, Colamonici OR. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene. 2007;26:7535–7543. doi: 10.1038/sj.onc.1210562. [DOI] [PubMed] [Google Scholar]
- 44.Schmit F, et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 45.Ito M, et al. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell. 2001;13:1891–1905. doi: 10.1105/TPC.010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito M. Conservation and diversification of three-repeat Myb transcription factors in plants. J Plant Res. 2005;118(1):61–69. doi: 10.1007/s10265-005-0192-8. [DOI] [PubMed] [Google Scholar]
- 47.Amatschek S, et al. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844–856. doi: 10.1158/0008-5472.can-03-2361. [DOI] [PubMed] [Google Scholar]
- 48.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 49.Clappier E, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 50.Persson M, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci USA. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuler GD, Altschul SF, Lipman DJ. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.