Abstract
The small GTPase Rab5 is a conserved regulator of membrane trafficking; it regulates the formation of early endosomes, their transport along microtubules, and the fusion to the target organelles. Although several members of the endocytic pathway were recently implicated in spindle organization, it is unclear whether Rab5 has any role during mitosis. Here, we describe that Rab5 is required for proper chromosome alignment during Drosophila mitoses. We also found that Rab5 associated in vivo with nuclear Lamin and mushroom body defect (Mud), the Drosophila counterpart of nuclear mitotic apparatus protein (NuMA). Consistent with this finding, Rab5 was required for the disassembly of the nuclear envelope at mitotic entry and the accumulation of Mud at the spindle poles. Furthermore, Mud depletion caused chromosome misalignment defects that resembled the defects of Rab5 RNAi cells, and double-knockdown experiments indicated that the two proteins function in a linear pathway. Our results indicate a role for Rab5 in mitosis and reinforce the emerging view of the contributions made by cell membrane dynamics to spindle function.
Mitosis coordinates the equal segregation of chromosomes into two daughter cells. The regulation, dynamics, and structure of chromosomes and spindle microtubules during mitosis have been studied in some detail, but the mechanisms that control the structural reorganization of endomembranes and their roles during cell division are still unclear. Membrane organelles like the Golgi, endoplasmic reticulum, and nuclear envelope (NE) are completely reorganized during cell division. In metazoans, the NE can be either completely or partially disassembled at mitotic entry, leading to open (e.g., mammals) and semiopen (e.g., Drosophila) mitoses. Increasing evidence supports the notion that endomembranes and their associated proteins have important functions during mitosis (1, 2). Many NE components released during mitosis have been found to also have a role during mitosis (3–7). Furthermore, several members of the endocytic and exocytic pathways were recently implicated in spindle organization (8, 9). Clathrin becomes spindle-associated during mitosis (10) and serves as an adapter that recruits Aurora-A–phosphorylated transforming acidic coiled-coil 3 (TACC3) protein (11). Its associated cyclin G-associated kinase (GAK) controls centrosome integrity, chromosome congression, and microtubule outgrowth from kinetochores (12, 13).
The Rab5 GTPase regulates the formation of early endosomes (14) as well as their transport along microtubules (15) and fusion of endocytic vesicles to the target organelles. Rab5 in its active form is able to recruit several downstream effectors onto endosomes, and it has been proposed to create distinct membrane domains (14). Recently, Rab5 has also been implicated in other cellular processes such as cell signaling, phagosome maturation (16, 17), and endoplasmic reticulum reorganization and nuclear lamin disassembly during mitosis in Caenorhabditis elegans (18).
Here, we show that Rab5 depletion affects chromosome movements before anaphase onset in Drosophila cells. Moreover, we found that Rab5 associated in vivo with Lamin and mushroom body defect (Mud), the Drosophila counterpart of nuclear mitotic apparatus protein (NuMA), which is known to be important for spindle formation and maintenance in vertebrate cells (19–21). Consistent with this finding, the NE did not disassemble properly at mitotic entry in Rab5 RNAi cells, and Mud failed to accumulate at spindle poles. Finally, Mud depletion recapitulated the chromosome segregation defects observed after Rab5 RNAi, and depletion of either protein reduced interkinetochore tension. Our results indicate that Rab5 regulates not only the dispersal of Lamin in prophase but also chromosome behavior during prometaphase; the latter is most likely regulated indirectly through its association with Mud.
Results and Discussion
Rab5 Depletion Affects Chromosome Alignment.
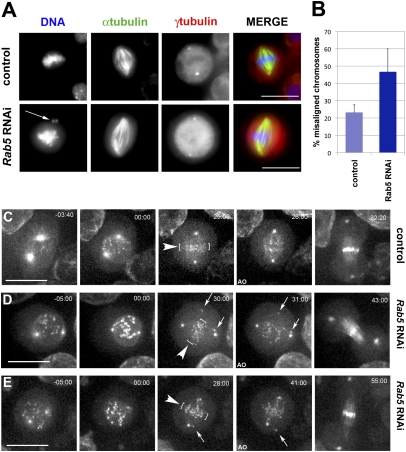
Small GTPases act as molecular switches in a large number of cell biological processes. To investigate the role of small GTPases in mitosis in cultured Drosophila cells, we systematically depleted all of the small GTPases present in the Drosophila genome by RNAi. We found that efficient Rab5 depletion (Fig. S1E) caused an increase in cells with chromosome alignment defects (Fig. 1A and Fig. S1 A and B). This surprising phenotype was not observed after depletion of any other Rab GTPases in the RNAi screen. To discount off-target effects, we used two different Rab5 dsRNAs and in both cases, observed ill-defined metaphase plates, often with chromosomes lagging at the poles (Fig. 1A). These lagging chromosomes were positive for the centromere identifier protein (CID) [centromere proteinA (CENPA) in human] indicating that they had intact centromeres (Fig. S1A). Moreover, the association of the spindle assembly checkpoint (SAC) component BubR1 (22) with kinetochores revealed that the SAC had not been satisfied and that these cells had not yet entered anaphase (Fig. S1B). In addition to a twofold increase in the number of misaligned chromosomes (Fig. 1B), Rab5 RNAi cells also showed a significant (2.6-fold) increase in the number of binucleate cells (ANOVA P value = 0.02) (Fig. S1 C and D). This binucleate phenotype was previously described (23).
Fig. 1.
Rab5 RNAi causes chromosome alignment defects. (A) Cells were treated with dsRNA directed against Rab5 and fixed and stained to detect DNA (blue in merged column), α-tubulin (green in merged column), and γ-tubulin (red in merged column). The arrow marks misaligned chromosomes. (Scale bars: 10 μm.) (B) Quantification of misaligned chromosomes. The results of at least three separate experiments were analyzed; bars indicate SD. At least 150 mitotic cells were counted for each experiment. (C) Selected images from a time-lapse recording of a Drosophila control cell expressing a Polo::GFP transgene. Time is in minutes to seconds relative to NEBD. The first frame is at a slightly earlier stage than the first frame of the RNAi-treated cells. The arrowheads and white brackets indicate the metaphase plate before anaphase onset (25:00-min time point). (Scale bar: 10 μm.) (D and E) Selected images from two videos of Polo::GFP cells depleted of Rab5. The arrows indicate misaligned chromosomes, and the white brackets mark the metaphase plate. Time is in minutes to seconds relative to NEBD. AO, anaphase onset. (Scale bar: 10 μm.)
To visualize centrosome and kinetochore dynamics after Rab5 RNAi, we performed time-lapse imaging using cells stably expressing Polo::GFP (24). In these cells, Polo::GFP accumulated at the centrosomes in prophase (Fig. 1C, −03:40 time point and Movie S1) and subsequently, on kinetochores at NE breakdown (NEBD) (Fig. 1C, 00:00 time point and Movie S1), allowing the simultaneous observation of spindle dynamics and chromosome behavior. In control cells, all kinetochores aligned properly at the metaphase plate (Fig. 1C, 25:00 time point and Movie S1), whereas after Rab5 RNAi, many kinetochores failed to congress at the metaphase plate even after numerous attempts (Fig. 1 D and E, 30:00 and 28:00 time points, and Movies S2 and S3). In all filmed cells, some chromosomes never congressed to the metaphase plate. However, despite the presence of misaligned chromosomes, all these cells entered anaphase, indicating that the SAC had been satisfied (Fig. 1 D and E, 31:00 and 41:00 time points). To determine whether Rab5 RNAi cells were delayed in mitotic progression, we measured the interval between NEBD and anaphase onset. Rab5 RNAi cells displayed a significant delay in entering anaphase (30 ± 8 min, n = 12) compared with controls (19 ± 6 min, n = 11; ANOVA P value = 0.001), a likely consequence of problems in chromosome alignment; 2 of 12 Rab5 RNAi cells also failed cytokinesis because of the presence of lagging DNA at the cleavage sites (Movie S4), suggesting that the presence of binucleate cells observed in fixed preparations of Rab5 RNAi cells could be a secondary consequence of abnormal chromosome behavior (Fig. S1 C and D). We also found that Rab5 mutant larval brains (25) had aneuploid neuroblasts and abnormal metaphase and anaphase figures (Fig. S1 F and G).
Rab5 Shows Dynamic Localization During Mitosis.
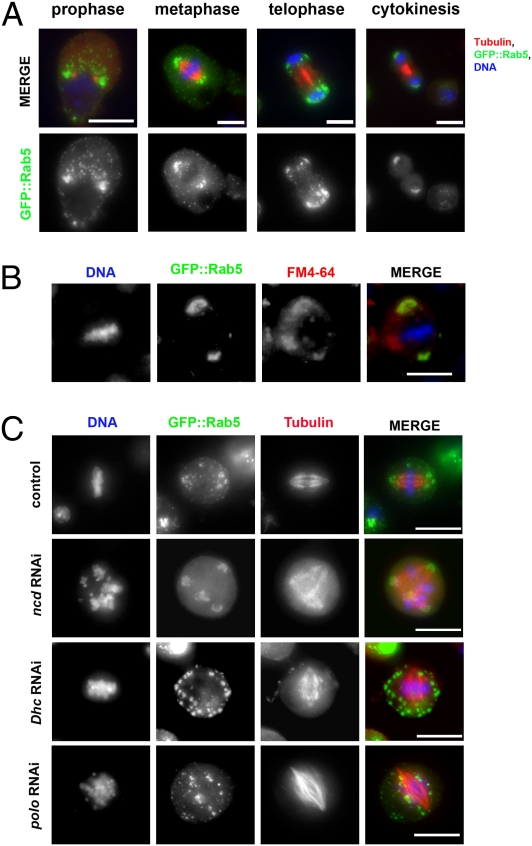
Rab5 localizes to cytoplasmic vesicles during interphase (26), but its distribution during mitosis is not characterized. Using a cell line stably expressing an N-terminal GFP-tagged version of this GTPase, we observed that, in prophase, GFP::Rab5 localized predominantly around the centrosomes (Fig. 2A). The association with spindle poles persisted throughout metaphase and subsequently, in telophase, and cytokinesis GFP::Rab5 accumulated around the newly reformed NEs (Fig. 2A). Rab5 dynamics during mitosis were also confirmed by time-lapse analysis using a cell line stably expressing GFP::Rab5 and Tubulin::mRFP (Movie S5). Quantitation of protein expression levels revealed that the GFP-tagged transgene was expressed at a similar level as the endogenous Rab5 gene (Fig. S2B). Moreover, we were able to confirm a similar localization pattern for the endogenous protein using an antibody against Drosophila Rab5 (Fig. S2A). Costaining GFP::Rab5 cells with a lipophilic styryl compound (FM4–64) indicated that a subset of FM4–64-stained membranes colocalized perfectly with GFP::Rab5 at metaphase, indicating that Rab5 was still associated with membranous vesicles during mitosis (Fig. 2B). Rab5 colocalizes with GFP::2xFYVE, a bona fide endosomal marker (25) (Fig. S2C), confirming that Rab5 GTPase is associated with membrane vesicles around the spindle poles (Fig. S2C).
Fig. 2.
Mitotic GFP::Rab5 localization is astral microtubule- and Dynein-dependent. (A) Drosophila S2 cells expressing GFP::Rab5 were fixed and stained to reveal GFP (green), tubulin (red), and DNA (blue). (Scale bars: 10 μm.) (B) Cells were fixed and stained to detect GFP::Rab5 (green), lipophilic dye FM4-64 (red), and DNA (blue). (Scale bar: 10 μm.) (C) Drosophila S2 cells expressing GFP::Rab5 were treated with dsRNAs against Ncd (ncd RNAi), Dhc64C (Dhc RNAi), or Polo (polo RNAi) for 72 h and then stained to reveal GFP (green), tubulin (red), and DNA (blue). (Scale bars: 10 μm.)
A previous study indicated that Rab5 vesicles could move along microtubules (15), and thus, we determined whether Rab5 mitotic vesicles exhibited motor-dependent minus end-directed movement along microtubules to the spindle poles. We depleted the only two minus end motors present in the Drosophila genome, the kinesin-14 nonclaret disjunctional (Ncd) and the Dynein heavy-chain 64C (Dhc64C), as part of the Dynein complex. Depletion of Dhc64C led to a complete disruption of GFP::Rab5 accumulation at the spindle poles (Fig. 2C), whereas ncd RNAi did not affect GFP::Rab5 localization, although it caused the expected multipolar spindle phenotype (27). Efficient depletion of Dhc64C and Ncd was confirmed by Western blot (Fig. S2D).
We next investigated whether aster microtubules were essential for the transport of Rab5 to centrosomes. The Polo mitotic kinase is required for centrosomes maturation and nucleation of astral microtubules, and its depletion leads to the formation of mono- or bipolar spindles that lack astral microtubules (28). We found that GFP::Rab5 failed to localize to spindle poles after efficient Polo depletion (Fig. 2C and Fig. S2D). This finding strongly suggests that astral microtubules are required for the Rab5 GTPase to travel to centrosomes. Taken together, these experiments indicate that, during mitosis, Rab5-positive vesicles move to the poles of the nascent spindle along astral microtubules in a dynein-dependent manner.
Rab5 Associates in Vivo with Lamin and Mud.
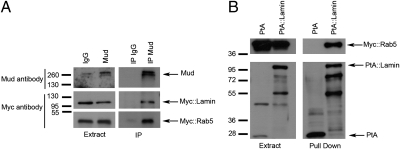
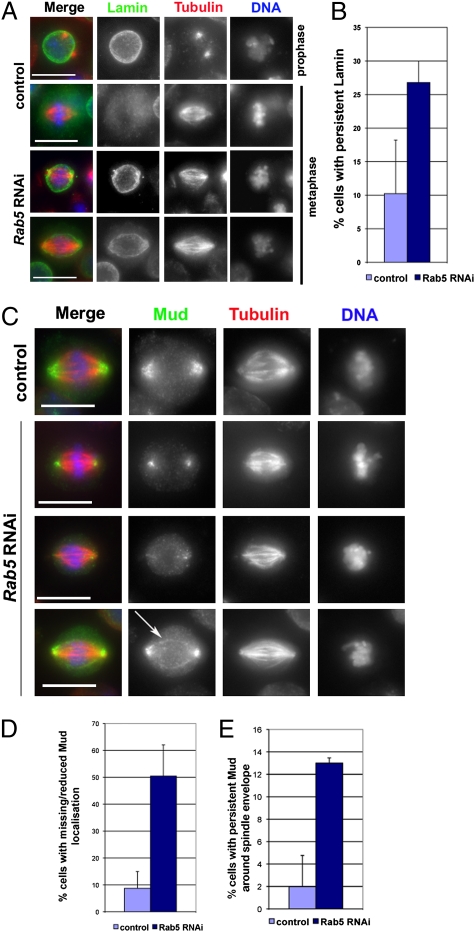
To gain insight into potential functional interactions of Rab5 with the mitotic apparatus, we sought to identify Rab5-interacting proteins. We tagged Rab5 with two IgG binding domains of Protein A (PtA) and expressed this construct in cultured Drosophila cells. We then affinity-purified PtA::Rab5 along with its associated partners and identified all of the components by MS. Drosophila cells cannot be synchronized, and we could only enrich for mitotic cells using the proteasome inhibitor MG132 (29). We also incubated the cell extracts with a nonhydrolysable GTP analog Guanosine 5′-O-(γ-thio) triphosphate before purification to lock Rab5 in its active form. As expected, we identified many endocytic proteins in our purifications, including dynamin (Shibire in Drosophila), Rab4 GTPase, and Clathrin (Fig. S3). However, we also isolated two unexpected proteins: the single Drosophila homolog of the two vertebrate B-type Lamins and Mud, the Drosophila counterpart of the vertebrate NuMA (30). Both NuMA and Mud play an important role in spindle orientation during asymmetric cell division (31–33), and NuMA has recently been implicated in spindle assembly and chromosome congress in mammalian cells (20, 21). We confirmed the association of Rab5 with Lamin in reciprocal pull-down experiments using a cell line stably expressing an N-terminal PtA-tagged version of Lamin. In addition to known Lamin interacting proteins such as Otefin (34) and histones (35) (Fig. S3), we also purified Mud and Rab5, confirming that these three proteins are present in a complex in vivo (Fig. S3). Immunoprecipitation (IP) experiments confirmed that the anti-Mud antibody could pull down both Myc::Rab5 and Myc::Lamin in DMel cells treated with MG132 and Guanosine 5′-O-(γ-thio) triphosphate (Fig. 3A). We could also pull down Myc::Rab5 from extracts of a stable cell line expressing PtA::Lamin (Fig. 3B). The above findings led us to ask whether depletion of Rab5 might affect the mitotic localization of Lamin and Mud. Lamin is the major component of the nuclear lamina that is dispersed on mitotic entry. We found that Rab5 RNAi caused a two- to threefold increase in prometaphase/metaphase cells showing persistent Lamin around the spindle envelope (Fig. 4 A and B), the highly fenestrated remnants of the nuclear envelope that encircles the mitotic apparatus in many Drosophila tissues (36, 37). Moreover, Lamin was not disassembled properly in prometaphase/metaphase cells, even in the presence of a clear bipolar spindle (Fig. 4A); this finding suggests that Rab5 promotes the disassembly of the nuclear lamina structure at mitotic entry, which is consistent with recent findings in C. elegans (18).
Fig. 3.
Co-IP between Rab5, Mud, and Lamin. (A) Drosophila DMel cells were transfected with Myc::Rab5 and Myc::Lamin for 48 h. Protein extracts were then used for IP using a rabbit anti-Mud antibody or rabbit IgG as control. Protein extracts and IP were analyzed by Western blot to detect Mud, Myc::Rab5, and Myc::Lamin. The numbers on the left indicate the sizes in kilodaltons of the molecular mass marker. (B) Drosophila DMel cells stably expressing either PtA::Lamin or PtA alone were transfected with a Myc::Rab5 construct for 48 h. Protein extracts were then used in a PtA pull-down assay. The extracts and pull-down assays were analyzed by Western blot to detect Myc::Rab5. The numbers on the left indicate the sizes in kilodaltons of the molecular mass marker.
Fig. 4.
Rab5 depletion impairs nuclear lamina disassembly and mitotic localization of Mud. (A) Cells were treated with Rab5 dsRNA (Rab5 RNAi) or GFP dsRNA (control) for 72 h and then fixed and stained to reveal Lamin (green), tubulin (red), and DNA (blue). (Scale bars: 10 μm.) (B) Quantification of cells showing persistent Lamin at the spindle envelope in metaphase after GFP (control) or Rab5 RNAi. At least 150 mitotic cells were counted for each experiment. Bars indicate SDs. (C) Cells were treated with Rab5 dsRNA (Rab5 RNAi) or GFP dsRNA (control) for 72 h and fixed and stained to reveal Mud (green), tubulin (red), and DNA (blue). The arrow marks Mud mislocalization. (Scale bars: 10 μm.) (D) Quantification of cells showing missing or reduced Mud localization around spindle poles in GFP (control) and Rab5 RNAi cells. Three independent experiments were carried out, and at least 150 mitotic cells were counted in each experiment. Bars indicate SDs. (E) Quantification of cells showing persistent Mud around the spindle envelope in GFP (control) and Rab5 RNAi cells. Three independent experiments were carried out, and at least 150 mitotic cells were counted in each experiment. Bars indicate SDs.
In untreated cells, Mud accumulated around the nuclear envelope in interphase and then translocated to the spindle poles in prophase and metaphase (Fig. S4A). We found that Rab5 accumulated in pericentrosomal regions in prophase where it colocalized with Mud (Fig. S4B). In prometaphase/metaphase, the two proteins were in close proximity; however, whereas Mud associated with the spindle poles, Rab5 showed a distribution around the poles (Fig. S4B). Depletion of Rab5 led to a clear reduction or even absence of Mud at the spindle poles in 50.4% (±11.6) of prometaphase/metaphase cells (n = 146) (Fig. 4 C and D), and Mud was often mislocalized around the spindle envelope in 13% (±0.46) of mitotic cells (n = 146) (Fig. 4 C and E). By contrast, Mud depletion did not prevent Rab5 recruitment to spindle poles (Fig. S5). Together, these data suggest that Rab5 is required for the proper disassembly of the nuclear lamina in mitosis to allow the release and polar transport of some mitotic proteins, including Mud, associated with this membranous structure in interphase.
Mud Depletion Causes Chromosome Misalignment Defects.
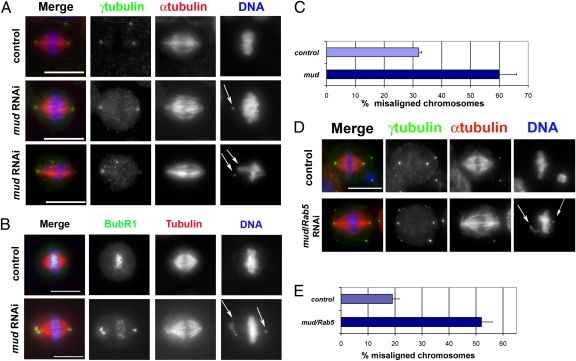
We reasoned that, if Rab5 and Mud were part of a functional mitotic complex, then Mud depletion should recapitulate at least some of the mitotic defects observed after Rab5 depletion. Consistent with this hypothesis, mud RNAi cells displayed chromosome congress defects very similar to those defects observed after Rab5 RNAi (Fig. 5 A and C). Mud-depleted cells showed a twofold increase in the number of misaligned chromosomes, which sometime localized close to the spindle poles (Fig. 5 A and C). These chromosomes exhibited a strong BubR1 signal at kinetochores (Fig. 5B), indicating that the SAC had not been satisfied.
Fig. 5.
mud RNAi causes chromosome misalignment defects similar to Rab5 knockdown. (A) Drosophila S2 cells were treated with GFP (control) or mud dsRNA for 3 d and fixed and stained to reveal γ-tubulin (green), α-tubulin (red), and DNA (blue). Arrows show misaligned chromosomes in cells depleted of Mud. (Scale bars: 10 μm.) (B) Cells were treated with GFP (control) or mud dsRNA for 3 d and fixed and stained to detect the SAC protein BubR1 (green), tubulin (red), and DNA (blue). (Scale bars: 10 μm.) (C) Quantification of cells showing misaligned chromosomes in control and Mud-depleted cells. Three independent experiments were carried out, and at least 150 mitotic cells were counted in each experiment. Bars indicate SDs. (D) Cells were treated with dsRNAs against either GFP (control) or mud and Rab5 for 3 d and fixed and stained to reveal γ-tubulin (green), α-tubulin (red), and DNA (blue). Arrows mark misaligned chromosomes in cells codepleted of Mud and Rab5. (Scale bar: 10 μm.) (E) Quantification of cells showing misaligned chromosomes in cells treated with dsRNAs against either GFP (control) or mud and Rab5. Three independent experiments were carried out, and at least 150 mitotic cells were counted in each experiment. Bars indicate SDs.
To assess if Rab5 and Mud function in a parallel or linear pathway, we performed a double knockdown experiment. Simultaneous depletion of both proteins did not exacerbate the chromosome alignment defect of the single knockdowns (Fig. 5 D and E), indicating that Rab5 and Mud act in the same pathway. These results are consistent with the association in vivo of the two proteins and the fact that Mud is abnormally localized in Rab5 RNAi cells but not vice versa, suggesting that Rab5 acts upstream of Mud.
Rab5 Is Required for Kinetochore Microtubule Tension.
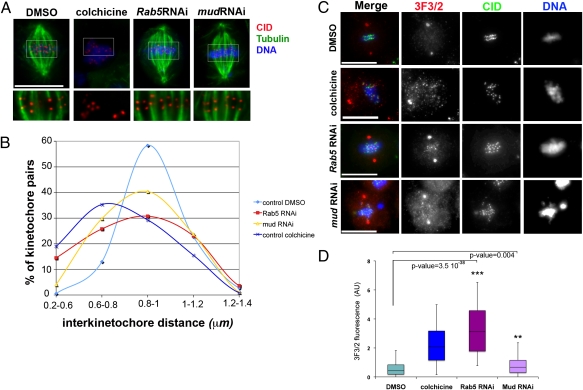
Recent studies in mammalian cells indicated that defects in chromosome congress and spindle maintenance after NuMA depletion were most likely caused by a lack of tension at kinetochore microtubules (20, 21). Thus, we asked whether a similar lack of kinetochore microtubule tension could be observed after depletion of either Rab5 or Mud. We treated cells with dsRNAs directed against Rab5, Mud, or GFP (control) for 72 h. Control GFP RNAi cells were also incubated with either the microtubule depolymerizing drug colchicine or its solvent DMSO for 3 h to provide samples of mitotic cells in which kinetochores were under normal tension or no tension at all in the absence of microtubules (Fig. 6A). We then measured and compared the interkinetochore distances of kinetochore pairs in all samples at late prometaphase/metaphase. Consistent with published observations (38), the interkinetochore distance in control cells treated with DMSO was 0.94 ± 0.15 mm, whereas when tension was removed after colchicine treatment, this distance was reduced to 0.78 ± 0.18 mm. In comparison, the distance across sister kinetochores was 0.88 ± 0.16 in mud RNAi cells and 0.85 ± 0.23 in Rab5 RNAi cells. The mean values of Rab5 or mud RNAi cells were significantly reduced compared with the DMSO control (two-tailed t test; control-Rab5 P value = 5.6 × 10−6, control-mud P value = 0.0001) (Fig. 6A). Importantly, the distributions of interkinetochore distances were skewed away from the mean in control cells to the mean of colchicines-treated cells (Fig. 6B). This finding is indicative of reduction in the interkinetochore tension for cells depleted of Rab5 or Mud that could contribute to the congress defects observed in our experiments.
Fig. 6.
Rab5 and Mud-depleted cells display reduced kinetochore tension. (A) Drosophila S2 cells were treated with colchicine, its solvent DMSO, or dsRNAs directed against Rab5 or mud on concanavalin-A–coated coverslips and then stained to reveal tubulin (green), CID (red), and DNA (blue). Insets show a 2× magnification of three optical sections of the metaphase plates stained to reveal CID (red) and tubulin (green). (Scale bar: 10 μm.) (B) Distributions of the distances of interkinetochore pairs (CID–CID distance) for cells treated with DMSO (n = 185), colchicine (n = 116), Rab5 RNAi (n = 221), and mud RNAi (n = 198). Note that, in control cells, 58% of kinetochore pairs have an interkinetochore distance ranging between 0.8 and 1 mm, whereas after Rab5 or mud RNAi, only 30% and 40% of the interkinetochore pairs were separated by a similar distance, respectively. The graph also shows an increase in the percentage of cells in the lower range of distance values (0.6–0.8 mm) in Rab5 and mud RNAi experiments, a trend similar to colchicines-treated cells. (C) Drosophila S2 cells were treated with colchicine, its solvent DMSO, or dsRNAs directed against Rab5 or mud and then stained to detect CID (green), 3F3/2 (red), and DNA (blue). (Scale bars: 10 μm.) (D) The intensity of 3F3/2 fluorescence at centromeres was calculated using the formula 3F3/2 fluorescence = (ICF − IC)/IC, where ICF is the fluorescence intensity at the centromere and IC represents the background intensity measured within an identical area inside the cytoplasm. More than 50 centromeres from two separate experiments were analyzed. P values were calculated using Student t test. AU, arbitrary units.
The 3F3/2 monoclonal antibody recognizes a phosphoepitope at the kinetochore that has been used as molecular readout of kinetochore tension, and therefore, we asked whether this epitope could be detected at kinetochores of Rab5 or mud RNAi cells. In control cells, the 3F3/2 signal at kinetochores was either very low or absent as expected (Fig. 6C) (39). In contrast, kinetochores in Rab5 RNAi cells were positive for 3F3/2, which was similar to colchicines-treated cells (Fig. 6 C and D), whereas mud RNAi kinetochores showed weaker, although significant, staining (Fig. 6 C and D). These results also support the evidence of a reduced interkinetochore tension after Rab5 or mud RNAi. Together, these experiments suggest that the abnormal chromosome behavior observed after Rab5 and Mud knockdown could result from a reduction in interkinetochore tension, which is in agreement with previous findings that NuMA is necessary in vertebrate cells for proper kinetochore tension and chromosome alignment at the metaphase plate (20, 21).
Conclusion
Our results indicate that Rab5 controls the mitotic behavior of Lamin and the NuMA-related protein Mud, which are both components of the interphase nuclear envelope in Drosophila. It is noteworthy that dynein, known to transport NuMA to the poles (40) and facilitate breakdown of the nuclear envelope (41), is also required for Rab5 to accumulate around the poles in mitosis. Increasing evidence suggests that Lamin and membranous structures contribute to a spindle matrix that is important for proper spindle formation and a mechanical support to hold the spindle structure in place (2). NuMA has been postulated to be part of this spindle matrix in the vicinity of the poles. Indeed, in the partially open mitoses of Drosophila embryo, residual Lamin in the persisting spindle envelope has been proposed to play an important role in buffering forces generated by microtubule motors (42). A similar function has been hypothesized for the membranous Lamin B network that is associated with the spindles of vertebrates (4). In this light, we propose that the chromosome alignment defects observed in both Rab5 RNAi cells and mutant Rab5 neuroblasts could reflect defective spindle function resulting from a combination of Mud/NuMA abnormal distribution and incomplete dispersion of Lamin at mitotic entry. We also cannot rule out other additional consequences of Rab5 depletion on spindle function or the possibility that other proteins could also be involved in Rab5 mitotic function.
Our results point out a key role for Rab5 in disassembling the nuclear envelope and regulating the distribution of spindle matrix components in Drosophila. Evidence that Rab5 is also required for chromosome alignment in human cells [discussed in the work by Serio et al. (43)] suggests that this role has been conserved during evolution. Indeed, this finding may be one part of a larger mechanism that coordinates the dissolution of the interphase endocytic machinery to put it to an alternative use during mitosis.
Materials and Methods
Drosophila Cells Culture and RNAi-Mediated Interference.
The DMel strain of Drosophila S2 cells (Invitrogen) was used in all experiments. These cells were cultured in serum-free medium (Express Five; GIBCO) supplemented with 1 mM glutamine, penicillin, and streptomycin at 25 °C. Generation of all dsRNAs was performed as previously described (44). We used two different oligonucleotide sequences targeting different regions of Rab5 and mud genes. All of the oligonucleotides used in this work are available on request.
Fly Stocks and Cytology.
The Oregon R strain was used as WT. Rab5 alleles were a gift from M. Gonzalez-Gaitan (Geneva, Switzerland) (25). After testing all of the possible heteroallelic combinations of different Rab5 alleles, we found that only Rab51/Rab53 animals reached the third instar larval stage required for the preparation of neuroblasts mitotic chromosome squashes as described previously (45).
Plasmid Constructs and Generation of Stable Cell Lines.
ORFs encoding the protein(s) of interest (Rab5 and Lamin) were amplified by PCR using Accuprime polymerase (Invitrogen) and primers containing attB flanking sequences for Gateway Technology. PCR products were purified and introduced into a pDONR221 plasmid (Invitrogen) to generate entry vectors. Expression vectors of the proteins of interest were generated with the ORF under the control of a constitutive (actin 5C) or inducible (metallothionein) promoter (29). The tubulin::mRFP plasmid was a gift of Matthew S. Savoian (University of Cambridge, Cambridge, United Kingdom). The GFP::2XFYVE stable cell line was a gift from T. Takeda (University of Cambridge, Cambridge, United Kingdom). Stable cell lines were generated using Blasticidin (Invitrogen) selection as previously described (29).
More detailed descriptions of immunocytochemistry, microscopy, live imaging, PtA affinity purification, MS, Western blot analysis, IP experiments, and statistical analysis are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Letizia Lanzetti for sharing unpublished results; T. S. Hays, H. A. Nash, P. G. Wilson, M. S. Savoian, T. Takeda, V. Korolchuk, and M. Gonzalez-Gaitan for reagents; P. Lió for statistical advice, T. Maresca for his 3F3/2 antibody staining protocol, and P. Almeida Coelho for help with image deconvolution; and M. S. Savoian, P. Almeida Coelho, T. Takeda, and E. Wegel for critical reading of the manuscript and all D.M.G. laboratory members (past and present) for invaluable discussions throughout this work. This research was supported in part by grants from the Biotechnology and Biological Sciences Research Council Link and Cancer Research United Kingdom Programme (to D.M.G.). V.A. was a Human Frontier Science Program fellow.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103720108/-/DCSupplemental.
References
- 1.Fürthauer M, González-Gaitán M. Endocytosis and mitosis: A two-way relationship. Cell Cycle. 2009;8:3311–3318. doi: 10.4161/cc.8.20.9700. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Y. A membranous spindle matrix orchestrates cell division. Nat Rev Mol Cell Biol. 2010;11:529–535. doi: 10.1038/nrm2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol. 2010;12(2):164–169. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai MY, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- 5.Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loïodice I, et al. The entire Nup 107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belgareh N, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Zheng Y. A requirement for epsin in mitotic membrane and spindle organization. J Cell Biol. 2009;186:473–480. doi: 10.1083/jcb.200902071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miserey-Lenkei S, et al. A role for the Rab6A’ GTPase in the inactivation of the Mad2-spindle checkpoint. EMBO J. 2006;25:278–289. doi: 10.1038/sj.emboj.7600929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–1157. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin CH, Hu CK, Shih HM. Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J Cell Biol. 2010;189:1097–1105. doi: 10.1083/jcb.200911120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimizu H, Nagamori I, Yabuta N, Nojima H. GAK, a regulator of clathrin-mediated membrane traffic, also controls centrosome integrity and chromosome congression. J Cell Sci. 2009;122:3145–3152. doi: 10.1242/jcs.052795. [DOI] [PubMed] [Google Scholar]
- 13.Tanenbaum ME, et al. Cyclin G-associated kinase promotes microtubule outgrowth from chromosomes during spindle assembly. Chromosoma. 2010;119:415–424. doi: 10.1007/s00412-010-0267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- 16.Miaczynska M, et al. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 17.Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453:241–245. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- 18.Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178(1):43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radulescu AE, Cleveland DW. NuMA after 30 years: The matrix revisited. Trends Cell Biol. 2010;20:214–222. doi: 10.1016/j.tcb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silk AD, Holland AJ, Cleveland DW. Requirements for NuMA in maintenance and establishment of mammalian spindle poles. J Cell Biol. 2009;184:677–690. doi: 10.1083/jcb.200810091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haren L, Gnadt N, Wright M, Merdes A. NuMA is required for proper spindle assembly and chromosome alignment in prometaphase. BMC Res Notes. 2009;2:64. doi: 10.1186/1756-0500-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahmani Z, Gagou ME, Lefebvre C, Emre D, Karess RE. Separating the spindle, checkpoint, and timer functions of BubR1. J Cell Biol. 2009;187:597–605. doi: 10.1083/jcb.200905026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16:1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 24.D'Avino PP, et al. Recruitment of Polo kinase to the spindle midzone during cytokinesis requires the Feo/Klp3A complex. PLoS One. 2007;2:e572. doi: 10.1371/journal.pone.0000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wucherpfennig T, Wilsch-Bräuninger M, González-Gaitán M. Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J Cell Biol. 2003;161:609–624. doi: 10.1083/jcb.200211087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 27.Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: Comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archambault V, Glover DM. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 29.D'Avino PP, et al. Isolation of protein complexes involved in mitosis and cytokinesis from Drosophila cultured cells. Methods Mol Biol. 2009;545:99–112. doi: 10.1007/978-1-60327-993-2_6. [DOI] [PubMed] [Google Scholar]
- 30.Bowman SK, Neumüller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Du Q, Stukenberg PT, Macara IG. A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- 32.Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 33.Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg M, et al. Interactions among Drosophila nuclear envelope proteins lamin, otefin, and YA. Mol Cell Biol. 1998;18:4315–4323. doi: 10.1128/mcb.18.7.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattout A, Goldberg M, Tzur Y, Margalit A, Gruenbaum Y. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J Cell Sci. 2007;120(1):77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- 36.Stafstrom JP, Staehelin LA. Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo. Eur J Cell Biol. 1984;34(1):179–189. [PubMed] [Google Scholar]
- 37.Katsani KR, Karess RE, Dostatni N, Doye V. In vivo dynamics of Drosophila nuclear envelope components. Mol Biol Cell. 2008;19:3652–3666. doi: 10.1091/mbc.E07-11-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bousbaa H, Correia L, Gorbsky GJ, Sunkel CE. Mitotic phosphoepitopes are expressed in Kc cells, neuroblasts and isolated chromosomes of Drosophila melanogaster. J Cell Sci. 1997;110:1979–1988. doi: 10.1242/jcs.110.17.1979. [DOI] [PubMed] [Google Scholar]
- 40.Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salina D, et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108(1):97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 42.Civelekoglu-Scholey G, Tao L, Brust-Mascher I, Wollman R, Scholey JM. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J Cell Biol. 2010;188:49–68. doi: 10.1083/jcb.200908150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serio G, et al. Small GTPase Rab5 participates in chromosome congression and regulates localization of the centromere-associated protein CENP-F to kinetochores. Proc Natl Acad Sci USA. 2011;108:17337–17342. doi: 10.1073/pnas.1103516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D'Avino PP, Savoian MS, Capalbo L, Glover DM. RacGAP50C is sufficient to signal cleavage furrow formation during cytokinesis. J Cell Sci. 2006;119:4402–4408. doi: 10.1242/jcs.03210. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez C, Glover DM. Techniques for studying mitosis in Drosophila. In: Fantes P, Brooks R, editors. The Cell Cycle: A Practical Approach. Oxford: IRL; 1993. pp. 163–168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.