Abstract
Recent studies have identified a number of transcriptional regulators, including E proteins, EBF1, FOXO1, and PAX5, that act together to orchestrate the B-cell fate. However, it still remains unclear as to how they are linked at the earliest stages of B-cell development. Here, we show that lymphocyte development in HEB-ablated mice exhibits a partial developmental arrest, whereas B-cell development in E2A+/−HEB−/− mice is completely blocked at the LY6D− common lymphoid progenitor stage. We show that the transcription signatures of E2A- and HEB-ablated common lymphoid progenitors significantly overlap. Notably, we found that Foxo1 expression was substantially reduced in the LY6D− HEB- and E2A-deficient cells. Finally, we show that E2A binds to enhancer elements across the FOXO1 locus to activate Foxo1 expression, linking E2A and FOXO1 directly in a common pathway. In summary, the data indicate that the earliest event in B-cell specification involves the induction of FOXO1 expression and requires the combined activities of E2A and HEB.
Steady state hematopoiesis relies on fine tuned transcriptional networks that are modulated by external signals from the surrounding microenvironment. Hematopoiesis is initiated in hematopoietic stem cells (HSCs). HSCs have the ability to generate the entire spectrum of hematopoietic cells as well as self-renew without losing their multilineage potential (1). HSCs initially differentiate into multipotent progenitors (MPPs). MPPs have lost their ability to self-renew and can only transiently support hematopoiesis on transplantation (2). The MPP compartment itself is heterogeneous and contains a subfraction of cells expressing high surface levels of fms-like tyrosine kinase 3 (FLT3), often referred to as lymphoid-primed multipotent progenitors (LMPPs) (3). These cells have largely lost their megakarycyte/erythocyte potential (3). Thus, LMPPs are considered to represent progenitor cells en route to a lymphoid cell fate. LMPPs are the likely precursor of the common lymphoid progenitors (CLPs), with the ability to develop into the entire spectrum of lymphoid cells (4, 5). However, recent studies have shown that the CLP compartment is also heterogeneous, containing immature cells along with a small subset of cells committed to the B-cell lineage (6). The CLPs can be segregated using lymphocyte antigen 6 complex, locus D (LY6D) as a marker (7, 8). Specifically, the LY6D− CLPs represent the more immature compartment, whereas the B lineage-committed cells can be found within the LY6D+ fraction.
The development of lymphoid progenitors requires the activities of an ensemble of transcriptional regulators (9–13). Prominent among these regulators are the E proteins E2A, E2-2, and HeLa E-box binding protein (HEB) (14). The E2A proteins maintain the HSC pool and promote the developmental progression of myelolymphoid and myeloerythorid progenitors during early hematopoiesis (15–17). At the CLP cell stage, they act upstream and in concert with Early B cell factor 1 (EBF1), forkhead box O1 (FOXO1) and Paired box gene 5 (PAX5) to promote specification and commitment to the B-cell lineage (10, 13, 18, 19). The E2A locus encodes for two isoforms, E12 and E47, that arise through differential splicing (14). Whereas E47 plays a critical role in early B-cell development, the activity of E12 is not essential (20). Although some T cells develop in E2A-deficient mice, the E2A proteins have also been shown to play critical roles in early thymocyte development (21). They induce the expression of genes involved in Notch and pre-T cell receptor signaling and in turn, act in concert with Notch signaling to promote T-lineage development (21). A unique role for the E2-2 proteins has also recently been established. Specifically, it was shown that plasmacytoid dendritic cell development is blocked in E2-2–deficient mice (22).
HEB is expressed at high levels in developing thymocytes, where it is required to promote efficient maturation beyond the pre-TCR checkpoint (21) and induce the invariant natural killer T cell fate (23). During fetal life, HEB is required to promote developmental progression of early thymocyte progenitors and pro-B cells (24, 25). However, the roles of HEB in adult hematopoesis and lymphocyte development and how HEB acts in concert with E2A to promote B-cell development have not yet been carefully examined. Here, we report that HEB is broadly expressed at substantial levels within the entire spectrum of adult hematopoietic progenitors. Whereas erythroid and myeloid development seemed normal, a developmental block was observed in the CLP compartment that was similar to the block described for E2A-ablated CLPs (7, 26). Interestingly, E2A and HEB seem to act in concert to promote B-cell development; HEB−/− and E2A+/− mice exhibit a partial block at the LY6D− CLP cell stage, whereas E2A+/−HEB−/− mice exhibit an arrest. Furthermore, HEB- and E2A-deficient LY6D− CLPs regulate an overlapping set of target genes, most notably, the transcription factor FOXO1. Finally, we identify enhancer elements, characterized by the presence of H3K4me1 islands, across the FOXO1 locus that are responsive to E2A activity. These observations directly link E2A, HEB, and FOXO1 in B-cell progenitors into a common framework that specifies the B-cell fate.
Results
Expression of E2A, HEB, Id2, and Id3 in Progenitor Cells.
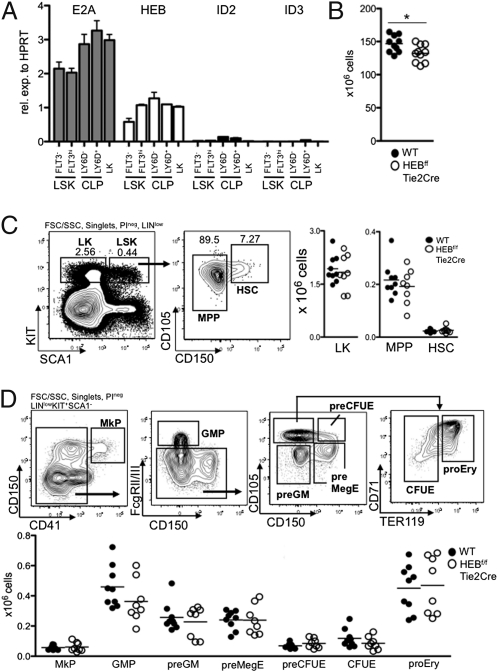
To characterize the patterns of E2A, HEB, Id2, and Id3 expression in bone marrow progenitor cells, mRNA was isolated from FACS-sorted progenitor populations and analyzed by real-time PCR. Both E2A and HEB were expressed in the Lineage−/low SCA1+KIT+ (LSK) compartment known to harbor the majority of the HSCs’ potential (1) as well as substantial levels within the entire spectrum of hematopoietic progenitors (Fig. 1A). Id2 was expressed at low levels in the CLP compartments, whereas Id3 expression was only detectable in the LY6D+ CLP compartment (Fig. 1A). Whereas the roles of E2A in early adult hematopoiesis have been well-documented, the function of HEB in adult hematopoietic development has remained to be determined. As a first approach to this question, mice in which the HEB DNA binding regions were flanked by loxP sites (27) were bred to transgenic mice carrying a Tie2-Cre transgenic construct to generate mice where HEB is deleted from the entire hematopoietic system (Fig. S1A). HEBf/fTie2Cre mice were viable and fertile, and they lacked obvious developmental defects. However, on examination of bone marrow cellularity, HEBf/fTie2Cre mice showed a small but significant reduction in the total number of nucleated cells (P = 0.025) (Fig. 1B).
Fig. 1.
HEB is expressed in bone marrow progenitors. (A) Indicated FACS-sorted progenitor populations were analyzed by real-time PCR for the abundance of E2A, HEB, Id2, and Id3 mRNA. Values were normalized to hypoxanthine guanine phosphoribosyl transferase (HPRT) expression and are shown as mean ± SEM using purified mRNA from two independent sorts. (B) Absolute numbers of bone marrow cells (from femurs, tibias, and cresta iliac) of sex- and age-matched WT and HEBf/fTie2Cre mice. Each dot represents the number from a single mouse. The horizontal lines indicate the mean in each group. The asterisk indicates a P value < 0.05. C Left represents FACS plots of the gating strategies used to identify LK, MPP, and HSC populations. Lineage (LIN) includes CD11B, GR1, CD3ε, and NK1.1. C Right displays the absolute cell numbers of each population in WT and HEBf/fTie2Cre mice. Each dot represents the number from a single mouse, and the horizontal lines are the mean in each group. Data shown are pooled from two independent experiments. D Upper shows representative FACS plots of the gating strategy to identify erythromyeloid progenitor populations within the LK population. D Lower shows the absolute cell number of each population in WT and HEBf/fTie2Cre bone marrow. Each dot represents the number from a single mouse, and the horizontal lines are the mean in each group. Data shown are pooled from two independent experiments. MkP, megakaryocyte progenitor; GMP, granulocyte macrophage progenitor; preGM, pregranulocyte macrophage; preMegE, premegakarycocyte erythrocyte; preCFUE, pre-CFU erythrocyte; CFUE, CFU erythrocyte; proEry, proerythrocyte.
HEB Activity Is Not Essential to Promote HSCs and Myelo-Erythroid Development.
Because HEB is expressed in both the primitive LSK compartment as well as hematopoietic progenitors derived from HSCs, we analyzed the hematopoietic compartment of HEB-ablated mice in the adult bone marrow by flow cytometry (Fig. 1C). The majority of the long-term HSCs can be found within a subpopulation of the LSKs expressing CD150 on the surface (28). The CD150+ LSK and the CD150− LSK compartments were not affected in the bone marrow of HEBf/fTie2Cre mice (Fig. 1C). In addition, no significant changes were found in the numbers of the Lineagelow/− SCA− KIT+ (LK) population known to harbor myelo-erythroid progenitors (29) (Fig. 1C). Furthermore, because HEB has been reported to be associated with T cell acute lymphocytic leukemia 1 (TAL1), E2A, and ETO2 in erythroid cells (30), we examined the heterogenous LK compartment (31). No statistically significant differences were observed among these subsets, and all subpopulations were present in normal numbers in the bone marrow of HEBf/fTie2Cre compared with WT mice (Fig. 1D). Taken together, these observations indicate that HEB, unlike E2A, is dispensable for the steady state maintenance of HSCs and the generation of myelo-erythroid progenitors.
HEB Promotes the B-Cell Fate.
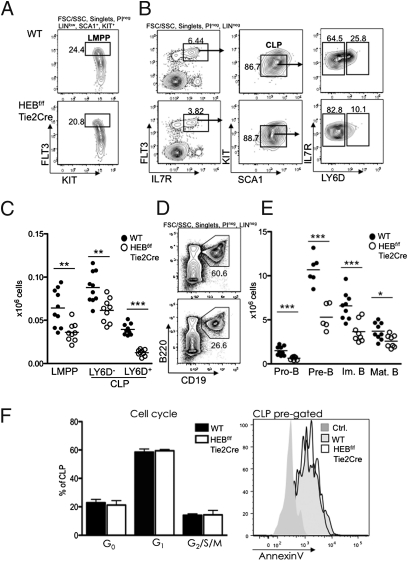
An early step to the development of lymphoid-restricted cells is the development of LMPPs that are defined by the up-regulation of FLT3 in a subset of the LSK compartment. Interestingly, the LMPP compartment was significant reduced in HEBf/fTie2Cre relative to WT mice (P = 0.0026) (Fig. 2 A–C). The CLP compartment was also significantly affected in HEBf/fTie2Cre mice. Specifically, both the LY6D− CLP (P = 0.0024) and LY6D+ CLP compartments (P < 0.0001) were substantially reduced (Fig. 2 B and C) and strongly resembled the E2A+/− mice (Fig. S1B). Cellularity of the CD19+ compartment in HEBf/fTie2Cre bone marrow was significantly altered compared with WT bone marrow (Fig. 2 D and E). The total reduction in CD19+ bone marrow B-cell progenitors corresponded well to the overall reduction seen in bone marrow cellularity, suggesting that in vivo HEBf/fTie2Cre progenitors did not adopt alternate cell fates at the expense of B-cell differentiation.
Fig. 2.
HEB is required to specify the B-cell fate. (A) Representative FACS plots of LMPPs from WT and HEBf/fTie2Cre bone marrow. Lineage (LIN) includes CD11B, GR1, TER119, CD3ε, LY6C, NK1.1, CD19, and CD11C. (B) Representative FACS plots of LY6D− CLPs and LY6D+ CLPs from WT and HEBf/fTie2Cre bone marrow. (C) Total cellularity of LMPP, LY6D− CLPs, and LY6D+ CLPs in WT and HEBf/fTie2Cre bone marrow. Each dot represents the number from a single mouse, and the horizontal lines indicate the mean in each group. Data shown are pooled from two independent experiments. **P < 0.01; ***P < 0.001. (D) Representative FACS plots of CD19+B220+ cells from WT and HEBf/fTie2Cre bone marrow. LIN includes CD11B, GR1, and TER119. (E) Total cell numbers of B-cell progenitor populations within the CD19+B220+ bone marrow B-cell populations. All populations were first gated as CD19+B220+. Pro-B, KIT+IgM−IgD−; Pre-B, KIT−IgM−IgD−; Im. B, KIT−IgM+IgD−; Mat. B, KIT−IgM+/low IgD+. Each dot represents the number from a single mouse, and the horizontal lines are the mean in each group. Data shown are pooled from two independent experiments. *P < 0.05; ***P < 0.001. F Left displays the cell cycle distribution (quantified by Ki67 and 7AAD FACS staining) of WT and HEB-deficient CLPs (n = 3 in each group). F Right shows a representative plot of Annexin V staining comparing WT and HEB-deficient CLPs (total n = 3 in each group). For F Left and F Right, CLPs are defined as depicted in B.
Previous studies have shown that E2A-ablated HSCs enforce a cell cycle checkpoint in HSCs (15, 16). To explore the possibility that HEB performs a similar role in progenitor cells, the fraction of cycling progenitors was analyzed in HEBf/fTie2Cre bone marrow. Lack of HEB in the CLP compartment did not alter the cell cycle distribution, and in contrast to E2A-deficient mice, no differences in cell cycle progression were found in the LSK and LK compartments of HEBf/fTie2Cre mice (Fig. 2F and Fig. S2 A and B). Furthermore, Annexin V staining revealed that HEB seemed to not be required for the viability of the cells in the CLP, LSK, and LK compartments (Fig. 2F and Fig. S2 A and B). These data indicate that the development of early hematopoietic progenitors in the absence of HEB resembles the development observed in E2A-deficient mice.
HEB Deficiency Interferes with the LY6D− CLP to LY6D+ CLP Transition.
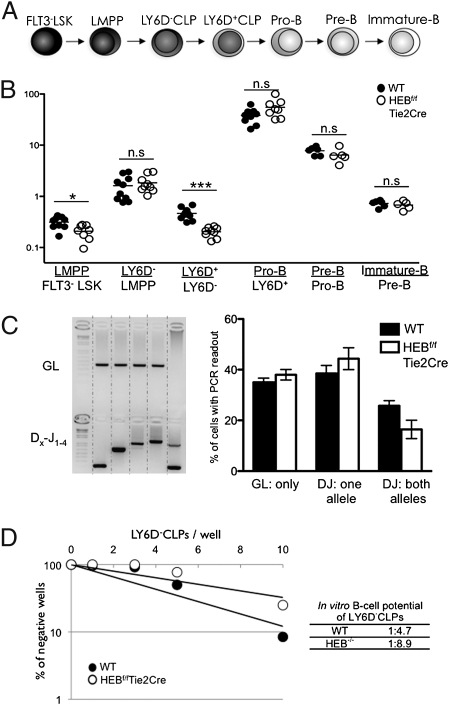
B-cell development can be separated into phenotypically defined stages based on surface markers and Ig rearrangement status (Fig. 3A). To identify the compartments where developmental progression was affected by a lack of HEB, the ratio of cell numbers of consecutive developmental steps were plotted (Fig. 3B). This analysis indicated that a critical function of HEB is exerted during transition from the LY6D− to LY6D+ cell stage. No significant changes were found in the pro-, pre-, and immature B-cell transitions, indicating that HEB is dispensable for developmental progression beyond the LY6D+ stage. Rather, we suggest that the reduced numbers of CD19+ bone marrow cells present in the bone marrow of HEBf/fTie2Cre originate from the developmental defects in the CLP compartment.
Fig. 3.
HEB promotes the transition from the LY6D− to LY6D+ CLP compartment. (A) Schematic overview of B-cell development in the bone marrow. (B) Plot of cell number ratios from the consecutive developmental stages depicted in A. The total cellularity of an early population for a mouse is divided by the total cell number of the proceeding population. Each dot represents the ratio from a single mouse, and the horizontal lines are the mean in each group. Data shown are pooled from two independent experiments. *P < 0.05; ***P < 0.001. n.s., not significant. C Left shows an example of DH–JH rearrangement PCR readouts from LY6D+ CLPs. C Right shows the distribution of cells within the LY6D+ CLP compartment with respect to DH–JH rearrangement status. A total of 192 cells/population from two independent sorts was assayed. Displayed are the mean ± SEM of cells producing at least one PCR product. (D) Result of limiting dilution assay for in vitro B-cell potential. CLPs from WT and HEB-deficient mice were sorted onto preplated OP9 stroma cells and cultured in conditions promoting B-lymphoid development. The number of cells plated per well is plotted vs. the percentages of wells negative for B-cell growth (CD19+ cells). Full lines represent the best fit regression. The frequency of cells with B-cell potential was determined as the cell concentration where 37% of the cells were not generating B cells. The frequency for WT and HEBf/fTie2Cre cells is indicated. Data shown are from a representative experiment from a total of three experiments.
A recent study using Rag1-GFP reporter mice identified functionally distinct low and high reporter-expressing cells within the LY6D+ CLP population (8). These observations raise the question as to whether the reduced frequency of LY6D+ CLPs is caused by a partial block in the LY6D− to LY6D+ transition and/or a transition within the LY6D+ fraction. To explore this question, single LY6D+ cells were examined for the presence of DHJH rearrangements (Fig. 3C). We found that the frequency of DHJH rearrangements was not altered in HEBf/fTie2Cre mice, indicating that the distribution of subpopulations within the LY6D+ CLP compartment remained unperturbed (Fig. 3C). These data indicate that, although the lack of HEB caused a partial block in the transition from LY6D− to LY6D+ CLPs, development within the LY6D+ compartment was not significantly altered in the absence of HEB.
The data described above indicate that HEB acts to promote specification to the B-cell fate. To directly evaluate how HEB acts in progenitors, we evaluated the in vitro B-cell developmental potential of HEBf/fTie2Cre LY6D− progenitors using a limiting dilution assay approach; 1, 3, 5, or 10 uncommitted LY6D− CLPs were plated on OP9 stroma cells in the presence of the cytokines FLT3-Ligand, IL-7, and stem cell factor. On culturing the cells for 14 d, the number of cells plated per well was plotted against the percentages of wells negative for B-cell growth, which were measured by CD19 expression. The frequency of LY6D− CLPs containing in vitro B-cell potential was determined as the concentration of cells in which 37% of the wells lacked B cells (32). One of five WT cells developed into B cells (Fig. 3D). In contrast, only one of nine cells of the HEB-deficient cells progressed to the committed B-cell stage (Fig. 3D). These data indicate a 50% reduction in the ability of HEB-ablated progenitors to differentiate into committed B cells. These data correspond well with the decrease observed in the HEB-deficient bone marrow. In contrast, HEBf/fTie2Cre LY6D− CLPs efficiently differentiated into early T-lineage cells on culture in the presence of OP9-DL1 cells (Fig. S3). Collectively, these data indicate that HEB acts in the CLP compartment to specify the B-cell fate.
HEB and E2A Act in Concert to Specify the B-Cell Fate.
The data described above indicate a critical role for HEB in promoting B-lineage specification in a manner similar to the role described for E2A. To determine whether E2A and HEB act together to specify the B-cell fate, E2A and HEB compound mice were generated and analyzed by flow cytometry for abnormalities in early hematopoiesis. Because Tie2Cre expression did not efficiently excise E2A alleles in E2Af/f mice (27), we used Vav-iCre transgenic mice to achieve a higher degree of excision. E2Af/fVaviCre and HEBf/fVav-iCre displayed the same hematopoietic phenotype as the E2A−/− and HEBf/fTie2Cre mice (Fig. 4A and Fig. S4). Next, we compared HEBf/fVav-iCre, E2Af/+Vav-iCre, and E2Af/+HEBf/fVav-iCre mice for abnormalities in CLP development (Fig. 4A). As expected, E2Af/+Vav-iCre and HEBf/fVav-iCre mice exhibited a partial block at the CLP cell stage (Fig. 4A). However, the LY6D+ compartment was close to absent in E2Af/+HEBf/fVav-iCre mice (Fig. 4A). These data indicate that E2A and HEB act in concert to initiate B-cell development.
Fig. 4.
HEB and E2A act in concert to specify the B-cell fate. (A) Representative FACS plots comparing bone marrow stained for LY6D+ CLPs in WT, HEBf/fVaviCre, E2Af/+VaviCre, HEBf/fE2Af/+VaviCre, and E2Af/fVaviCre mice. (B) Microarrays analysis displaying genes that are changed by a factor of at least twofold between WT and HEBf/fTie2Cre LY6D− CLPs. E2A−/− displays how this subset of genes is affected in E2A−/− LY6D− CLPs. Displayed data are derived from the means from two or more microarray replicas using material from independent sorts. (C) Graph shows the number of CCR9+ LY6D− CLPs in WT and HEBf/fTie2Cre mice. Each dot represents the number from a single mouse, and the horizontal lines are the mean in each group. Representative data from one of a total of two experiments are shown. **P < 0.01. D Left, right column shows the result of microarray analysis, displaying genes that are changed by a factor of at least twofold between WT and E2A−/− LY6D− CLPs. In addition, the center column displays how these genes are affected in HEBf/fTie2Cre LY6D− CLPs. Highlighted in the left column is the location of genes of interest for lymphocyte development (a full gene list is in Table S1). D Right displays box plots of cluster analysis showing how genes patterns in D Left are changed in WT and HEB- and E2A-deficient LY6D− CLPs. ***P < 0.001. Displayed data are the means derived from two or more microarray replicas using material from independent sorts.
To determine how E2A and HEB act in the CLP compartment to prime the B-cell fate, we performed microarray analysis using mRNA from sorted LY6D− CLPs derived from HEBf/fTie2Cre mice. Notably, only 18 genes were altered by a factor of more than twofold in HEBf/fTie2Cre vs. WT LY6D− CLPs (Fig. 4B). To verify that chemokine (C-C motif) receptor 9 (CCR9), a previously identified E2A target, is also regulated by HEB, HEB-ablated CLPs were analyzed for the expression of Ccr9 by flow cytometry (17). As predicted, Ccr9 expression was substantially reduced in HEBf/fTie2Cre LY6D− CLPs (Fig. 4C). To directly evaluate the extent to which E2A and HEB share a common set of target genes, mRNA from E2A−/− LY6D− CLPs was next examined by microarray analysis. The majority of the genes altered in the absence of HEB exhibited a similar change as depletion of E2A activity (Fig. 4B).
To further explore the possibility that HEB and E2A share a common set of target genes in the LY6D− compartment, we examined the entire spectrum of genes changed in the absence of E2A; 197 transcripts were up- or down-regulated more than twofold in E2A-deficient LY6D− CLPS compared with the corresponding WT cells (Fig. 4D Left and Table S1). We next examined how these genes were changed in the absence of HEB. This analysis revealed a striking pattern of E-protein dose dependency (Fig. 4D Right). Genes up- or down-regulated by E2A were, in general, also affected by the absence of HEB, albeit to a lower degree. Among the changed transcripts were Ccr9, Id2, Blnk, Dntt, Notch1, IL-7ra, and most notably, Foxo1. These data indicate that E2A and HEB share a large set of common target genes and that it is the dosage of E proteins that promotes the development of LY6D+ CLPs.
Regulation of Foxo1 Expression by the E Proteins.
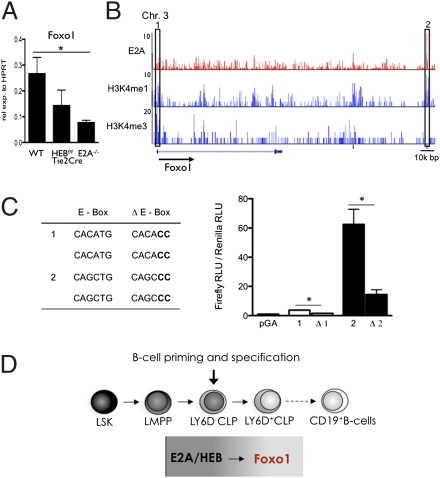
The microarray data described above indicate that both E2A and HEB are required to activate normal FOXO1 expression in the CLP compartment. To quantitatively compare FOXO1 transcript levels in WT and HEB- and E2A-deficient CLPs, mRNA derived from each of the populations was examined by real-time PCR. Consistent with the microarray data, FOXO1 transcript levels were reduced in both HEBf/fTie2Cre and E2A−/− CLPs (Fig. 5A).
Fig. 5.
Direct regulation of Foxo1 expression in CLPs by the E proteins. (A) LY6D− CLPs from WT, HEBf/fTie2Cre, and E2A−/− mice were analyzed by real-time PCR for the abundance of Foxo1 mRNA. Values were normalized to HPRT expression and are shown as mean ± SEM using purified mRNA from two independent sorts. (B) E2A (red; top row) occupancy, H3K4me1 (blue; middle row), and H3K4me3 (blue; bottom row) epigenetics mark across the Foxo1 locus in EBF1-deficient cells identified by ChIP followed by genome-wide deep sequencing. Numbers on the left indicate the number of tags observed. The box on the left indicates a Foxo1 locus promoter region with E-box sites; the box on the right indicates a region where E2A occupancy is associated with H3K4me1. (C) Transcriptional activity of H3K4me1 islands (corresponding to the boxes in B) with associated E2A occupancy and presence of E boxes. Δ, deletion within the E-box sequences. Data shown are the mean ± SEM derived from two independent experiments. *P < 0.05. (D) Schematic diagram depicting B-cell development and the activities of E2A and HEB in B-cell specification.
To explore the possibility that E proteins directly regulate FOXO1 expression, we examined the regulatory regions of the Foxo1 locus for E2A occupancy within putative enhancer regions as characterized by H3K4 monomethylation (H3K4me1). We previously reported the distribution of H3K4me1 and H3K4me3 as well as E2A occupancy in EBF1−/− hematopoietic progenitors (10). On inspection of epigenetic marks across the FOXO1 locus, we identified high-affinity E2A binding sites in putative enhancers as well in the promoter region (Fig. 5B) (10). Subsequently, the putative enhancer and promoter regions, containing the identified WT as well as mutated E-box sites, were inserted into an enhancer luciferase reporter construct, transfected into pro-B cells, and assayed for transcriptional activity. Notably, deletion of the E-box sites substantially interfered with reporter activity (Fig. 5C) (Ppeak1 = 0.0178, Ppeak2 = 0.047). Thus, these data indicate that E-protein activity directly regulates Foxo1 expression (Fig. 5D).
Discussion
Previous observations have indicated that the E2A proteins play critical roles throughout hematopoiesis. In the HSC compartment, the E2A proteins maintain the hematopoietic stem cell pool (15, 16), and in developing progenitors, the E2A proteins promote the developmental progression of erythroid/megakaryocytic and myeloid progenitors (15). However, perhaps most prominent is the block observed in the CLP compartment in E2A-deficient mice at a stage in which B-cell development is initiated (7, 26). Specifically, although the LY6D− CLP compartment in E2A-deficient mice is present, albeit in reduced numbers, the LY6D+ compartment is complete absent (7). Here, we show that E2A and HEB mechanistically act together to specify the B-cell fate.
Whereas HSCs were not significantly perturbed in HEB-deficient mice, we observed a clear reduction in B-cell progenitors in the adult bone marrow. The defect in B-lymphoid differentiation was initiated in the LMPP compartment but manifested predominantly during the LY6D− to LYD6+ CLP transition. However, committed B-lineage cells derived from the LY6D+ CLP compartment developed in expected ratios. Thus, these data indicate that the partial block in the LY6D− to LY6D+ CLP transition is the major cause for the observed reduction in B-cell progenitor numbers in HEB-deficient bone marrow. Furthermore, HEBf/fVaviCre and E2Af/+VaviCre mice show a partial block at the CLP cell stage, whereas HEBf/fE2Af/+VaviCre mice display a more complete arrested block in the LY6D− compartment. Consistent with these observations, using microarray analysis, we show that the transcription signatures of E2A- and HEB-deficient CLPs substantially overlap.
We are now faced with the question as to how HEB and E2A act at the LY6D− CLP cell stage to initiate a B lineage-specific program of gene expression. Here, we show that Foxo1 expression is reduced in the absence of E proteins. A genome-wide screen for E2A binding sites revealed that the E2A proteins bind to regulatory elements present in the FOXO1 locus (10), and we show that these E2A binding sites indeed play a role in modulating FOXO1 expression. In addition to the E proteins, an ensemble of transcriptional regulators has been shown to play critical roles in early B-cell development. Among these regulators are EBF1, PU.1, FOXO1, and PAX5 (13, 33). Furthermore, IL-7R–mediated signaling also has been proposed to modulate EBF1 expression (34–36). How is this spectrum of transcriptional regulators connected to orchestrate the B-cell fate? Based on previous data as well as the observations described here, we would like to propose that, in LY6D− CLPs, E2A and HEB act in concert to induce the expression of FOXO1. FOXO1, in turn, together with E2A, HEB, and PU.1 as well as IL-7R–mediated signaling, then activates the expression of EBF1 in LY6D+ CLPs. E2A, FOXO1, EBF1, and IRF4 as well as IRF8 subsequently induce the expression of PAX5 (10, 13, 37, 38). These data bring into question how the E proteins are activated in the CLP LY6D− compartment. E2A protein levels do not change significantly during the developmental progression from the HSC to the CLP cell stage (15). In addition, we show here that Id2 and Id3 mRNA levels do not change substantially during the developmental progression from the LSK to the CLP cell stage. The question then arises as to why a B lineage-specific program gene expression is not induced at the HSC or LMPP cell stage. We would like to suggest that transcriptional repression rather than activation regulates the expression of FOXO1 during early hematopoiesis, because the low levels of FOXO1 expression are not affected in E2A-ablated LMPPs (17). Thus, before reaching the CLP compartment, FOXO1 levels are kept low by active repression. On reaching the CLP compartment, the activities of transcriptional repressors are blocked, permitting E2A and HEB to induce FOXO1 transcription and specify the B-cell fate. The critical problem now will be to determine how the activation of a B lineage-specific program of gene expression by the E proteins is suppressed during the earlier stages of hematopoiesis.
Materials and Methods
Mice.
HEBf/f and E2Af/f mice were a gift from Yuan Zhuang (Duke University, Durham, NC). Tie2Cre and VaviCre mice were purchased from The Jackson Laboratory. E2A-deficient mice have been previously described (9). Mice were analyzed at 10–15 wk of age. Animal studies were approved by the Institutional Animal Care and Use Committee of the University of California at San Diego.
FACS Staining and Purification of Bone Marrow Cells.
Bone marrow DNA was prepared as previously described (26) cells were Fc-blocked (CD16/CD32; 2.4G2) and stained with combinations of the antibodies FLT3(A2F10), CD11B(M1/70), GR1(RB6-8C5), TER119 (Ter119), CD3e(145-2c11), CD11C(N418), LY6C (AL-21), NK1.1(PK136), SCA1 (D7), CD19(1D3), KIT(2B8), IL-7R(A7R34), B220(RA3-6B2), IgD(11-26c), IgM(11/41), CD41(MWReg30), CD16/32(2.4G2), CD150(TC15-12F12.2 (BioLegend), and CD105(MJ7/18) followed by streptavidin-conjugated Qdot655 (Invitrogen) to visualize biotinylated antibodies and propidium iodine to exclude dead cells. For CLP detection, cells where stained with LY6D(49-H4) (BD Bioscience) followed by Qdot605 goat anti-rat IgG (Invitrogen) before Fc blocking. Antibodies were purchased from eBioscience unless otherwise noted. For progenitor isolation, cells were subjected to magnetic-activated cell sorting enrichment of CD27+ or KIT+ immunomagnetic beads (Miltenyi Biotec) before antibody staining. Analysis was performed using an LSRII, and cell sorting was performed on FACSAria II (BD Biosciences).
RNA Analysis.
Quantitative RT-PCR analysis of sorted cells was performed as previously described (6). Assays on Demand (Applied Biosystems) probes used were Hprt (Mm00446968_m1), E2A (Mm01175598_m1), HEB (Mm00441699_m1), Id2 (Mm00711781_m1), and Id3 (Mm00492575_m1). All experiments were performed in duplicate at least two times using cells from independent sorts.
IgH D–J Rearrangements in Single Cells.
Nested single-cell PCR has been previously described (26) and was performed with slight modifications. Briefly, single-cell LY6D+ CLPs were sorted directly into 96-well plates containing PCR buffer containing 20 mM Tris⋅HCl 50 mM KCl (pH 8.4), snap-frozen, and stored in −80 °C. In the first round, PCR amplification was performed with, 0.2 mM dNTP, 1 μL homemade Taq, 0.5 mg/mL BSA, and 0.4 μM of each primer in a total of 50 μL/well; 1 μL first-round PCR was used as a template in the second round of PCRs. Second-round primers were used at 2 μM concentrations. Only samples producing either a germline product and/or a specific rearrangement product are included in the analysis.
Annexin V Staining.
After staining with appropriate surface markers, cells were stained using Annexin V Apoptosis detection kit (eBioscience) according to manufacturer's instructions.
Cell Cycle Analysis.
Cells were fixed with the BD Cytofix/Cytoperm solution kit (BD Bioscience) followed by incubation with anti-human Ki67 (BD Bioscience) and 7AAD according to manufacturer's instructions.
In Vitro Evaluation of B- and T-Cell Potential by OP9 Coculture.
For evaluation of in vitro B-cell potential, 1, 3, 5, or 10 cells/well were deposited (using a FACSAria) directly into 96-well plates containing preplated OP9 cells. In each experiment, 10 or more wells were analyzed per cell concentration. For evaluation of in vitro T-cell potential, single cells were deposited on preplated OP9DL1 cells in similar conditions as the B-cell cultures. Culture conditions and FACS evaluations have been previously described (6). Frequencies from limiting dilution assays were calculated as previously described (32).
Affymetrix Gene Expression and Data Analysis.
Gene expression assays have been previously described (8).
Luciferase Assays.
Luciferase assays were performed as previously described (10).
Supplementary Material
Acknowledgments
We thank Yuan Zhuang for providing the HEBf/f and E2Af/f mice, Dr. Chris Brenner and Annamaria Kauzlaric for help with statistical analyses, Dr. Yin Lin for help with accessing chip-sequencing data, and members of the laboratories of D.B., M.S., and C.M. for helpful discussions. We are grateful to Gerd Sten and Liselotte Lenner for technical help. C.M. is supported by the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE27402).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111766108/-/DCSupplemental.
References
- 1.Ikuta K, Weissman IL. Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc Natl Acad Sci USA. 1992;89:1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, et al. Identification of Lin(-)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 3.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 5.Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–5570. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansson R, et al. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112:1048–1055. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- 7.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mansson R, et al. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 2010;115:2601–2609. doi: 10.1182/blood-2009-08-236398. [DOI] [PubMed] [Google Scholar]
- 9.Bain G, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 10.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 12.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 13.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Murre C. Developmental trajectories in early hematopoiesis. Genes Dev. 2009;23:2366–2370. doi: 10.1101/gad.1861709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci USA. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, et al. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dias S, Månsson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier H, et al. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 19.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med. 2009;206:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braunstein M, Anderson MK. HEB-deficient T-cell precursors lose T-cell potential and adopt an alternative pathway of differentiation. Mol Cell Biol. 2011;31:971–982. doi: 10.1128/MCB.01034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borghesi L, et al. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J Exp Med. 2005;202:1669–1677. doi: 10.1084/jem.20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 29.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 30.Meier N, et al. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 31.Pronk CJ, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Cumano A, Paige CJ. Enrichment and characterization of uncommitted B-cell precursors from fetal liver at day 12 of gestation. EMBO J. 1992;11:593–601. doi: 10.1002/j.1460-2075.1992.tb05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9:1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsapogas P, et al. IL-7 mediates Ebf-1-dependent lineage restriction in early lymphoid progenitors. Blood. 2011;118:1283–1290. doi: 10.1182/blood-2011-01-332189. [DOI] [PubMed] [Google Scholar]
- 35.Roessler S, et al. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decker T, et al. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Treiber T, et al. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity. 2010;32:714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.