Abstract
Background
To date, 11 HIV-1 subtypes and 48 circulating recombinant forms have been described worldwide. The underlying reason why their distribution is so heterogeneous is not clear. Host genetic factors could partly explain this distribution. The aim of this study was to describe HIV-1 strains circulating in an unexplored area of Mekong Delta, Vietnam, and to assess the impact of optimal epitope mutations on HLA binding.
Methods
We recruited 125 chronically antiretroviral-naive HIV-1-infected subjects from five cities in the Mekong Delta. We performed high-resolution DNA typing of HLA class I alleles, sequencing of Gag and RT-Prot genes and phylogenetic analysis of the strains. Epitope mutations were analyzed in patients bearing the HLA allele restricting the studied epitope. Optimal wild-type epitopes from the Los Alamos database were used as reference. T-cell epitope recognition was predicted using the immune epitope database tool according to three different scores involved in antigen processing (TAP and proteasome scores) and HLA binding (MHC score).
Results
All sequences clustered with CRF01_AE. HLA class I genotyping showed the predominance of Asian alleles as A*11:01 and B*46:01 with a Vietnamese specificity held by two different haplotypes. The percentage of homology between Mekong and B consensus HIV-1 sequences was above 85%. Divergent epitopes had TAP and proteasome scores comparable with wild-type epitopes. MHC scores were significantly lower in divergent epitopes with a mean of 2.4 (±0.9) versus 2 (±0.7) in non-divergent ones (p<0.0001).
Conclusions
Our study confirms the wide predominance of CRF01_AE in the Mekong Delta where patients harbor a specific HLA pattern. Moreover, it demonstrates the lower MHC binding affinity among divergent epitopes. This weak immune pressure combined with a narrow genetic diversity favors immune escape and could explain why CRF01_AE is still predominant in Vietnam, particularly in the Mekong area.
Introduction
Since its introduction in humans, HIV-1 has undergone dramatic diversification, extensively spreading in all the continents and rapidly diversifying in Caucasian, African and Asian populations. The radial evolution of group M has led to multiple clades or subtypes (A1, A2, B, C, D, F1, F2, G, H, J and K), all of which are circulating in African populations. To date, 11 HIV-1 subtypes and 48 circulating recombinant forms have been described worldwide. HIV-1 subtype B predominates in the Caucasian populations of Europe and the Americas, HIV-1 subtype C in South Asia and CRF01_AE in South East Asia. In particular, a large number of HIV-1 recombinants between subtypes B and C (including CRF07_BC and CRF08_BC) are largely responsible for the current AIDS epidemic in China [1], [2]. The underlying reason why the distribution of the different HIV-1 subtypes is so heterogeneous and unbalanced is not clear. Viral lineage, defined as the founder effect, is certainly an important driver of HIV-1 evolution [3] but cannot explain all virus-clade-specific differences, which are also the consequence, at least in part, of the worldwide HLA distribution. The rapid spread of some subtypes and/or recombinants in Oriental populations could be due to the relative genetic homogeneity of these populations, which contrasts with the extreme genetic diversity in Africans [4], [5], [6].
HLA class I molecules play a critical role in defining the epitopes of cytotoxic T lymphocytes (CTLs), which are essential in host viral defense against HIV [7]. Large cohort studies have shown that certain HLA class I alleles correlate with HIV control, HLA B*57:01 being the most consistently recognized association with immune control of infection [8], [9], [10], [11]. Moreover, homozygosity is associated with faster progression to AIDS, suggesting that heterozygosity at each HLA class I allele allows the presentation of a greater range of epitopes and a more effective control of HIV replication [12], [13].
Mutations affecting HLA-restricted epitopes may lead to immune escape if they impact on epitope antigen processing, HLA binding or TCR recognition. Several studies have identified a growing number of epitope mutations leading to immune escape, especially in Caucasian populations infected with subtype B or African populations infected with subtype C [14], [15], [16].
In Vietnam, we and others have demonstrated that CRF01_AE is widely predominant, but little is known about the impact of epitope mutations observed among Asian HIV-1 infected patients on CTL recognition [17], [18], [19], [20].
The aim of this study was (i) to describe HIV-1 subtypes circulating among antiretroviral (ART)-naive patients in an unexplored area, the Mekong Delta, Vietnam; (ii) to analyze the polymorphism of Gag and RT-Prot sequences; and (iii) to study the impact of amino acid divergences within CRF01_AE epitopes on antigen processing and HLA binding among this host Vietnamese population.
Methods
Ethical statement
This study was approved by the Ethics Committee of the Vietnamese Ministry of Public Health and was conducted in accordance with the set guidelines for research. All patients provided their written informed consent for the collection of the samples and their subsequent analysis.
Population and samples
From June to October 2008, 125 chronically HIV-1-infected individuals including 80 men and 45 women were recruited from 5 centers for preventive medicine located in the provincial cities of the Mekong Delta (Vietnam): Dong Nai, Vung Tau, Tien Giang, An Giang and Dong Thap ( Figure 1 ). Most (95.2%) of these individuals are drug users or sexually transmitted disease patients. The median age was 30 (20–50) years. All patients except for 3 individuals who were receiving treatment are antiretroviral therapy (ART)-naïve.
Figure 1. Vietnamese cities of Mekong Delta taking part to the study.
Blood samples were collected from each patient using ethylenediaminetetraacetic acid (EDTA) tubes. CD4 T lymphocytes were counted with a FACScan flow cytometer (BD Biosciences, San Jose, CA). The plasma samples were separated and stored at −80°C until use.
The median CD4+ cell count in our study population was 147 cells/µl (1–1032) and the plasma viral load was unknown. Clinical staging was as follows according to the WHO classification: 33% stage I, 27% stage II, 35% stage III, 5% stage IV.
HLA class I typing
Genomic DNA was extracted from the frozen white blood cell pellets using the MagNA Pure automatic system (Roche) according to the manufacturer's specifications, and quantitated by UV optical density measurement. Intermediate-to-high resolution was performed by reverse Polymerase Chain Reaction-Sequence Specific Oligonucleotide (PCR-SSO) hybridization using the Luminex® flow beads LabType® assay (InGen, Chilly-Mazarin, France) for the A, B and Cw loci. Allelic ambiguities were solved with PCR-Sequence Specific Primer (SSP) amplification using Olerup assays (BioNoBis, Montfort L'Amaury, France). The manufacturers' recommendations were strictly followed. Allele assignment was performed by comparison with the official nomenclature of April 2010 and validated by the WHO committee for HLA system factors [21].
Allele frequencies and linkage disequilibrium
Allele and haplotype frequencies as well as linkage disequilibrium between all pairs of HLA loci were obtained with the HLA Frequency Analysis Tool of the Los Alamos HIV Immunology Database (http://www.hiv.lanl.gov/content/immunology/tools-links.html). Frequency data for other populations were obtained from previous studies: Kinh Vietnamese [22], Chinese and European [23].
PCR amplification of HIV-1 Prot, Gag and RT, and sequencing
Viral RNA was extracted from 200 µl of plasma samples using the MagNA Pure LC Total Nucleic Acid Isolation-High Performance kit with the MagNA Pure LC system ((Roche Diagnostics, Mannheim, Germany). HIV regions were amplified by RT-PCR using the Titan One Tube kit (Roche Diagnosis, Mannheim, Germany) followed by nested PCR using the Amplitaq Gold with GeneAmp Kit (Applied Biosystem, Foster City, CA). The protease gene was amplified and sequenced according to the ANRS procedure and Gag gene as previously described [18]. The whole RT gene was obtained by using 3 sets of primers, corresponding to polymerase (outer primers: MJ3 5′-AGTAGGACCTACACCTGTCA-3′ and MJ4 5′-CTGTTAGTGCTTTGGTTCCTCT-3′; inner primers: A(35) 5′-TTGGTTGCACTTTAAATTTTCCCATTAGTCCTATT-3′ and NE1(35) 5′-CCTACTAACTTCTGTATGTCATTGACAGTCCAGCT-3′), thumb-connection (outer primers: 02TCRT5 5′-GGATGGAAAGGATCACCAGCAAT-3′ and 02TCRT3 5′-TGATTTGTTGTCTCAGTTAGGGAA-3 inner primers: 02TCnes5 5′-TGGATGGGATATGAACTCCATCCTGA-3′ and 02TCnes3 5′-TTAGCTGCCCCATCTACATCG-3′ and RNase H domains (outer primers: 02RNRT5 5′-AAATATGCAAAAAGGAGGTCTGC-3′ and 02RNRT3 5′-CAGTCTACTTGTCCATGCATGGC-3′; inner primers: 02RNnes5 5′-GAAACATGGGAAGCATGGTGGATGGAGTA-3′ and 02RNnes3 5′-TCTTCTTGGGCTTTATCTATGCCATCTA-3′. PCR products were sequenced on both strands using a CEQ DTCS Quick Start kit on an automated sequencer Beckman CEQ 2000 DNA analyzer system (Beckman Coulter, Fullerton, CA) as previously described [18]. Genotypic resistance was interpreted with the ANRS algorithm v18.
Phylogenetic analysis
The derived nucleotide sequences of RT, Prot and Gag regions were aligned by the Clustal W 1.74 alignment program with known reference strains of M, N and O pooled from the HIV Database (http://www.hiv.lanl.gov/). Phylogenetic trees were inferred by the neighbor-joining method from matrix distances calculated after gap stripping of alignments, according to a Kimura two-parameter algorithm. The circular trees were obtained using the on-line tool ITOL (http://itol.embl.de/) [24].
GenBank accession numbers for the sequences reported in this study are HQ542709 to HQ542803, HQ542613 to HQ542708 and HQ542545 to HQ542612 for RT, Prot and Gag sequences respectively.
Sequence alignment and consensus
Protein sequence analyses were performed using tools available at the Institut de Biologie et Chimie des Protéines (IBCP) Network Protein Sequence Analysis (NPSA) website (http://npsa-pbil.ibcp.fr) [25]. Multiple-sequence alignments of protein sequences obtained from patients were performed with ClustalW using default parameters. A primary consensus Mekong sequence was generated for Gag, RT and Prot and was used for alignment with consensus B and HxB2 strains. Identity was determined as a percentage of strictly conserved amino acids between the two sequences compared.
Immune recognition tools
The immune epitope database (www.immuneepitope.org) was used to predict T-cell epitope recognition. The different steps involved in the MHC class I antigen presentation pathway were evaluated by three scores: the proteasomal score reflects the efficiency of antigen-processing by the total amount of cleavage site usage releasing the peptide C-terminus; the TAP score predicts transporter associated with antigen processing (TAP) transport by estimating the binding of a peptide or its N-terminal prolonged precursors to TAP, with highest affinity for a peptide meaning the highest transport rates; the MHC score defines the epitope affinity for the MHC molecule. All scores are logarithmic values with higher values indicating higher predicted efficiency.
Statistical analysis
Comparisons between the two groups were performed with the paired t-test using GraphPad Prism version 4.00 for Macintosh (GraphPad Software, San Diego, CA, USA). A p≤0.05 was considered as statistically significant.
Results
Sequence analysis
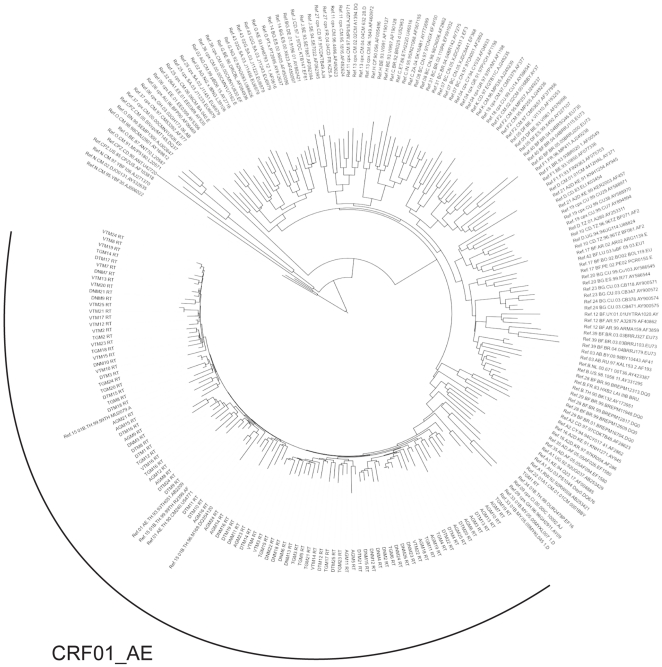
Sequence genotyping was performed for 105 samples. All RT, Prot and Gag sequences clustered with subtype CRF01_AE (depicted for RT gene in Figure 2 , data not shown for the other genes). The detailed analysis of these sequences allowed us to define (i) general polymorphisms of CRF01_AE sequences, (ii) specific polymorphisms located in intra-epitopic regions, (iii) drug resistance mutations.
Figure 2. Phylogenic analysis of RT sequences of 125 Vietnamese HIV-1 patients.
General polymorphisms
A CRF01_AE Vietnamese natural consensus including polymorphisms present in more than 50% of the sequences was established for Gag, RT and Prot proteins. The percentage of homology found between the Mekong consensus reference and the B or CRF01_AE consensus reference (as defined in Los Alamos database) is shown in Table 1 . Our Mekong consensus sequence was in agreement with the CRF01_AE consensus observed in untreated Southeast Asian patients and described in previous studies, with a percentage of homology above 98%. The percentage of homology between the Mekong and B consensus sequences was 93.2%, 92.9% and 85.1% for RT, Prot and Gag amino acid sequences, respectively (92.5%, 90.9% and 85.3% respectively with the HxB2 strain).
Table 1. Percentage of homology between CRF01_AE Vietnamese consensus sequences of RT, Prot and Gag aligned with the consensus CRF01_AE or B strain.
| Mekong RT | Mekong Prot | Mekong Gag | |
| CRF01_AE consensus | 99.1% | 99.0% | 84.6% |
| B consensus | 93.2% | 92.9% | 85.1% |
Amino Acid substitutions located in intra-epitopic regions
Using this consensus sequence, we focused our analysis on polymorphisms present in optimal CTL epitopes as defined in the Los Alamos database. Figure 3 depicts the location and HLA restriction elements of CTL epitopes on Gag and RT sequences on HxB2 aligned with the CRF01_AE Vietnamese consensus sequence. As there are only very few differences between Mekong and whole CRF01_AE consensus sequences and these differences are not relevant in terms of HLA binding predictions, the consensus amino acid of CRF01_AE was not added in the figure.
Figure 3. Comparison of Gag (A) and RT (B) Vietnamese consensus sequences with HxB2 strains.
Optimal CTL epitopes are highlighted by boxes. HLA restriction is indicated on the corresponding epitopes. Amino acid substitutions are indicated in bold. Shaded vertical bars separate blocks of 10 amino acids.
A detailed analysis of amino acid substitutions in epitopic regions was assessed for RT, Gag and Prot sequences and compared to the reference strain HxB2. A high number of amino acid polymorphisms were detected in optimal CTL epitopes as defined in the Los Alamos database. The analysis was accurately performed for 106 optimal CTL epitopes displayed by Gag, RT and Prot proteins. The proportion of divergent epitopes was 45.2% (19/42) in RT-Prot protein and 57.8% (37/64) in Gag protein. Thirteen RT epitopes presented more than 60% of identity with HxB2 epitopes, while 20 RT epitopes exhibited less identity including 13 highly divergent epitopes, i.e. epitopes with conserved divergence among all patient sequences (proportion of 60.6% divergent RT epitopes). With regard to Gag, more than 60% of identity between Mekong and HxB2 epitope sequences was observed for 20 epitopes. Among the 44 epitopes with low identity, 24 were highly divergent (corresponding to a proportion of 68.7% of the Gag epitopes).
Drug resistance mutations
According to the international list of surveillance drug resistance mutations (SDRMs) updated in 2009 [26], we identified some drug resistance mutations (DRMs) from 7 HIV-1 infected individuals. None of them had received any antiretroviral treatment. The DRMs in the RT are listed in Table 2 . These DRMs are demonstrated to be unrelated to polymorphism, and are associated with nucleoside reverse transcriptase inhibitors (NRTIs) (184V, 65R) and non-nucleoside reverse transcriptase inhibitors (NNRTIs) (98G, 103N, 106I, 181C, 181I, 188V, 190A, 230L).
Table 2. Drug resistance mutation (DRM) in RT sequences according to the international list of surveillance drug-resistance mutations (SDRMs) updated in 2009 [26].
| Sample ID | RT DRMs | NRTI and NNRTI resistance |
| DN-M3 | 230L | NVP, EFV |
| DT-M8 | 181C | NVP, EFV |
| DT-M9 | 65R, 181C | TDF, NVP, EFV |
| DT-M12 | 103N, 190A | NVP, EFV |
| DT-M15 | 184V, 181I | 3TC, NVP, EFV |
| DT-M24 | 103N | NVP, EFV |
| DT-M19 | 98G, 106I, 181C, 188V | NVP, EFV, ETV |
NVP = nevirapine, EFV = efavirenz, TDF = tenofovir, 3TC = lamivudine, ETV = etravirine.
In addition, the protease-coding region carried minor resistance mutations to protease inhibitors (PIs) (for example 36I) reflecting natural polymorphism in non-B subtypes. The major DRM 46I was observed in one patient but this mutation can be considered as a polymorphism in CRF01_AE [26].
HLA Class I typing
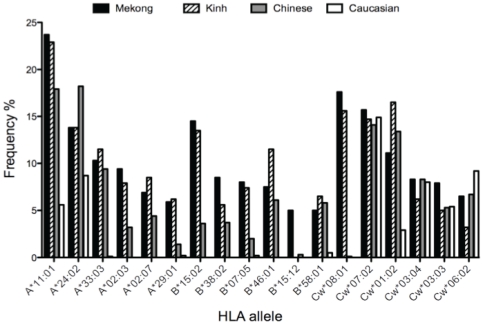
HLA class I typing was performed for 116 patients. The number of different HLA-A, HLA-B and HLA-Cw alleles detected in this study was 21, 42 and 21 respectively. The five most frequent HLA class I alleles in the Mekong population are represented in Figure 4 and are compared to three other populations: Vietnamese, Chinese and Caucasian people.
Figure 4. HLA allele frequencies in the Vietnamese Mekong Delta population (n = 116) compared to Kinh Vietnamese (dashed bars), Chinese (grey bars) and European (white bars) populations.
The 6 most representative HLA alleles of the Mekong Delta population are represented.
The most frequent HLA-A allele was A*11:01, followed by A*24:02, A*33:03 and A*02:03, with a frequency of 23.7%, 13.8%, 10.3%, and 9.4%, respectively. A*02:07 and A*29:01 were also common with a frequency of 6.9% and 5.9%, respectively. The most frequent HLA-B allele was B*15:02, followed by B*38:02, B*07:05 and B*46:01 present in 14.5%, 8.5%, 8% and 7.5% of the population, respectively. B*15:12 and B*58:01 were found at a frequency of 5%. Among HLA Cw alleles, Cw*08:01, Cw*07:02, Cw*01:02, Cw*03:04 and Cw*03:03 were predominant with a frequency of 17.6%, 15.7%, 11.1%, 8.3% and 7.9%, respectively. Moreover, we calculated the linkage disequilibrium in this population and found that most of the pairwise associations were statistically significant with a p value<0.001. The most frequent two-locus haplotypes were B*15:02-Cw*08:01 (23.3%−), B*38:02-Cw*07:02 (14.7%) and B*46:01-Cw*01:02 (12.1%). The two most common three-locus haplotypes were A*29:01-B*07:05-Cw*15:04 and A*29:01-B*07:05-Cw*15:05 with a frequency of 9.5%.
Epitope Processing and HLA-binding predictions
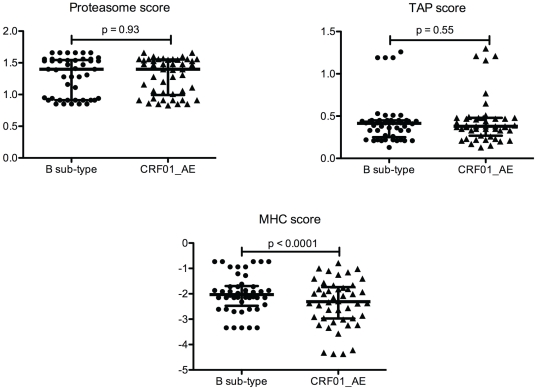
We then investigated amino acid divergences observed within CTL epitopes in the 105 Vietnamese RT-Prot and Gag sequences according to each individual's HLA restriction elements. We found 50 epitopes of interest presenting one to three divergences. The processing into the cell as well as the binding to HLA molecules of each divergent CTL epitope described in the Vietnamese sequences was predicted through the proteasome, TAP and MHC scores ( Figure 5 ). The scores for the divergent epitopes were matched and compared with those obtained with the CTL epitopes described for subtype B. The analysis was performed extensively for each HLA restriction element whenever the HLA allele was available on the immune epitope database.
Figure 5. Comparisons of proteasome, TAP and MHC scores between wild-type (B sub-type) and divergent epitopes (CRF01_AE sequences).
All scores are logarithmic values, high values corresponding to highly predicted efficiency.
Forty-seven different HLA-restricted epitopes could be analyzed ( Table 3 ). Comparisons of the three scores in each group of epitopes are presented in Figure 4 . Proteasome and TAP scores were comparable in divergent and wild-type epitopes, with a mean (± SD) of 1.29 (±0.3) in each group for the proteasome score and a mean (± SD) of 0.43 (±0.26) for wild-type epitopes vs 0.44 (±0.27) for divergent epitopes. The differences were not significant. MHC scores were significantly lower in divergent epitopes with a mean of 2.4 (±0.9) vs 2 (±0.7) in wild-type epitopes (p<0.0001). To refine the results, we analyzed the data according to the IC50 of each epitope (provided on the IEDB database) based on the rough guideline that peptides with IC50 values <50 nM are considered as having high affinity, between 50 and 500 nM as having intermediate affinity and above 500 nM as having low affinity. A strong change in ranking affinity was obtained for 15 epitopes presenting amino acid changes compared to wild-type (highlighted in grey in Table 3 ). The binding affinity evaluated by the MHC score was decreased in 11 out of the 15 epitopes with a mean of −3.18 (±0.85) for the divergent epitopes vs −2.04 (±0.57) for the wild-type epitopes (p<0.0001). In 4 out of the 15 epitopes, the MHC score was slightly increased with a mean of −1.6 (±0.15) for the divergent epitopes vs −1.57 (±0.31) for the wild-type epitopes (p not significant).
Table 3. Proteasome, TAP and MHC score of CTL epitopes.
| HLA | Protein location | Wild Type Epitope | Prot. Score | TAP Score | MHC Score | MHC IC50[nM] | Mutated Epitope | Prot. Score | TAP Score | MHC Score | MHC IC50[nM] |
| A*24:02 | p17 | KYKLKHIVW | 1,66 | 0,45 | -2,15 | 142,8 | KYHMKHIVW | 1,6 | 0,46 | −2,15 | 142 |
| KYRLKHIVW | 1,66 | 0,47 | −2,33 | 212,9 | |||||||
| KYRMKHIVW | 1,6 | 0,47 | −2,38 | 237,4 | |||||||
| KYRMKHLVW | 1,56 | 0,47 | −2,08 | 121 | |||||||
| QYKLKHIVW | 1,66 | 0,38 | −2,63 | 422,3 | |||||||
| RYRMKHLVW | 1,56 | 0,48 | −1,99 | 98,4 | |||||||
| B*35:01 | p17 | HSSQVSQNY | 1,31 | 1,26 | −1,98 | 95,2 | SSSKVSQNYb | 1,31 | 1,3 | −3,23 | 1690,9 |
| B*40:01 | p17 | IEIKDTKEAL | 1,56 | 0,44 | −2,61 | 402,8 | IDIKDTKEALb | 1,56 | 0,38 | −4,22 | 16458 |
| IDVKDTKEALb | 1,56 | 0,38 | −4,32 | 20736 | |||||||
| IEVKDTKEALb | 1,56 | 0,44 | −2,91 | 819 | |||||||
| IQVKDTKEALb | 1,56 | 0,49 | −4,37 | 23631,4 | |||||||
| A*02:01 | p17 | SLYNTVATL | 1,54 | 0,51 | −1,72 | 53 | SLFNLVATLa | 1,55 | 0,48 | −1,4 | 25,1 |
| SLFNTIATLa | 1,55 | 0,48 | −1,57 | 37 | |||||||
| SLFNTVATLa | 1,54 | 0,48 | −1,66 | 45,9 | |||||||
| A*26:01 | p24 | EVIPMFSAL | 1,41 | 0,39 | −1,21 | 16,3 | EVIPMFTALb | 1,48 | 0,39 | −1,93 | 85,5 |
| B*07:02 | p24 | TPQDLNTML | 1,58 | 0,27 | −2,72 | 526,8 | TPQDLNMML | 1,57 | 0,27 | −2,96 | 920,8 |
| B*53:01 | p24 | QASQEVKNW | 1,47 | 0,36 | −2,32 | 209,7 | QATQEVKNW | 1,47 | 0,35 | −2,29 | 193,1 |
| B*57:01 | p24 | QASQEVKNW | 1,47 | 0,36 | −2,43 | 270,5 | QATQEVKNW | 1,47 | 0,35 | −2,56 | 360,5 |
| B*57:01 | p24 | TSTLQEQIGW | 1,4 | 0,38 | −1,94 | 87,2 | TSNLQEQIGWb | 1,4 | 0,35 | −3,24 | 1719,2 |
| B*58:01 | p24 | TSTLQEQIGW | 1,4 | 0,38 | −1,62 | 41,4 | TSNLQEQIGW | 1,4 | 0,35 | −2,01 | 102,3 |
| A*02:01 | RT | ALVEICTEM | 0,94 | 0,22 | −2,14 | 138,1 | ALTEICKEMb | 1,06 | 0,17 | −2,96 | 907,6 |
| ALIEICTEM | 0,94 | 0,22 | −1,81 | 64 | |||||||
| A*02:01 | RT | VIYQYMDDL | 1,16 | 0,53 | −2,7 | 500,1 | IIYQYMDDL | 1,16 | 0,51 | −2,64 | 439 |
| A*02:01 | RT | ILKEPVHGV | 0,95 | 0,13 | −2,12 | 132 | ILKTPVHGV | 1,03 | 0,13 | −2,45 | 281 |
| A*03:01 | RT | AIFQSSMTK | 0,91 | 0,33 | −1,28 | 18,9 | AIFQCSMTK | 0,91 | 0,33 | −1,4 | 25,2 |
| A*11:01 | RT | AIFQSSMTK | 0,91 | 0,33 | −0,94 | 8,6 | AIFQCSMIK | 0,83 | 0,33 | −1,15 | 14,1 |
| AIFQCSMTK | 0,91 | 0,33 | −1,1 | 12,5 | |||||||
| AIFQSSMTR b | 1,06 | 0,77 | −1,89 | 77,1 | |||||||
| A*11:01 | RT | IYQEPFKNLK | 0,85 | 0,21 | −3,34 | 2179,4 | IYQEPFRNLK | 0,86 | 0,21 | −3,23 | 1699,2 |
| IYQEPFKNLR | 1,01 | 0,65 | −4,37 | 23411 | |||||||
| INQEPFKNLK | 0,85 | 0,15 | −3,57 | 3751,1 | |||||||
| IFQEPFKNLK | 0,85 | 0,2 | −3,12 | 1303,6 | |||||||
| IYQETFKNLK | 0,85 | 0,21 | −3,34 | 2209,6 | |||||||
| A*11:01 | RT | QIIEQLIKK | 0,91 | 0,22 | −1,91 | 81,2 | QIIEELIKK | 0,91 | 0,22 | −2,01 | 102,6 |
| QIVEELIKK | 0,91 | 0,23 | −2,37 | 234,4 | |||||||
| QVIEELIKK | 0,91 | 0,23 | −2,37 | 234,4 | |||||||
| A*26:01 | RT | ETKLGKAGY | 1,28 | 1,19 | −1,87 | 73,6 | ENKQGKAGYb | 1,2 | 1,16 | −3,01 | 1023,6 |
| ETRLGKAGY | 1,28 | 1,21 | −1,82 | 66,2 | |||||||
| ETRLGKAGY | 1,28 | 1,21 | −1,82 | 66,2 | |||||||
| B*08:01 | RT | GPKVKQWPL | 1,11 | 0,26 | −2,06 | 115 | GPKVRQWPLa | 1,11 | 0,26 | −1,76 | 57,1 |
| B*57:01 | RT | IVLPEKDSW | 1,39 | 0,41 | −2 | 100,7 | IELPEKDSWb | 1,39 | 0,39 | −2,86 | 718,8 |
| B*58:01 | RT | IAMESIVIW | 1,53 | 0,42 | −0,73 | 5,4 | IATESIIIW | 1,48 | 0,37 | −1,02 | 10,5 |
| IATESIVIW | 1,53 | 0,37 | −1 | 10 | |||||||
| IVTESIVIW | 1,53 | 0,38 | −1,67 | 46,4 | |||||||
| IAIESIIIW | 1,48 | 0,42 | −0,79 | 6,2 | |||||||
| IATEGIIIW | 1,48 | 0,37 | −1,25 | 17,6 |
Wild type epitope scores are matched with the corresponding divergent epitope score (Prot. score = proteasome score).
Epitopes for which amino acid divergence induces improved MHC binding.
Epitopes for which amino acid divergence decreases MHC binding.
Discussion
HIV-1 diversity is modulated at the population level by host immune pressure, which induces a high rate of divergence in immunodominant epitopes recognized by the prevalent HLA class I alleles in these populations. CTL escape mutations occur at critical sites within HLA-restricted CTL epitopes. Indeed, an amino acid substitution may abrogate epitope-HLA binding, reduce T-cell receptor recognition, impair antigen processing or generate antagonistic CTL responses [27]. These mutations result in CTL immune escape but could lead to a severe fitness cost to the virus [28]. Thus, determining the exact kinetics and dynamics of the duel between host and virus is always a challenge. When HIV is transmitted from person to person, mutational escape and reversion rapidly shape HIV evolution. The present global HIV-1 diversity is the result of cumulative infections followed by intra-host viral evolution. The gold standard to analyze it is to provide a comprehensive immunologic and virologic analysis in the context of an acute infection [29]. Due to the difficulty of identifying very early cases of HIV transmission, this type of study is rare.
The present study focused on MHC class I antigen presentation of optimal CTL HIV-1 epitopes across a chronically infected HLA-diverse host population. The patients were HIV-1 infected, all but 3 were ART-naïve, and they all lived in five provincial cities in the Mekong Delta, Vietnam. As expected, our phylogenetic analyses confirmed that HIV-1 CRF01_AE is the unique strain circulating in Southern Vietnam. In addition, DRMs could be detected even in a naive population. The analysis of DRMs on RT-Prot sequences showed 7 ARV-resistant mutants (mostly to NNRTI) among 122 ART-naive patients (6.7%). These results are consistent with a study performed in Northern Vietnam in 2008 showing a prevalence of 4.4% detected for RTI mutations in contrast with a prevalence of 1.7% for PI. This percentage may be explained by the still limited use of PI in Vietnam [19]. We had no information regarding NNRTI use by the corresponding patients for prevention of mother-to-child transmission, which raises the hypothesis that these mutations are SDRMs transmitted from treated to untreated individuals. Since the prevalence is above 5% corresponding to the threshold of surveillance for WHO, our results should be an argument for initiating longitudinal surveys of resistance in the southern part of the country.
The predominance of the CRF01_AE strain is not surprising. Indeed, since the first case of HIV-1 infection detected in Ho Chi Minh City in 1990, the CRF01_AE strain is still largely predominant in Vietnam. Our study is the first to provide data on HIV-1 subtypes in the Mekong area where we identified only the CRF01_AE circulating form. However, there have been a few other studies describing HIV-1 genotypes in other regions of Vietnam such as Ho Chi Minh city, Hanoi and Hai Phong among similar cohorts. In all of them, CRF01_AE predominates although a few other subtypes have been identified such as B' [20] and some recombinants as CRF01_AE/B' [30], CRF02_AG/D [30] and CRF01_AE/C [31].
This wide predominance of CRF01_AE despite individual genetic variability and continuous human influx and efflux remains to be understood. One possible explanation could be a more efficient transmission of the virus. Interestingly, a study in a longitudinal cohort of injection-drug users in Thailand conducted from 1995 through 1998 found an increased probability of transmission of CRF01_AE as compared with subtype B, though it was unclear whether epidemiologic, virologic or host factors were affecting viral spread [32]. In this study, our goal was to evaluate the importance of host factors in the predominance of this viral strain.
The genetic factors of the Vietnamese population living around the Delta Mekong were analyzed according to the distribution of HLA-A, B, and Cw alleles and haplotypes by high-resolution DNA typing. Although there have been some studies on HLA typing in the Vietnamese population, there is only one comprehensive four-digit typing report of HLA class I alleles harbored by the Kinh population in Vietnam, the most prevalent ethnic group in the country [22]. Our results are consistent with the latter study. We clearly show that HLA A*11:01 and B*46:01, which are depicted as Asian alleles, are widely present in our cohort with a high predominance of HLA A*11:01 among HLA-A alleles, whereas the prevalence of both alleles is about 5.6% and 0% in European people, respectively [23]. Other alleles such as A*29:01, B*07:05 and B*15:12 appear to be more specific to the Vietnamese population with a frequency above 5% compared to a frequency below 0.5% in Chinese and European populations [23]. The two major three-digit haplotypes are A*29:01-B*07:05-Cw*15:04 (9,5%) and A*29:01-B*07:05-Cw*15:05 (9,5%), which are considered by Hoa and colleagues as a Kinh signature [22].
Interestingly, even if our population is very similar to the Kinh one, it differs with regard to one specific allele, HLA B*15:12. This allele is absent in the Kinh population and other Vietnamese ethnic populations like the Uyghur [33], Jinua and Wa [34]. It has been described as having a very low frequency (lower than 2.5%) in other Asian populations such as the Chinese, Yi and Hani populations [35]. As this is the first paper reporting such a prevalence for this allele in a defined area, it might be considered as a genetic signature of this population living in the Mekong Delta of Vietnam.
This remarkable diversity and specificity of the HLA class I molecules clearly shape immune responses individually, as the HLA-class I molecules play an important role in CD8+ T-cell recognition. However, the relative impact of these forces on the evolution of HIV at the population level is difficult to evaluate. It has been suggested that virus-clade-specific differences could result, at least in part, from the impact of HLA differences between populations living in distinct regions of the world. Indeed, escape mutations within CD8+ T-cell epitopes can be transmitted relatively frequently and persist in HIV-mismatched recipients, thereby accumulating over time to ultimately represent the most prevalent form of the virus [14], [36], [37], [38].
In our study, we aimed to analyze the impact of divergences within CTL epitopes located in Gag and RT-Prot among Vietnamese HIV-1 strains on HLA class I binding recognition. Results were compared to HLA class I binding recognition of wild-type CTL epitopes from the reference HxB2. Our analysis shows that there is a high degree of polymorphism of CRF01_AE strains compared to the HxB2 reference strain or subtype B strain. Divergences within CRF01_AE CTL epitopes did not modify antigen processing as evaluated by TAP and proteasome scores. However, MHC binding was drastically reduced in two thirds of the divergent epitopes identified, and when the amino acid divergence increased it, the rise was modest. So far, viral escape via mutations within CTL epitopes has been well documented. In most cases, the mutation occurred within the epitope, leading either to reduced binding to the MHC class I molecule and/or an alteration of T-cell receptor recognition. A few mutations affecting antigen processing have also been reported in portable flanking sequences [39].
In our study, all predictions of HLA class I presentation were conducted using T-cell predictions from the Immune Epitope DataBase, but the impact of the divergences we identified on the CTL response could not be assessed, as patients' peripheral blood lymphocytes were not available for such in vitro experiments [39]. Therefore, even if these predictions were made using experimental data, they should be interpreted with extreme caution. Proteasome score is the logarithm of the total amount of cleavage site usage releasing the peptide C-terminus. It does not take into account other factors such as the amount of source protein degraded, the type of cell involved in the protein degradation [40] or the amount of immunoproteasome. Moreover, as this software was set up with a limited number of HLA alleles A and B, we could not analyze several alleles that were common in our Vietnamese population, such as: A*33:03, A*02:07, A*29:01, B*38:02, B*07:05 and B*15:12. No data were available for HLA-Cw alleles.
Furthermore, in the absence of in vitro analysis, notably ELISpot, compensatory and new CTL responses could not be detected [41] nor could cross-reactive immune responses, as has been shown with subtype A, CRF01_AE and CRF02_AG [42]. Finally, we cannot rule out that the divergences observed in epitopes are chance occurrences, are compensatory, or that they could be due to co-variance and not only result from immune pressure.
However, despite these limitations and in the context of a chronic infection, we demonstrate that in a homogeneous HLA diverse population from the Mekong Delta, Vietnam, MHC binding of known CTL epitopes seems to be strongly reduced due to intraepitopic mutations, thereby facilitating immune evasion of Vietnamese HIV-1 strains. This weak immune pressure combined with a low genetic diversity could explain why CRF01_AE has emerged and spread so rapidly in Southern Asia since 1990.
This finding emphasizes the importance of the immune system in shaping HIV-1 evolution in vivo at a population level, as already demonstrated in previous papers. Moore et al. elegantly showed that particular host HLA class I alleles are clearly associated with polymorphisms in HIV-1 at sites of least functional or structural constraint, whereas absence of polymorphism is also HLA allele–specific [43]. Other studies suggest that escape mutations within CD8+ T-cell epitopes are frequently transmitted and persist in HIV-mismatched recipients, thereby accumulating over time to ultimately represent the most prevalent form of the virus [37]. More recently, Kawashima et al. demonstrated that the frequency of the mutations present in epitopes associated with viral control is strongly correlated with the prevalence of the restricting HLA allele in 9 distinct extensive cohorts [44]. All these data confirm that HIV can adapt to HLA control in order to evade CD8+ T-cell responses and highlight the challenge to find a vaccine to keep pace with the evolving virus.
Acknowledgments
We thank the health authorities of the Socialist Republic of Vietnam and professor F Barré Sinoussi who is the head of the South Asian site of ANRS (for the northern part). The authors are indebted to the technicians of the HLA laboratory for their contribution to this work and to Ray Cooke for linguistic assistance. HF dedicates this work to the memory of Dr Jean Gérard Guillet.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by Agence Nationale de Recherches Sur le sida et les hepatites virales (ANRS) (ANRS 12159). PB was supported by a fellowship from the European Community's Seventh Framework Program (FP7/2007–2013) under the “Collaborative HIV and Anti-HIV Drug Resistance Network” project (grant no. 223131). The funders had no role in study design, data collection, decision to publish or preparation of the manuscript.
References
- 1.Kilmarx PH. Global epidemiology of HIV. Curr Opin HIV AIDS. 2009;4:240–246. doi: 10.1097/COH.0b013e32832c06db. [DOI] [PubMed] [Google Scholar]
- 2.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, et al. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 4.Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tishkoff SA, Kidd KK. Implications of biogeography of human populations for ‘race’ and medicine. Nat Genet. 2004;36:S21–27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- 6.Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, et al. Mapping human genetic diversity in Asia. Science. 2009;326:1541–1545. doi: 10.1126/science.1177074. [DOI] [PubMed] [Google Scholar]
- 7.Bangham CR. CTL quality and the control of human retroviral infections. Eur J Immunol. 2009;39:1700–1712. doi: 10.1002/eji.200939451. [DOI] [PubMed] [Google Scholar]
- 8.Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS. 2003;17:2581–2591. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 9.Bailey JR, Williams TM, Siliciano RF, Blankson JN. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med. 2006;203:1357–1369. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambotte O, Boufassa F, Madec Y, Nguyen A, Goujard C, et al. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis. 2005;41:1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 12.Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 13.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 14.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, et al. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One. 2009;4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright JK, Brumme ZL, Carlson JM, Heckerman D, Kadie CM, et al. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J Virol. 2010;84:10820–10831. doi: 10.1128/JVI.01084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolland M, Carlson JM, Manocheewa S, Swain JV, Lanxon-Cookson E, et al. Amino-acid co-variation in HIV-1 Gag subtype C: HLA-mediated selection pressure and compensatory dynamics. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan NT, Recordon-Pinson P, Hung PV, Uyen NT, Lien TT, et al. HIV type 1 isolates from 200 untreated individuals in Ho Chi Minh City (Vietnam): ANRS 1257 Study. Large predominance of CRF01_AE and presence of major resistance mutations to antiretroviral drugs. AIDS Res Hum Retroviruses. 2003;19:925–928. doi: 10.1089/088922203322493111. [DOI] [PubMed] [Google Scholar]
- 18.Lazaro E, Theodorou I, Legrand E, Recordon-Pinson P, Boucher S, et al. Sequences of clustered epitopes in Gag and Nef potentially presented by predominant class I human leukocyte antigen (HLA) alleles A and B expressed by human immunodeficiency virus type 1 (HIV-1)-infected patients in Vietnam. AIDS Res Hum Retroviruses. 2005;21:586–591. doi: 10.1089/aid.2005.21.586. [DOI] [PubMed] [Google Scholar]
- 19.Phan TT, Ishizaki A, Phung DC, Bi X, Oka S, et al. Characterization of HIV type 1 genotypes and drug resistance mutations among drug-naive HIV type 1-infected patients in Northern Vietnam. AIDS Res Hum Retroviruses. 2010;26:233–235. doi: 10.1089/aid.2009.0206. [DOI] [PubMed] [Google Scholar]
- 20.Ishizaki A, Cuong NH, Thuc PV, Trung NV, Saijoh K, et al. Profile of HIV type 1 infection and genotypic resistance mutations to antiretroviral drugs in treatment-naive HIV type 1-infected individuals in Hai Phong, Viet Nam. AIDS Res Hum Retroviruses. 2009;25:175–182. doi: 10.1089/aid.2008.0193. [DOI] [PubMed] [Google Scholar]
- 21.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoa BK, Hang NT, Kashiwase K, Ohashi J, Lien LT, et al. HLA-A, -B, -C, -DRB1 and -DQB1 alleles and haplotypes in the Kinh population in Vietnam. Tissue Antigens. 2008;71:127–134. doi: 10.1111/j.1399-0039.2007.00982.x. [DOI] [PubMed] [Google Scholar]
- 23.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68:779–788. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 25.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 26.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature. 2001;410:980–987. doi: 10.1038/35073658. [DOI] [PubMed] [Google Scholar]
- 28.Kent SJ, Fernandez CS, Dale CJ, Davenport MP. Reversion of immune escape HIV variants upon transmission: insights into effective viral immunity. Trends Microbiol. 2005;13:243–246. doi: 10.1016/j.tim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 29.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caumont A, Lan NT, Uyen NT, Hung PV, Schvoerer E, et al. Sequence analysis of env C2/V3, gag p17/p24, and pol protease regions of 25 HIV type 1 isolates from Ho Chi Minh City, Vietnam. AIDS Res Hum Retroviruses. 2001;17:1285–1291. doi: 10.1089/088922201750461357. [DOI] [PubMed] [Google Scholar]
- 31.Tran TT, Maljkovic I, Swartling S, Phung DC, Chiodi F, et al. HIV-1 CRF01_AE in intravenous drug users in Hanoi, Vietnam. AIDS Res Hum Retroviruses. 2004;20:341–345. doi: 10.1089/088922204322996581. [DOI] [PubMed] [Google Scholar]
- 32.Hudgens MG, Longini IM, Jr, Vanichseni S, Hu DJ, Kitayaporn D, et al. Subtype-specific transmission probabilities for human immunodeficiency virus type 1 among injecting drug users in Bangkok, Thailand. Am J Epidemiol. 2002;155:159–168. doi: 10.1093/aje/155.2.159. [DOI] [PubMed] [Google Scholar]
- 33.Shen CM, Zhu BF, Deng YJ, Ye SH, Yan JW, et al. Allele polymorphism and haplotype diversity of HLA-A, -B and -DRB1 loci in sequence-based typing for Chinese Uyghur ethnic group. PLoS One. 2010;5:e13458. doi: 10.1371/journal.pone.0013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, Ogata S, Yu JK, Ohashi J, Yu L, et al. Distribution of HLA alleles and haplotypes in Jinuo and Wa populations in Southwest China. Hum Immunol. 2008;69:58–65. doi: 10.1016/j.humimm.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Shi L, Yao YF, Matsushita M, Yu L, Huang XQ, et al. Genetic link among Hani, Bulang and other Southeast Asian populations: evidence from HLA -A, -B, -C, -DRB1 genes and haplotypes distribution. Int J Immunogenet. 2010;37:467–475. doi: 10.1111/j.1744-313X.2010.00949.x. [DOI] [PubMed] [Google Scholar]
- 36.Draenert R, Le Gall S, Pfafferott KJ, Leslie AJ, Chetty P, et al. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, et al. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brumme ZL, Walker BD. Tracking the culprit: HIV-1 evolution and immune selection revealed by single-genome amplification. J Exp Med. 2009;206:1215–1218. doi: 10.1084/jem.20091094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. J Clin Invest. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazaro E, Godfrey SB, Stamegna P, Ogbechie T, Kerrigan C, et al. Differential HIV epitope processing in monocytes and CD4 T cells affects cytotoxic T lymphocyte recognition. J Infect Dis. 2009;200:236–243. doi: 10.1086/599837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maisnier-Patin S, Andersson DI. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res Microbiol. 2004;155:360–369. doi: 10.1016/j.resmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 42.Aidoo M, Sawadogo S, Bile EC, Yang C, Nkengasong JN, et al. Viral, HLA and T cell elements in cross-reactive immune responses to HIV-1 subtype A, CRF01_AE and CRF02_AG vaccine sequence in Ivorian blood donors. Vaccine. 2008;26:4830–4839. doi: 10.1016/j.vaccine.2008.06.097. [DOI] [PubMed] [Google Scholar]
- 43.Moore CB, John M, James IR, Christiansen FT, Witt CS, et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 44.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]