Abstract
RMRP is a non-coding RNA that is ubiquitously expressed in both humans and mice. RMRP mutations that lead to decreased RMRP levels are found in the pleiotropic syndrome Cartilage Hair Hypoplasia. To assess the effects of deleting RMRP, we engineered a targeting vector that contains loxP sequences flanking RMRP and created hemizygous mice harboring this engineered allele (RMRP conditional). We found that insertion of this cassette suppressed RMRP expression, and we failed to obtain viable mice homozygous for the RMRP conditional allele. Furthermore, we were unable to obtain viable homozygous RMRP null mice, indicating that RMRP is essential for early embryonic development.
Introduction
RMRP is a non-coding RNA that is highly expressed in a wide range of human and murine tissues [1]. Mutations in RMRP have been detected in individuals afflicted with Cartilage Hair Hypoplasia (CHH) [2], a syndrome characterized by short stature, sparse hair, immunodeficiency and in a subset of patients severe combined immunodeficiency or life threatening anemia. Cell-based reporter assays have shown that RMRP mutations result in decreased RMRP stability, which may account for the severe phenotypes seen in CHH [3]. Individuals that carry only a single RMRP mutation do not exhibit phenotypes associated with CHH [2]; however, affected individuals harbor mutations in both RMRP alleles [4] suggesting that these mutations inactivate the RMRP gene product.
The biological function(s) of RMRP remain incompletely understood. Biochemical studies have demonstrated that RMRP RNA binds to the mitochondrial posttranscriptional modification complex RNase MRP [5]. However, no apparent mitochondrial defects have been found in CHH patients. In addition, RMRP is also found in the nucleolus. We recently reported that together with the catalytic subunit of telomerase (hTERT), RMRP forms an RNA dependent RNA polymerase that converts single stranded RMRP RNA into double stranded RMRP [6].
To gain further insight into the biological functions of RMRP, we generated a genetically engineered mouse that lacks RMRP.
Results and Discussion
We created a targeting vector specific for murine RMRP using the pEasyflox backbone [7]. The targeting vector contains the RMRP gene and promoter (800 bp up stream of murine RMRP [1]) flanked by two loxP sequences. A neomycin selectable marker flanked by two loxP sequences was placed upstream of RMRP (Figure 1A).
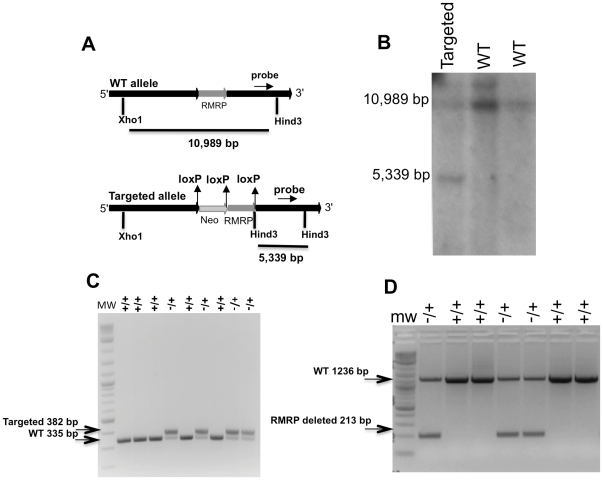
Figure 1. Targeting of murine RMRP.
A. Murine targeting vector (MTV) B. Southern blot of ES cells following selection for alleles with integrated MTV (the southern probe is shown in figure 1a) C. PCR analysis of RC (RMRP conditional) pups D. PCR analysis of pups derived from the interbreeding of RC mice and mice expressing CMV-Cre.
This targeting vector was introduced into mouse embryonic cells and individual clones containing the integrated targeting vector were selected by treatment with G418. Using southern blot analysis with a probe that can detect both the WT and targeted alleles, we found that 10% of the clones had integrated the RMRP targeting vector into the endogenous RMRP locus (Figure 1B). One of these clones was injected into female donor blastocysts producing 10 pups, 6 of which were chimeric, based on coat color. The chimeric mice were bred to FVB/N mice and the resulting pups were genotyped using a PCR based assay (Figure 1C). These mice contain the RMRP gene flanked by two loxP sequences and an insert coding for neomycin resistance upstream (RMRP conditional, RC) (Figure 1A).
We failed to obtain homozygous RC mice by crossing the hemizygous RC mice. Despite multiple attempts, we were unable to separate embryos earlier then E6.5 from the placenta. The RC mice harbor the neomycin resistance gene upstream of the RMRP gene, suggesting that insertion of DNA elements upstream of RMRP results in early embryonic lethality (Table 1). Thus, we hypothesized that the neomycin insertion impairs critical genomic elements that are essential for RMRP expression. Since prior work has confirmed that a subset of CHH patients harbor mutations in the RMRP promoter and these mutations decrease RMRP expression (1, 2), these observations suggest that the RMRP promoter is particularly sensitive to nucleotide changes.
Table 1. RMRP depletion is embryonic lethal.
| Mating | N (male/female) | +/+ | +/− | −/− | |
| RMRP +/−×RMRP +/− | 45 (20/25) | Expected | 11.25 | 22.5 | 11.25 |
| Observed | 19 | 26 | 0 | ||
| RC+/−×RC+/− | 47 (23/24) | Expected | 11.75 | 23.5 | 11.75 |
| Observed | 15 | 31 | 0 |
To confirm these findings, we tested whether complete deletion of RMRP would lead to a different phenotype. To this end, RC hemizygous mice were crossed to a mouse that ubiquitously and constitutively expresses the Cre recombinase (CMV-Cre). Using PCR with primers that are specific for the predicted engineered RMRP allele after recombination, we confirmed that the RMRP was deleted in the offspring of the hemizygous mice (Figure 1D). Similar to what we observed in RC mice, we failed to obtain pups harboring homozygous deletion of RMRP (Table 1). These observations suggest that that insertion of exogenous DNA sequences upstream of RMRP results in aberrant RMRP expression and results in embryonic lethality.
The levels of RMRP may be critical for RMRP function. Specifically, Nakashima et al. have proposed a model by which RMRP mutations found in CHH patients leads to destabilization of RMRP [3]. When we assessed total levels of RMRP in murine embryonic fibroblasts (MEFs) obtained from RMRP or RC hemizygous mice, we found that RMRP was expressed at 50% of the level found in wild type MEFs (Figure 2A). RC and RMRP +/− mice were monitored from birth to 18 months of age and no abnormality in size, fur or behavior was detected. This is in consonance with what has been observed in human carriers of RMRP mutations [2].
Figure 2. RMRP depletion leads to reduced levels of RMRP transcript.
Total RNA was produced from E13.5 MEFs and RMRP level was measured by A. qRT-PCR B. Northern blot using either a sense or antisense RMRP probe. Error bars represent SD of three replicas.
We previously found that two species of RMRP are present in human cells: single stranded RMRP RNA and a double stranded RMRP RNA composed of a single RNA containing both the sense and antisense strands [6]. The double stranded version of RMRP requires the presence of the catalytic subunit of telomerase, TERT. Using Northern blot analysis with probes designed to detect sense or antisense RMRP, we detected decreased levels of both species of RMRP in total RNA extracted from RMRP +/− E13.5 MEFs as compared to WT MEFs (Figure 2B). The RMRP antisense probe detects both single and double stranded RMRP and the RMRP sense probe detects only double stranded RMRP. These observations demonstrate that reduction of RMRP reduces the function of the TERT-RMRP RdRP.
To assess the embryonic stage that requires RMRP we isolated embryos from breeding of RMRP +/− mice at several early time points. We failed to identify homozygous embryos as early as E6.5 (Table 2) indicating, that RMRP is required early in embryonic development.
Table 2. RMRP depletion is lethal in early stage embryos.
| N | +/+ | +/− | −/− | ||
| E13.5 | 27 | Expected | 6.75 | 13.5 | 6.75 |
| Observed | 11 | 16 | 0 | ||
| E10.5 | 8 | Expected | 2 | 4 | 2 |
| Observed | 1 | 7 | 0 | ||
| E6.5 | 20 | Expected | 5 | 10 | 5 |
| Observed | 4 | 16 | 0 |
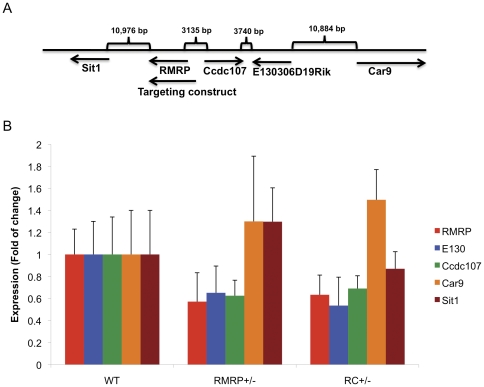
The RMRP gene is located in close proximity to several other genes including Sit1, Ccdc107, E130 and Car9 (Figure 3A). To assess whether the expression of these genes are affected by RMRP depletion, we measured the expression of these genes in E13.5 MEFs derived from WT, RMRP +/− or RC+/− mice. We confirmed that RMRP levels were decreased by 50% in both RMRP +/− and RC+/− MEFs. We also found that the expression of Ccdc107 and E130 were also decreased in RMRP +/− and RC+/− MEFs while the expression of Sit1 and Car9 was not affected (Figure 3B). Although the targeting of RMRP involved the insertion of a small number of nucleotides, RMRP, Ccdc107 and E130 are located closely together, and deletion of RMRP affects the expression of all of these genes.
Figure 3. The effect of RMRP depletion on neighboring genes.
A. Map of the genomic structure and genes surrounding RMRP. B. Expression of RMRP, Sit1, Ccdc107, E130 and Car9 in E13.5 MEFs from the indicated genotypes. Error bars represent SD of three replicas.
Based on these observations, we tested whether Ccdc107 or E130 are essential for survival and thus may also contribute to the observed embryonic lethal phenotype. We used MEFs from E13.5 WT or RMRP +/− mice. Using RNAi, we reduced the expression of these genes to 5–20% of levels found in cells transfected with a control siRNA (Figure 4A and B). Seven days post transfection the cells were tested for viability, by monitoring the ATP content of the cells (Figure 4C). We failed to observe any alteration in cell proliferation/viability after suppression of either Ccdc107 or E130 indicating that these genes are not essential. However, we cannot eliminate the possibility that the residual low levels of Ccdc107 or E130 (5–20% of control) are enough to sustain viability and when completely deleted will lead to embryonic lethality. Due to the very close proximity between these genes it is very difficult to target RMRP without disrupting Ccdc107 and E130 expression and further attempts to target RMRP should take this into consideration.
Figure 4. Genes near RMRP are not essential for cellular viability.
MEFs from E13.5 mice of either A. WT or B. RMRP+/− were transfected with siRNAs targeting Ccdc107 or E130. Three days later RNA was extracted from the cells and qRT-PCR was preformed using primers for RMRP, Ccdc107 or E130. C. The same cells as in A and B were plated (5000 cells/well) in a 96 well plate and 7 days post transfection cell number was assessed by Cell titer glow. Error bars represent SD of three replicas.
RMRP is a ubiquitously expressed non-coding RNA [1] that has critical functions both in mice and humans. We found that RMRP is essential for development at early stages of embryogenesis. We further demonstrated that insertion of DNA elements upstream of the RMRP promoter cause a decrease in RMRP expression in hemizygous mice and are lethal in homozygous mice. These observations suggest that expression of RMRP is tightly regulated and essential for early developmental processes. We conclude that future attempts to target RMRP must consider the tight regulation and early requirement of RMRP.
Materials and Methods
All laboratory animals will be cared for in the animal quarters of the Dana-Farber Cancer Institute under the direct supervision of the Dana-Farber Cancer Institute Animal Care and Use Committee (ACUC) under assurance number A3023-01. The Dana-Farber Cancer Institute is an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) accredited institution meeting or exceeding all standards for animal care and use. This work presented herein is has been approved by the ACUC under animal protocol 10-004.
Construction of RMRP mouse targeting vector
The pEasyflox vector [7] was used as a backbone to create the murine targeting vector. A 1097 bp fragment corresponding to RMRP and 800 bp of the upstream promoter sequence was PCR amplified from RP23-207P5 (http://bacpac.chori.org) using Xba1-RMRP and RMRP-Sal1 primers (Table 3). The PCR product was digested with Xba1 and Sal1 and ligated to the same sites in the pEasyflox to create pEasyflox-RMRP. Next a 4064 bp fragment upstream of the RMRP gene was PCR amplified from RP23-207P5 using F5′Cla1 and R5′Not1 primers (Table 3). Following digestion with Not1 and Cla1 the PCR product was ligated to the same sites in pEasyflox-RMRP to create pEasyflox-5′-RMRP. The downstream sequence of the targeting vector was amplified from RP23-207P5 using F3′Hind3 and R3′Xho1 primers (Table 3) and the 4029 bp fragment was digested with Hind3 and Xho1 and ligated into the same sites in pEasyflox-5′-RMRP to create the RMRP mouse targeting vector. The sequence of the vector was confirmed by direct sequencing.
Table 3. primers used in this study.
| Name | Seq (5→3) |
| Xba1-RMRP | ATATTCTAGATCCATGGGTGTTTTGTTCCCAAATC |
| RMRP-Sal1 | ATATGTCGACGCAGCTCGCTCTGAAGGCCTGT |
| F5′Cla1 | ATATATCGATGGATCCTGAAATTTGAAGGCAAATGGCAAATGG |
| R5′Not1 | ATATGCGGCCGCAGGAGTTACAGAGACAAAGTTTGGAG |
| F3′Hind3 | ATATAAGCTTCTTAGCTTCTAGGCGCGACTAATTT |
| R3′Xho1 | ATATCTCGAGGGATCCGGGCCCAAAATATACTTGAAGTAGTT |
| FRMRP | TGCTGAAGGCCTGTATCCT |
| RRMRP | TGAGAATGAGCCCCGTGT |
| FRCgeno | TGAGCCCCGTGTGGTTGGTGC |
| RRCgeno | AGACCAATTTTCTCACCATAACCAAA |
| FRMRPgeno | TTGCTAGTGTATGCAATGGTGTCAG |
| RRMRPgeno | TTGTAGAGTCATAAATTAGTCGCGC |
| FSouthern | GATTTCCCATCTACTACTTACACTGA |
| RSouthern | CTGTCTCCCATGGAATGTACAGTGGCC |
| FCcdc107 | GGCACACCCAGAACGGGGCTC |
| RCcdc107 | CGCAGTCAGAGCAACAGCTGGT |
| FE130 | CTGGTGGCCACGTACTTCCTGC |
| RE130 | TACCCTTCAGGCGCAGCTGAAG |
| FSit1 | TACAACTGTACCACTGACGATCA |
| RSit1 | ATCCCCTCCATAAGCCACG |
| FCar9 | CCGGAACTGAGCCTATCCAAC |
| RCar9 | CAGGAATACCGGGCCTTGC |
Northern blot
RNA was extracted from MEFs using an RNeasy kit (Qiagene). 10 µl of sample buffer (95% formamide 5% bromophenolblue) was added to 5 µg of total RNA in a total volume of 20 µl. The samples were applied to a 6% TBE-urea acrylamide gel (invitrogen cat number: EC6865BOX) and then blotted onto a Hybond-N+membranes (GE Healthcare). Following cross-linking with a UV crosslinker the membrane was pre-incubated for 1 hour at 60°C with hybridization buffer (0.5 M NaHPO4 PH = 7.2, 1 mM EDTA, 7% SDS). A radiolabeled RNA probe corresponding to the full length sense or antisense murine RMRP was added and incubated with the membrane overnight at 60°C. Following 4 washes with 2×SSC the gel was exposed and visualized.
Southern blot
DNA was prepared from ES cells targeted with the murine RMRP targeting vector using the Qiaamp kit (Qiagene). 10 µg of DNA was digested with Hind3, loaded onto a 1% agarose fomaldhyde gel and blotted onto a Hybond-N+membranes (GE Healthcare). Following UV crosslinking the membrane was pre-incubated for 1 hour at 65°C with hybridization buffer (0.5 M NaHPO4 PH = 7.2, 1 mM EDTA, 7% SDS). Next the probe was added to the blot and allowed to incubate overnight at 65°C. Following 3 washes with 1×SSC the membrane was exposed. The probe used for detection of targeted cells was generated by PCR amplification from a BAC clone (RP23-207P5 (http://bacpac.chori.org)) using FSouthern and RSouthern primers (Table 3).
Quantitative RT-PCR
Total RNA extracted from MEFs was reverse transcribed using the advantage RT-PCR kit (Clontech). Following reverse transcription the quantity of the transcript was determined using specific primers (Table 3) and syber green PCR mix (applied biosystems).
Genotyping
Tails were clipped from three week old mice and DNA was prepared. Genotyping was done RMRP null mice were genotyped using FRMRPgeno and RRMRPgeno (Table 3) and RC mice were genotyped using FRCgeno and RRCgeno (Table 3).
siRNA transfection and viability assay
For silencing the expression of Ccdc107 or E130, we used three independent siRNA duplexes targeting each gene. The siRNAs were purchased from IDT Ccdc107 cat number: MMC.RNAI.N001037913.12.5, MMC.RNAI.N001037913.12.2, MMC.RNAI.N001037913.12.1 and E130 cat number: MMC.RNAI.N001013377.12.8, MMC.RNAI.N001013377.12.3, MMC.RNAI.N001013377.12.1. For the negative control Ambion siRNA negative control 1 was used (cat number AM4635) 100,000 MEFs were plated in a 6 well plate and were transfected with 20 ng of the duplex mix. Three days later RNA was extracted from a fraction of the transfected MEFs and expression was assessed by qRT-PCR. The same cells were plated onto a 96 well plate (5000 cells/well, 4 wells/sample) and 7 days post transfection viability was measured using cell titer glow (Promega) 15 µl/well.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by a US National Institutes of Health postdoctoral fellowship to J.R. F32GM090437-01, and R01 AG23145 (WCH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hermanns P, Bertuch AA, Bertin TK, Dawson B, Schmitt ME, et al. Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum Mol Genet. 2005;14:3723–3740. doi: 10.1093/hmg/ddi403. [DOI] [PubMed] [Google Scholar]
- 2.Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 3.Nakashima E, Tran JR, Welting TJ, Pruijn GJ, Hirose Y, et al. Cartilage hair hypoplasia mutations that lead to RMRP promoter inefficiency or RNA transcript instability. Am J Med Genet A. 2007;143A:2675–2681. doi: 10.1002/ajmg.a.32053. [DOI] [PubMed] [Google Scholar]
- 4.Hermanns P, Tran A, Munivez E, Carter S, Zabel B, et al. RMRP mutations in cartilage-hair hypoplasia. Am J Med Genet A. 2006;140:2121–2130. doi: 10.1002/ajmg.a.31331. [DOI] [PubMed] [Google Scholar]
- 5.Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 6.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenten D, Gerlach VL, Guo C, Velasco-Miguel S, Hladik CL, et al. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur J Immunol. 2002;32:3152–3160. doi: 10.1002/1521-4141(200211)32:11<3152::AID-IMMU3152>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]