Abstract
TA (tail-anchored) proteins utilize distinct biosynthetic pathways, including TRC40 (transmembrane domain recognition complex of 40 kDa)-mediated, chaperone-dependent and/or unassisted routes to the ER (endoplasmic reticulum) membrane. We have addressed the flexibility of cytosolic components participating in these pathways, and explored the thermodynamic constraints of their membrane insertion, by exploiting recombinant forms of Sec61β and Cytb5 (cytochrome b5) bearing covalent modifications within their TA region. In both cases, efficient membrane insertion relied on cytosolic factors capable of accommodating a surprising range of covalent modifications to the TA region. For Sec61β, we found that both SGTA (small glutamine-rich tetratricopeptide repeat-containing protein α) and TRC40 can bind this substrate with a singly PEGylated TA region. However, by introducing two PEG [poly(ethylene glycol)] moieties, TRC40 binding can be prevented, resulting in a block of subsequent membrane integration. Although TRC40 can bind Sec61β polypeptides singly PEGylated at different locations, membrane insertion is more sensitive to the precise location of PEG attachment. Modelling and experimentation indicate that this post-TRC40 effect results from an increased energetic cost of inserting different PEGylated TA regions into the lipid bilayer. We therefore propose that the membrane integration of TA proteins delivered via TRC40 is strongly dependent upon underlying thermodynamics, and speculate that their insertion is via a phospholipid-mediated process.
Keywords: cytochrome b5 (Cytb5), PEGylation, Sec61β, small glutamine-rich tetratricopeptide repeat-containing protein α (SGTA), tail-anchored protein (TA protein), transmembrane domain recognition complex of 40 kDa (TRC40)
Abbreviations: BMH, bis(maleimido)hexane; BODIPY®, boron dipyrromethene (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene); Cytb5, cytochrome b5; EndoH, endoglycosidase H; ER, endoplasmic reticulum; Get, guided entry of tail-anchored proteins; HisTrx, histidine–thioredoxin; Hsp, heat-shock protein; IASD, 4-acetamido-4′-[(iodoacetyl)amino]stilbene-2,2′-disulfonate; mPEG, methoxypoly(ethylene glycol); OPG, opsin glycosylation tag; PDI, protein disulfide-isomerase; PEG, poly(ethylene glycol); SGTA, small glutamine-rich tetratricopeptide repeat-containing protein α; SRP, signal recognition particle; TA, tail-anchored; TMS, transmembrane segment; TRC40, transmembrane domain recognition complex of 40 kDa
INTRODUCTION
TA (tail-anchored) proteins constitute a group of integral membrane proteins characterized by the presence of a single C-terminally localized stretch of hydrophobic amino acids that acts as both the subcellular targeting signal and membrane anchor [1]. Although TA proteins are found in most, if not all, intracellular membranes, the ER (endoplasmic reticulum) acts as the entry site for TA proteins destined for the various compartments of the secretory pathway [2–5]. Importantly, the C-terminal location of the membrane-spanning region precludes its co-translational recognition by the SRP (signal recognition particle), ensuring that TA protein targeting to, and insertion into, the ER membrane is post-translational [4].
The precise route of TA protein delivery to the ER membrane is precursor-dependent and correlates with the relative hydrophobicity of the TMS (transmembrane segment) of the tail-anchor region [6–8]. Thus, comparatively hydrophilic tail anchors, such as those of Cytb5 (cytochrome b5) and protein tyrosine phospatase 1B, define a pathway(s) that is either mediated by Hsp70 (heat-shock protein 70)/Hsp40 chaperones [8,9] and/or does not utilize any cytosolic components [10]. This pathway(s) does not rely on a specific receptor at the ER membrane; hence protease-treated microsomes are still fully capable of accepting the TA protein substrate [6,11].
The majority of TA proteins, such as RAMP4 (ribosome-associated membrane protein 4) and Sec61β, have a TA region predicted to be more hydrophobic than that of Cytb5 [12]. These precursors use a specialized targeting factor, known as TRC40 (transmembrane domain recognition complex of 40 kDa; Asna1) in mammalian cells [13,14] and Get3 (guided entry of tail-anchored proteins 3) in Saccharomyces cerevisiae [15,16], for their delivery to the ER membrane. The TRC40/Get3 pathway requires additional cytosolic factors, such as Get4/Get5 and Sgt2 in yeast [17–20], and a membrane-bound receptor that has been defined as the Get1–Get2 complex in S. cerevisiae [16]. Recent structural analyses of the Get3 protein [21–25] and associated components [26,27] have provided new insights into the relationship between its ATPase cycle and TA protein delivery to the ER membrane and identified the likely substrate-binding site. However, since none of the current Get3 structures were obtained as a complex with a TA protein substrate, several aspects of substrate recognition by Get3 remain to be elucidated. Moreover, the precise details of TA protein recognition by upstream components, such as Sgt2 and its mammalian equivalent SGTA (small glutamine-rich tetratricopeptide repeat-containing protein α), are also currently unclear (see [20,28]).
To date, many biochemical studies addressing the biogenesis of TA proteins at the ER membrane have relied upon substrates generated by in vitro translation using cell lysates that contain essential cytosolic factors (see [7]). However, a bacterial system has recently been exploited to co-express model TA proteins with TRC40/Get3 [21,29], and the resulting complexes were shown to be sufficient for facilitating membrane insertion, indicating that TRC40/Get3 acts at a very late stage of TA protein delivery to the ER. Studies using purified recombinant TA proteins expressed in the absence of known cytosolic interacting partners are limited, and have largely focused on Cytb5 [10,30]. The use of recombinant TA proteins allows polypeptide synthesis to be physically and temporally separated from the ER delivery and membrane insertion steps, facilitating studies of the components involved in these processes. Indeed, we have recently taken this approach to study Sec61β biogenesis, and identified a novel role for Bat3 during TRC40-dependent ER delivery [28]. Notwithstanding the potential advantages of using recombinant forms of TA proteins [31], this approach has not been exploited extensively to date.
In the present study, we describe the successful production of recombinant TA protein models for each of the two major biosynthetic pathways, namely Sec61β and Cytb5. By chemically modifying these recombinant TA protein substrates, we show that both routes are surprisingly unconstrained, and can accommodate substrates with significantly modified transmembrane domains. Remarkably, PEGylated forms of both Sec61β and Cytb5 can also be efficiently integrated into the ER membrane, albeit via different routes. We find that the cytosolic factors SGTA and TRC40 can both accommodate substrates with a PEGylated TA region, and provide a model for the ER delivery and membrane integration of PEGylated Sec61β. By analysing additional variants of Sec61β, we show that the location of the PEG [poly(ethylene glycol)] moiety within the TA region strongly influences membrane integration in a manner that correlates with the predicted thermodynamic cost of this process. On this basis, we speculate that the membrane insertion step of the TRC40-dependent pathway for TA protein biogenesis may be a purely lipid-mediated event that does not require a dedicated integrase.
EXPERIMENTAL
Materials
Bacterial expression vector pHisTrx was a gift from Richard Kammerer (Paul Scherrer Institute, Villigen, Switzerland). Rabbit polyclonal antiserum recognizing TRC40 was a gift from Bernhard Dobberstein [ZMBH (Zentrum für Molekulare Biologie der Universität Heidelberg), Heidelberg, Germany]. All detergents were from Anatrace, except Triton X-100, which was from Sigma. Cross-linking reagents and EZ-link-biotin were from Pierce, whereas BODIPY® [boron dipyrromethene (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene)] and IASD {4-acetamido-4′-[(iodoacetyl)amino]stilbene-2,2′-disulfonate} were from Molecular Probes. mPEG [methoxypoly(ethylene glycol)]-5000 and mPEG-20000 maleimides were from Nektar. Nuclease-treated rabbit reticulocyte lysate was supplied by Promega.
Chemical modification of recombinant TA proteins
Recombinant TA proteins were expressed and purified as described previously [28], with the various cysteine residue mutants generated by site-directed mutagenesis and confirmed by DNA sequencing. Thiol-reactive probes, as indicated, were mixed with solutions of single cysteine residue mutants of Sec61βOPGS77C (where OPG is opsin glycosylation tag) (77 μM) in buffer B [50 mM Tris/HCl, pH 7.4, 300 mM NaCl, 10 mM imidazole, 10% (v/v) glycerol, 0.75% (w/v) OG (octyl β-D-glucopyranoside)] and Cytb5OPGS119C (25 μM) in buffer Q [50 mM Hepes/KOH, pH 7.9, 50 mM potassium acetate, 200 mM NaCl, 10% (v/v) glycerol, 0.1% LDAO (dodecanoyldimethylamine-N-oxide)] (see Supplementary Table S1 at http://www.BiochemJ.org/bj/436/bj4360719add.htm for details of the mutants) using the reagents at a final concentration of 1–3 mM and incubating for 2 h at room temperature (21 °C) in the dark. Reactions were quenched by addition of 20 mM 2-mercaptoethanol and further incubation for 10 min. Modified proteins were resolved by SDS/PAGE and then stained with Coomassie Brilliant Blue or immunoblotted with the anti-opsin tag antibody, or they were used for membrane integration reactions. All other Sec61βOPG cysteine mutants (see Supplementary Table S1) were purified and labelled as described using 1.5 mM mPEG-5000 maleimide, pre-quenched with glycine, for 1 h at 25 °C. After quenching any unchanged maleimides, labelled proteins were analysed as above or used for integration assays. Membrane integration assays of recombinant TA proteins were performed using sheep pancreatic microsomes as described previously [28].
Binding of PEGylated Sec61βOPG to recombinant SGTA and endogenous TRC40
Human SGTA was expressed in Escherichia coli BL21 Gold (DE3) pLysS cells and purified essentially as described above for TA proteins. HisTrx (histidine–thioredoxin)–SGTA, or BSA control, was then immobilized on UltraLink Biosupport beads (ThermoScientific) according to the manufacturer's instructions. Recombinant PEGylated Sec61βOPG variants were mixed with beads in buffer S [50 mM Hepes/KOH, pH 7.5, 40 mM potassium acetate, 5 mM MgCl2 and 0.05% pMAL-C12 amphiphile (Anatrace)], to give a final concentration of 3.3 μM and incubated for 1 h at 4 °C. Beads were washed with buffer S plus 0.5 M NaCl and any bound material eluted first with buffer S supplemented with 0.5% (v/v) Triton X-100, and then mixed with Laemmli sample buffer.
Association of PEGylated Sec61βOPG variants with endogenous TRC40 was tested by incubating 1–1.9 μM of recombinant modified Sec61βOPG with 140 μl of rabbit reticulocyte lysate for 30 min at 30 °C. Reaction mixtures were then split into two and each half was added to 20 μl [50% (v/v) suspension] of Protein A–Sepharose coated with 7.5 μl of rabbit anti-TRC40 or anti-PDI (protein disulfide-isomerase) antisera. The lysate was incubated with anti-Protein A–Sepharose antibody beads for 1 h at 4 °C, the resin was then washed extensively with buffer R (50 mM Hepes/KOH, pH 7.5, 40 mM potassium acetate and 5 mM MgCl2) supplemented with 0.5 M NaCl, and bound proteins were eluted with buffer R containing 0.5% (v/v) Triton X-100 (cf. [13]). The eluate was mixed with Laemmli sample buffer, and samples were resolved on SDS/PAGE and immunoblotted with the anti-opsin tag antibody. Quantitative analysis was carried out using a LiCor Biosciences system (described below). The non-specific binding of the indicated Sec61βOPG variants to anti-PDI antibody-coated Protein A–Sepharose beads was subtracted from the results obtained using anti-TRC40 antibody-coated Protein A–Sepharose, and the resulting values were normalized to the amount of each Sec61βOPG species present in the starting material (see Supplementary Figure S1 for a full worked example).
Quantitative immunoblotting
Following transfer of the samples on to a low-fluor PVDF membrane and incubation with a primary anti-opsin epitope tag antibody, a secondary anti-mouse fluorescent-dye-conjugated antibody was added at 1:10000 dilution. The membrane was incubated for 1 h at room temperature, scanned using a LiCor Biosciences system and the results were quantified with Odyssey 2.1 software.
Protease protection of membrane-integrated Sec61βOPG variants
Two membrane integration reaction mixtures were combined. Microsomes were isolated as described previously [28] and resuspended in 68 μl of LSS (low-salt sucrose) buffer [250 mM sucrose, 100 mM potassium acetate, 5 mM magnesium acetate and 50 mM Hepes/KOH, pH 7.9]. The suspension was then split into three and each 20 μl aliquot received 2 μl of water, 2 μl of proteinase K (2.5 mg/ml) or 2 μl of proteinase K (2.5 mg/ml) and 2.2 μl of 10% (v/v) Triton X-100. Digestion was carried out on ice for 30 min and proteinase K was inhibited by the addition of 2.5 mM PMSF and further incubation on ice for 10 min. Laemmli buffer was then added and the samples were immediately heated to 95 °C for 10 min. Samples were resolved on a 16% (v/v) Tris/Tricine polyacrylamide gel and immunoblotted with the anti-opsin epitope tag antibody.
RESULTS
Recombinant TA proteins are inserted into the ER membrane
Preliminary analysis indicated that a HisTrx-based fusion was superior to a glutathione transferase tag for the high-level expression and purification of the recombinant TA proteins used in the present study (results not shown). We used this approach to purify and characterize two model recombinant TA proteins, Sec61β and Cytb5, each of which favours a distinct pathway for their biogenesis at the ER membrane (see the Introduction; also [7,8,13,14,32]). Cells expressing recombinant TA proteins were lysed in the presence of detergent, soluble fractions were incubated with Ni-NTA (Ni2+-nitrilotriacetate)–agarose and, after washing the beads with buffer containing a low concentration of detergent, the various proteins were released from the tag by thrombin-mediated cleavage ([10,28,30]; see also the Experimental section and Supplementary Table S1). Coomassie Blue staining showed an acceptable level of protein purity, with only a fraction of Sec61βOPG and Cytb5OPG present in truncated forms (see Supplementary Figure S2A at http://www.BiochemJ.org/bj/436/bj4360719add.htm).
The membrane integration capacity of the recombinant proteins was investigated using a well-established N-glycosylation assay that relies on a short C-terminal opsin-derived epitope tag [4,8,11,13,33]. Crucially, this modification can only take place when the protein spans the membrane and the tag has entered the ER lumen, consistent with authentic membrane integration. Substantial levels of EndoH (endoglycosidase H)-sensitive protein species for both Sec61βOPG and Cytb5OPG were observed (Supplementary Figure S2B, cf. lanes 1 and 2, 3 and 4), clearly indicating that these two precursors are efficiently integrated into the ER membrane in the presence of cytosol.
Chemically modified TA proteins are efficiently inserted into the ER membrane
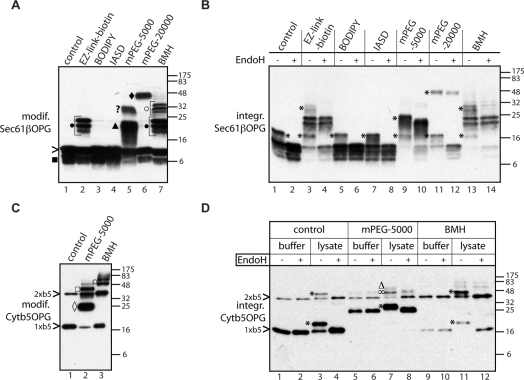
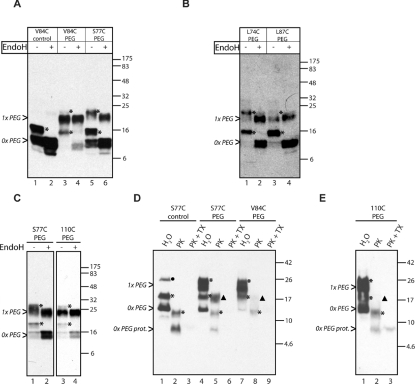
Purified Sec61βOPG and Cytb5OPG, both bearing single cysteine residues within their TA regions (Sec61βOPGS77C and Cytb5OPGS119C; see Supplementary Table S1), were treated with a variety of thiol-reactive probes differing in size and physicochemical properties (Figures 1A and 1C). In the case of PEGylation, the modification was confirmed by a shift in electrophoretic mobility (see Figures 1A and 1C), whereas for BODIPY®, the fluorescently tagged protein could be visualized after membrane integration (see Supplementary Figure S3 at http://www.BiochemJ.org/bj/436/bj4360719add.htm). In the case of IASD and biotinylation, we have not directly confirmed labelling, and cannot rule out the possibility that little if any protein was modified. However, the biotinylation reagent also generated covalently linked dimers of Sec61βOPG, most likely resulting from trace amounts of bifunctional cross-linkers present in the commercial reagent (Figure 1A, lane 2, •). This conclusion is strongly supported by the fact that we can generate apparently identical species upon treatment with a bifunctional maleimide (Figure 1A, lane 7, •). Treatment with mPEG-5000 maleimide and BMH [bis(maleimido)hexane] also generated even larger species (Figure 1A, lanes 5 and 7, ? and ○ respectively), but since these derivatives were not recovered in the membrane fraction after further analysis (cf. Figure 1B, lanes 9 and 10, 13 and 14), they were not further characterized. In the case of Cytb5OPG, a small amount of SDS-stable dimer was observed in the absence of any additional treatment (Figure 1C, lane 1). A similar species became more prominent after incubation with both mPEG-5000 maleimide and BMH, presumably due to covalent cross-linking of the dimer (Figure 1C). Both the monomeric and dimeric forms of Cytb5OPG appeared to be modified with mPEG-5000 (Figure 1C, lane 2, ◇ and □ respectively). Additional high-molecular-mass species were also observed with BMH (Figure 1C, lane 3, ○), but as for Sec61βOPG, little if any of these unassigned species were recovered in association with ER-derived membranes (cf. Figure 1D). Hence our analysis showed that cysteine residues introduced into the TA regions of both recombinant proteins can be covalently modified by PEGylation and cross-linking reagents, whereas Sec61βOPG can also accept a number of other thiol-reactive probes.
Figure 1. Chemically modified TA proteins are integrated into ER-derived membranes.
Sec61βOPGS77C (A) and Cytb5OPG (C) with single cysteine residues located in their TMSs (see Supplementary Table S1 at http://www.BiochemJ.org/bj/436/bj4360719add.htm) were modified (modif.) with thiol-reactive probes or the homobifunctional crosslinking reagent BMH, as indicated, and the products were analysed by immunoblotting. Full-length Sec61βOPG (>), a truncated form (■) and cross-linked dimers (●) are indicated, as are the monomer and dimer of Cytb5OPG (1xb5 and 2xb5 respectively). PEG-5000- and PEG-20000-modified Sec61βOPG (▲ and ◆ respectively), and PEGylated Cytb5OPG monomer (◇) and dimer (□) are indicated. Other products assumed to represent abberant cross-linked species (? and ○) were not competent for membrane integration (integr.) and their precise identity was not pursued. (B and D) N-glycosylated species are indicated with an asterisk (*), except the N-glycosylated, PEG-modified Cytb5OPG dimer (Δ) (D, lane 7). A product corresponding to the PEGylated Cytb5OPG dimer, and an N-glycosylated, non-PEGylated dimer that co-migrate is also indicated (∞). Molecular mass markers in kDa are also indicated on the right-hand side of the Western blots.
A role for cytosolic components during the membrane integration of Cytb5 is controversial [7,8,10]. Our previous analysis of recombinant Cytb5OPG found that immunodepletion of TRC40 or Bat3 from cytosol had no effect on its insertion, clearly distinguishing its integration from that of Sec61βOPG [28]. This difference is further underlined when the kinetics of membrane insertion for recombinant Cytb5OPG and Sec61βOPG are compared (see Supplementary Figures S2C and S2D), with Cytb5OPG integration reaching completion substantially more rapidly (cf. [34]). We therefore established whether the membrane integration of recombinant Cytb5OPG is cytosol-dependent in our assay. We can only detect the N-glycosylation of Cytb5OPG in the presence of both cytosol and ER-derived membranes (Figure 1D, lanes 1 to 4), using experimental conditions where the efficient N-glycosylation of recombinant TA proteins has been observed in the presence of buffer alone [21,29]. We therefore conclude that the efficient membrane integration of the recombinant Cytb5OPG used in our experimental system is cytosol-dependent.
To examine the constraints on the cellular components involved in the biogenesis of Sec61βOPG and Cytb5OPG, we tested the cytosol-dependent integration of the two modified recombinant polypeptides into ER-derived membranes. Remarkably, none of the treatments with thiol-reactive reagents tested resulted in a complete block of membrane integration, despite the location of the target cysteine residue within the TA region of both proteins (Figure 1B, lanes 1–12 and Figure 1D, cf. lanes 3 and 4, 7 and 8). In fact, even homodimers of both Sec61βOPG and Cytb5OPG were still measurably inserted into the lipid bilayer (Figure 1B, cf. lanes 3, 4, 13 and 14 and Figure 1D, cf. lanes 3, 4, 11 and 12). Membrane integration of a Cytb5OPG dimer occurred both with and without prior BMH treatment, suggesting that the close proximity of the two TA regions of the protein monomers does not preclude membrane integration of a Cytb5OPG dimer (Figure 1D, cf. lanes 3 and 4, 11 and 12). A more detailed analysis of Sec61βOPG dimers, generated by using cross-linking reagents of different length spacers, revealed that membrane integration was not qualitatively affected by the distance between individual subunits (see Supplementary Figure S4A at http://www.BiochemJ.org/bj/436/bj4360719add.htm). Moreover, partial EndoH digestion of the membrane-associated material showed that both subunits of Sec61βOPG and Cytb5OPG dimers are N-glycosylated, indicating that both polypeptides are fully inserted into the ER membrane (Supplementary Figures S4B and S4C). Remarkably, even the addition of large hydrophilic ER membrane-impermeable probes [35], including PEG-5000 (Figure 1B, cf. lanes 9 and 10, *, and Figure 1D, lanes 7 and 8, ∞) and, for Sec61βOPG, PEG-20000 (Figure 1B, cf. lanes 11 and 12), to the TA-regions did not prevent authentic membrane insertion of the recombinant TA proteins. Perhaps most strikingly, a Cytb5OPG dimer modified with mPEG-5000 was still capable of being efficiently inserted into the ER membrane (Figure 1C, lane 2, and Figure 1D, cf. lanes 7 and 8, Δ). It is conceivable that the introduction of a PEG moiety into the TA region of a protein might alter its choice of biosynthetic route. In particular, we wished to confirm that the requirements for PEGylated Sec61βOPG were the same as those previously defined for the unmodified protein. Sec61β is known to exploit the TRC40-dependent pathway for ER integration, a route that is characterized by a protease-sensitive membrane component [11,14]. We therefore investigated the effect of trypsin treatment upon the membrane integration of Sec61βOPG and found that the membrane association and N-glycosylation of both PEGylated and non-PEGylated forms of the protein were identically sensitive to protease treatment (see Supplementary Figure S5A at http://www.BiochemJ.org/bj/436/bj4360719add.htm). In contrast, the TRC40-independent membrane integration of Cytb5 was unaffected by trypsin treatment, confirming both the resilience of the N-glycosylation machinery and the specificity of the assay (see Supplementary Figure S5A, and also [11,14]). We also discovered that prequenching of mPEG-5000 maleimide with glycine prevented the formation of aberrant products (cf. Figure 1A, lane 5 and Supplementary Figure S5B), and used this approach for all subsequent experiments.
PEGylation of Sec61βOPG affects distinct stages of its membrane integration
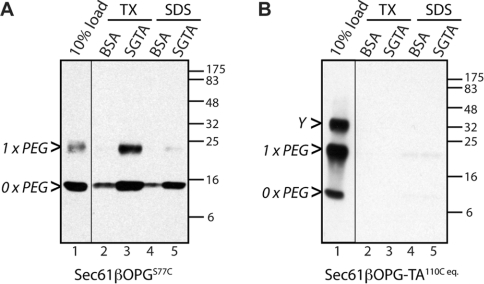
Delivery of Sec61β to the ER membrane is mediated by the TRC40 protein [14], and if this cytosolic factor is removed, the TA protein fails to reach its target destination efficiently [10,28]. Several studies also suggest a role for yeast Sgt2p [18,26], and its mammalian equivalent SGTA [28,36], acting on the same pathway but upstream of Get3/TRC40 [17,20]. To address the potential role of mammalian SGTA in TA protein biogenesis, we analysed the interaction of our recombinant TRC40 substrate, Sec61βOPG, with immobilized recombinant SGTA. We find that Sec61βOPG binds efficiently to SGTA, but not to a BSA control, in a Triton X-100-sensitive manner (Figure 2A, lanes 2 and 3). PEGylated Sec61βOPG appears to bind at least as efficiently as the unmodified protein (Figure 2A, lanes 2 and 3), and is also released by Triton X-100, consistent with a hydrophobic interaction via the TA region as previously observed for TRC40 binding (cf. [13]). In contrast, an alternative version of Sec61β that lacks the hydrophobic TA region but also bears a single PEG residue shows no association with immobilized SGTA, confirming that the association of a PEG moiety alone is not capable of mediating a Triton-sensitive interaction with SGTA (Figure 2B, lanes 2 and 3).
Figure 2. Recombinant Sec61βOPG binds to immobilized SGTA.
Sec61βOPGS77C (A) or Sec61βOPG-TA110C eq. (see Supplementary Table S1) (B) were PEGylated as described in the Experimental section and mixed with immobilized recombinant SGTA or a BSA control. The Triton X-100 (TX)-or Laemmli buffer (SDS)-eluted fractions were resolved by SDS/PAGE, and Sec61βOPG variants were then detected by immunoblotting. Each panel shows samples resolved on a single blot and exposed for the same time with irrelevant lanes removed. An uncharacterized, but PEGylation-dependent, product of Sec61βOPG-TA110C eq. is indicated (Y). Molecular-mass markers in kDa are also indicated on the right-hand side of the Western blots.
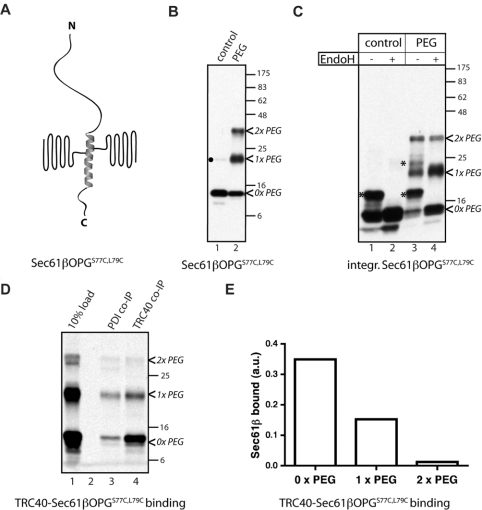
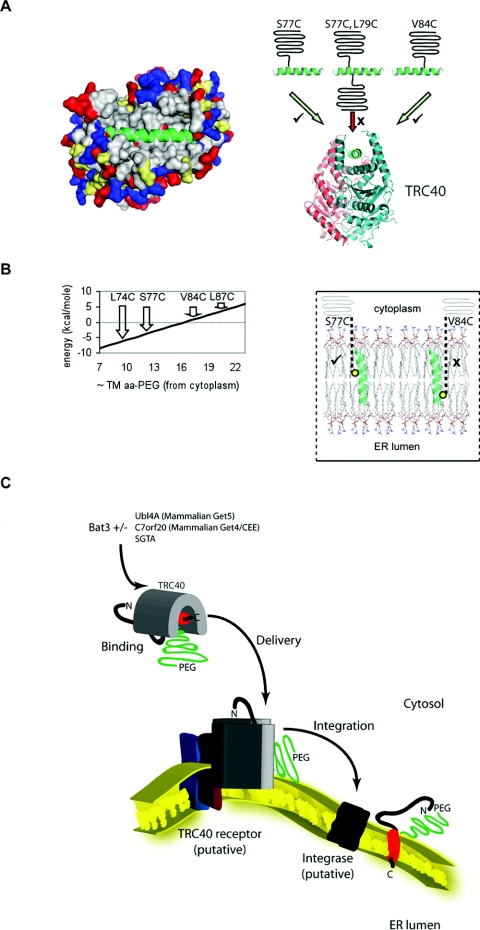
Crystal structures of Get3 [21,23,24], the yeast homologue of TRC40, reveal that it has a deep groove at the dimer interface that most likely acts as a TA protein recognition site. On the basis of our results (cf. Figure 1B), similar to SGTA, TRC40 must presumably accommodate a Sec61β polypeptide modified with a single mPEG-5000 moiety. To investigate further the apparent flexibility of substrate binding by TRC40, we mutated both Ser77 and Leu79 of the Sec61βOPG TA-region into cysteine residues (Sec61βOPGS77C,L79C) and modified this polypeptide with mPEG-5000. Assuming that the transmembrane region adopts an α-helix-like conformation, this double modification would yield a TA protein with two PEG-5000 molecules extending from opposite sides of the polypeptide chain (Figure 3A). Immunoblotting analysis confirmed the efficient labelling of Sec61βOPGS77C,L79C, yielding populations bearing both one and two mPEG-5000 molecules (Figure 3B, cf. lanes 1 and 2). When membrane insertion was tested, both unmodified and singly PEGylated forms of Sec61βOPGS77C,L79C were visibly N-glycosylated, in agreement with our previous results (Figure 3C, cf. lanes 3 and 4; cf. Figure 1B, lanes 1 and 2, 9 and 10). However, no N-glycosylation of Sec61βOPGS77C,L79C labelled with two mPEG-5000 molecules was seen, even after prolonged exposure of the immunoblot (Figure 3C, cf. lanes 3 and 4; and results not shown).
Figure 3. PEGylation of Sec61βOPGS77C,L79C prevents TRC40 binding.
Sec61βOPGS77C,L79C was generated (see Supplementary Table S1 at http://www.BiochemJ.org/bj/436/bj4360719add.htm) and the purified protein was modified with mPEG-5000. A schematic diagram of the doubly labelled polypeptide is presented assuming an α-helix-like conformation of the transmembrane region (A). Covalent attachment of PEG-5000 was confirmed by immunoblotting (B) and membrane integration by EndoH sensitivity (C). Small amounts of Sec61βOPG species most likely corresponding to a dimer formed during protein purification were observed (B, ●). Binding of PEG-modified Sec61βOPGS77C,L79C to TRC40 was addressed by pre-incubating the protein with rabbit reticulocyte lysate followed by co-immunoprecipitation with anti-TRC40 or control anti-PDI antibodies. Bound Sec61βOPG was eluted and detected by quantitative immunoblotting (D), allowing relative binding to TRC40 to be compared (E, see Supplementary Figure S1 at http://www.BiochemJ.org/bj/436/bj4360719add.htm for a full worked example). The values shown are the average from two independent experiments. * indicates N-glycosylated species. Molecular-mass markers in kDa are also indicated on the right-hand side of the Western blots.
The lack of any membrane-inserted Sec61βOPGS77C,L79C that had been modified with two PEG-5000 species could result from either its inefficient delivery to the ER membrane or an inability to be integrated into the phospholipid bilayer. To address this issue, we tested the association of the PEG-labelled Sec61βOPGS77C,L79C with TRC40 by immunoisolation and quantitative immunoblotting. We found that the addition of two PEG probes to a single TMS of Sec61βOPGS77C,L79C almost completely abolished TA protein binding to TRC40 (Figure 3D, cf. lanes 3 and 4, see also Supplementary Figure S1 at http://www.BiochemJ.org/bj/436/bj4360719add.htm). Also, we revealed that the attachment of even a single PEG probe to Sec61βOPGS77C,L79C reduced its binding to TRC40 to approximately half the level observed with the unmodified protein (Figure 3E). This is consistent with a qualitative reduction in the membrane integration of the singly modified chain when compared with the unmodified version of Sec61βOPG (Figure 3C, cf. lanes 3 and 4, marked 0×PEG and 1×PEG).
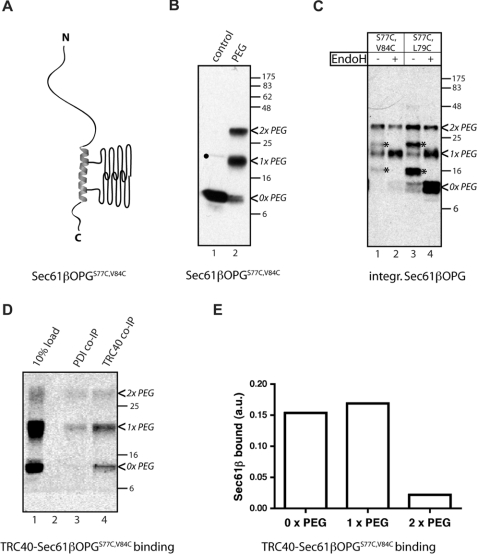
We next established whether a protein with a TA region modified with two PEG-5000 molecules predicted to be located on the same side of an α-helix-like conformation can be bound by TRC40 and delivered to the ER membrane. To this end, a third derivative of Sec61βOPG with both Ser77 and Val84 mutated to cysteine residues (Sec61βOPGS77C,V84C) (Figure 4A) was purified and modified with mPEG-5000 (Figure 4B, cf. lanes 1 and 2). When the membrane integration of the two double cysteine residue variants of Sec61βOPG was compared, surprisingly little if any N-glycosylation of even singly PEGylated Sec61βOPGS77C,V84C could be detected under conditions where the singly modified Sec61βOPGS77C,L79C was reproducibly integrated (Figure 4C). When the binding of Sec61βOPGS77C,V84C to TRC40 was analysed, the interaction of the singly PEGylated-Sec61βOPGS77C,V84C species was unperturbed, although association of the doubly PEGylated species was clearly diminished (Figures 4D and 4E). Hence, although TRC40 binding is relatively tolerant of the addition of a single PEG-5000 moiety to the Sec61β TA-region, a subsequent step of the membrane insertion process appears much more sensitive to the precise location of such a probe and PEGylation of Cys84 appears not to be tolerated.
Figure 4. PEGylation of Sec61βOPGS77C,V84C inhibits membrane integartion.
(A) Purified recombinant Sec61βOPGS77C,V84C (see Supplementary Table S1 at http://www.BiochemJ.org/bj/436/bj4360719add.htm) was modified with mPEG-5000 and is shown schematically as described for Figure 3(A). (B–D) PEGylation was confirmed by immunoblotting (B) with a presumptive Sec61βOPG dimer indicated (●). Membrane integration was determined by EndoH sensitivity (C), and TRC40 binding (D and E) was analysed as detailed in the legend to Figure 3 (see also Supplementary Figure S1 at http://www.BiochemJ.org/bj/436/bj4360719add.htm). * indicates N-glycosylated species. Molecular-mass markers in kDa are also indicated on the right-hand side of the Western blots.
In order to test this hypothesis further, we generated a single cysteine residue mutant, Sec61βOPGV84C, and studied its membrane integration compared with the Sec61βOPGS77C mutant. Cys84 was more efficiently PEGylated than Cys77 (Figure 5A and Supplementary Figure S1A), but the resulting Sec61β derivative showed little if any membrane-dependent N-glycosylation, whereas Sec61βOPGS77C was again efficiently modified (Figure 5A, lanes 3 to 6), as previously observed (Figure 1B, lanes 9 and 10). Furthermore, the effect of PEGylation of residue 84 could not be correlated to a reduction in TRC40 binding (Supplementary Figure S1). Hence the PEGylation of a single cysteine residue located at amino acid 84 of Sec61βOPG effectively prevents membrane integration as judged by N-glycosylation. One potential explanation for this location-specific effect of PEGylation is the thermodynamic cost of integrating the modified TA region into a phospholipid bilayer (see also Figure 6B, the Supplementary online data and Supplementary Table S2 at http://www.BiochemJ.org/bj/436/bj4360719add.htm and the Discussion section). We therefore explored the consistency of this positional effect by analysing the membrane integration of PEGylated forms of three further single cysteine residue mutants, L74C, L87C and a novel cysteine residue, added to the C-terminus of Sec61βOPG, denoted 110C. We observe that the membrane integration of PEGylated Sec61βOPGL74C and Sec61βOPGS77C is clearly more efficient than that of PEGylated Sec61βOPGV84C, Sec61βOPGL87C and Sec61βOPG110C (Figures 5A, 5B and 5C). To confirm that the location-specific effect of PEGylation was due to a perturbation of membrane integration, and not N-glycosylation itself, we also carried out a protease protection analysis (cf. [10]). We could identify a discrete membrane-dependent protease-protected fragment of PEGylated Sec61βOPGS77C (Figure 5D, lane 5, filled triangle). In contrast, no such product was observed with Sec61βOPGV84C or Sec61βOPG110C (Figure 5D, lane 8 and Figure 5E, lane 2, predicted location indicated by ▲). On this basis, we conclude that the PEGylation of amino acid residues located towards the C-terminus of Sec61βOPG preferentially inhibits membrane integration.
Figure 5. Distinct effects of PEGylation at different locations within the Sec61β TA-region.
Additional single cysteine Sec61βOPG variants were generated (see Supplementary Table S1 at http://www.BiochemJ.org/bj/436/bj4360719add.htm) and PEGylated. The membrane integration of PEGylated Sec61βOPGS77C and Sec61βOPGV84C (A), Sec61βOPGL74C and Sec61βOPGL87C (B) and Sec61βOPGS77C and Sec61βOPG110C (C) were determined by EndoH sensitivity. In addition, the membrane integration status of Sec61βOPGS77C and Sec61βOPGV84C (D) and Sec61βOPG110C (E) was determined by protease protection (see the Experimental section). The filled triangle indicates the actual (D, lane 5) or predicted (D, lane 8 and E, lane 2) location of a protease protected, PEGylated, fragment of Sec61βOPG. In (C), the Sec61βOPG110C samples were overexposed in order to best visualize any N-glycosylated PEGylated products. * indicates N-glycosylated species; ▲ indicates predicted location of membrane-dependent protease-protected fragment of PEGylated Sec61βOPG. Molecular-mass markers in kDa are also indicated on the right-hand side of the Western blots. PK, proteinase K; TX, Triton X-100.
Figure 6. Membrane delivery and integration of PEG-modified Sec61β.
(A) Left-hand panel, human TRC40 was modelled using the S. cerevisiae Get3 dimer structure as a template (PDB code 2WOJ) and with the 23-amino-acid-residue α-helical TMS of Sec61β (green) shown in the proposed binding groove. The surface of the TRC40 dimer is colour-coded by amino acid polarity: blue, positively charged; grey, non-polar; red, negatively charged; yellow, polar-uncharged. Right-hand panel, a schematic representation for the association of PEGylated Sec61β with TRC40 showing binding of polypeptides PEGylated at residue 77 or 84, but not of a doubly modified Sec61βS77C,L79C variant. (B) Left-hand panel, estimated energy changes (see Supplementary online data at http://www.BiochemJ.org/bj/436/bj4360719add.htm) for the incorporation of different singly PEG-modified TA-regions of Sec61βOPG into a lipid bilayer. The relative ΔG values for PEGylation at residues 74, 77, 84 and 87 are indicated. Right-hand panel, representation of PEGylated Sec61β TA regions within a model lipid bilayer using membrane co-ordinates taken from [47]. Experiments confirm the efficient membrane integration of a Sec61βOPG PEGylated on Cys74 and Cys77, but not Cys84, Cys87 or Cys110. (C) Working model for the delivery of PEGylated Sec61β to the ER membrane. A variety of upstream components including Bat3, SGTA and mammalian Get4 and Get5 [27,28,37] facilitate TRC40-mediated delivery to the mammalian ER membrane via its receptor [42]. Any requirement for a membrane integrase remains hypothetical (cf. [7]). Covalent modification of the TA region of Sec61β could in principle perturb any one of the steps illustrated.
DISCUSSION
We have exploited recombinant TA proteins as a tool for better understanding their biogenesis at the ER. Both Sec61βOPG [28] and Cytb5OPG (the present study) are efficiently integrated into ER-derived membranes, and in both cases this process is dependent upon the presence of cytosol. The role of cytosolic factors in the membrane integration of Cytb5 remains controversial [7,8,10]. Nevertheless, in the present study we can only detect an N-glycosylated membrane integrated form of the protein in the presence of lysate (cf. [7]). In the case of Sec61β, the requirement for cytosol correlates to a need for TRC40/Asna-1 [13,14], and other cytosolic factors including Bat3 [28] and the mammalian homologues of Get4 and Get5 [37]. We now provide the first evidence for a direct interaction between Sec61β and SGTA, the mammalian equivalent of yeast Sgt2. Sgt2 has recently been implicated in the transfer of TA protein substrates to Get3 [20]. These studies support our previous observation of a specific association between SGTA and TA regions [28], and we now conclude that this represents a direct interaction of SGTA with a hydrophobic substrate [38], and speculate that SGTA normally plays a role in TA protein transfer to TRC40.
Sec61β can tolerate the covalent attachment of a variety of reagents to a single cysteine residue located within its TA region, and retain its ability to be integrated into the ER membrane. Likewise, the membrane integration of Cytb5 is not blocked by the addition of a PEG-5000 moiety close to the middle of its TA region or by the formation of a protein dimer. In the case of Sec61β, polypeptides with various attachments including PEG-5000, and even PEG-20000, can all be post-translationally integrated. This suggests that the cellular components that mediate its delivery to, and integration into, the ER membrane can accommodate a range of non-physiological modifications without a substantial perturbation of their function. Our analysis of the binding of Sec61βOPG to cytosolic factors implicated in its delivery to the ER showed that the direct binding of recombinant Sec61βOPGS77C to SGTA was unaffected by PEGylation. Since the PEG moiety is attached to the hydrophobic TA region, this would suggest that SGTA has a hydrophobic groove-like binding site capable of accommodating this unnatural substrate. Interestingly, both TRC40 and the 54 kDa subunit of the SRP each have flexible hydrophobic clefts capable of accommodating a range of hydrophobic polypeptide regions [21,23,39], further supporting this notion (see also [20]).
To analyse the effects of PEGylation upon TRC40 binding, we engineered double cysteine residue mutants of Sec61βOPG that generated a mixture of singly and doubly modified species upon PEGylation and determined their association by co-immunoprecipitation. This analysis showed that the singly PEGylated form of Sec61βOPGS77C,L79C can still bind to TRC40 with substantial efficiency, whereas a doubly modified form cannot. When this PEGylated TA region is modelled into the substrate-binding site of TRC40, our data are in good agreement with the proposed deep pocket [21–24] that could accommodate the addition of one centrally localized PEG-5000 to the substrate, but not two (see Figure 6A). Hence, we conclude that the doubly PEGylated form of Sec61βOPGS77C,L79C is incapable of membrane integration due to its inability to bind TRC40. When an alternative cysteine residue mutation was used to generate a doubly PEGylated variant, Sec61βOPGS77C,V84C, although the singly modified protein could bind TRC40, it was no longer membrane-integrated. Subsequent analysis of the single cysteine residue mutant Sec61βOPGV84C showed that PEGylation of Cys84 specifically perturbs biosynthesis at a step occurring after TRC40 binding; for example, substrate release by TRC40 or the subsequent membrane integration step (Figure 6C). Our calculations of the free energy changes resulting from the insertion of a PEG polymer into a lipid bilayer favour the introduction of a constraint at the membrane insertion step. Hence, the free energy associated with the transfer of the unmodified Sec61βOPG TMS into a lipid bilayer can be estimated as −11.5 kcal (1 kcal=4.184 kJ)/mol (see Supplementary online data and Supplementary Table S2), assuming a non-Sec61 translocon-mediated pathway for insertion [7,40,41]. By the same criteria, the integration of Sec61βOPG PEGylated at Cys77 has an estimated energetic cost of −4 kcal/mol, and hence remains favourable (Figure 6B, S77C). In contrast, the energetic cost of integrating Sec61βOPG PEGylated at Cys84 is +1 kcal/mol (Figure 6B, V84C; see Supplementary online data for detailed calculations). Thus our modelling supports the idea that the membrane integration efficiency of a PEGylated Sec61β TA region is dependent on the precise location of the probe relative to the lipid bilayer (Figure 6B).
To further challenge this model, we created three further single cysteine residue mutants of Sec61βOPG, Sec61βOPGL74C, Sec61βOPGL87C and Sec61βOPG110C and analysed the membrane integration of their PEGylated species. As predicted, the modification of residue 74 towards the N-terminal side of the TA region did not prevent membrane integration, whereas the alteration of residue 87 towards the C-terminal end, and the addition of a novel cysteine at the extreme C-terminus, both did. Taken together, these observations suggest that thermodynamic constraints are a major factor in the membrane integration of TRC40-dependent TA proteins. Although we cannot exclude the possibility that the membrane insertion of Sec61β requires a specialized integrase, we believe that the distinct behaviour of the differently PEGylated Sec61β TA regions is more consistent with a simple partitioning model for membrane insertion. In this scenario, the recently identified membrane-bound receptor for TRC40 [42] would act to deliver TA proteins to the surface of the ER membrane, but subsequent integration would rely solely on the favourable thermodynamics of TA protein partitioning into the lipid bilayer (Figure 6C, cf. [7]).
In the case of Cytb5, the precise identity of any cellular components that mediate its membrane delivery and integration are uncertain [10,13,14,28,29]. Nevertheless, we see an equally flexible pathway for Cytb5 biogenesis that can tolerate both PEGylation and enforced dimerization. We conclude that any cytosolic factors that facilitate Cytb5 integration can tolerate these species without substantial perturbation (Figure 1). Once at the membrane, the rate at which Cytb5OPG can be N-glycosylated appears to be substantially higher than that of Sec61βOPG (Supplementary Figure S2), and Cytb5OPG integration is also insensitive to trypsin treatment of the membranes (Supplementary Figure S5), further supporting the use of different pathways for integration into the ER [7,33,34]. Whether the flexibility of the post-translational pathways that underlie TA protein biogenesis is of physiological significance remains to be determined. However, our results are in good agreement with the finding that certain viral proteins are palmitoylated within their TMSs prior to membrane integration [43]. They are also consistent with the proposal that soluble SNAP-25 (25 kDa synaptosome-associated protein) is initially targeted to the ER membrane in a complex with the TA protein syntaxin [44], presumably via the binding of the complex to TRC40 prior to ER delivery [14]. It may even be the case that SGTA and TRC40 can bind to covalently modified TA proteins that are generated in vivo such as ubiquitinated species, consistent with reports of the co-translational ubiquitination of membrane proteins [45] and known links between the yeast homologue of TRC40, Get3 and the ubiquitin–proteasome system [46].
Online data
AUTHOR CONTRIBUTION
Pawel Leznicki conceived and carried out the experiments, analysed the data and contributed to the preparation of the manuscript. Jim Warwicker carried out the structural and thermodynamic modelling work and contributed to the preparation of the manuscript. Stephen High contributed to experimental design and the preparation of the manuscript for submission.
ACKNOWLEDGEMENTS
We thank Richard Kammerer for the pHisTrx expression vector, and Bernhard Dobberstein, Vincenzo Favaloro and Fabio Vilardi for the anti-TRC40 serum. We are grateful to Lisa Swanton and Martin Pool for their comments during manuscript preparation, and all of our other colleagues who provided reagents and advice.
FUNDING
This work was supported by the Wellcome Trust [grant number GR080877/Z/06/Z (Ph.D. studentship to P.L.)].
References
- 1.Kutay U., Hartmann E., Rapoport T. A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 2.Behrens T. W., Kearns G. M., Rivard J. J., Bernstein H. D., Yewdell J. W., Staudt L. M. Carboxyl-terminal targeting and novel post-translational processing of JAW1, a lymphoid protein of the endoplasmic reticulum. J. Biol. Chem. 1996;271:23528–23534. doi: 10.1074/jbc.271.38.23528. [DOI] [PubMed] [Google Scholar]
- 3.Borgese N., Gazzoni I., Barberi M., Colombo S., Pedrazzini E. Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell. 2001;12:2482–2496. doi: 10.1091/mbc.12.8.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B., Rapoport T. A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linstedt A. D., Foguet M., Renz M., Seelig H. P., Glick B. S., Hauri H. P. A C-terminally-anchored Golgi protein is inserted into the endoplasmic reticulum and then transported to the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5102–5105. doi: 10.1073/pnas.92.11.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brambillasca S., Yabal M., Makarow M., Borgese N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J. Cell Biol. 2006;175:767–777. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabu C., Schmid V., Schwappach B., High S. Biogenesis of tail-anchored proteins: the beginning for the end? J. Cell Sci. 2009;122:3605–3612. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabu C., Wipf P., Brodsky J. L., High S. A precursor-specific role for Hsp40/Hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J. Biol. Chem. 2008;283:27504–27513. doi: 10.1074/jbc.M804591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abell B. M., Rabu C., Leznicki P., Young J. C., High S. Post-translational integration of tail-anchored proteins is facilitated by defined molecular chaperones. J. Cell Sci. 2007;120:1743–1751. doi: 10.1242/jcs.002410. [DOI] [PubMed] [Google Scholar]
- 10.Colombo S. F., Longhi R., Borgese N. The role of cytosolic proteins in the insertion of tail-anchored proteins into phospholipid bilayers. J. Cell Sci. 2009;122:2383–2392. doi: 10.1242/jcs.049460. [DOI] [PubMed] [Google Scholar]
- 11.Abell B. M., Pool M. R., Schlenker O., Sinning I., High S. Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 2004;23:2755–2764. doi: 10.1038/sj.emboj.7600281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalbfleisch T., Cambon A., Wattenberg B. W. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic. 2007;8:1687–1694. doi: 10.1111/j.1600-0854.2007.00661.x. [DOI] [PubMed] [Google Scholar]
- 13.Favaloro V., Spasic M., Schwappach B., Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefanovic S., Hegde R. S. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 15.Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 16.Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H. D., Schwappach B., Weissman J. S. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battle A., Jonikas M. C., Walter P., Weissman J. S., Koller D. Automated identification of pathways from quantitative genetic interaction data. Mol. Syst. Biol. 2010;6:379. doi: 10.1038/msb.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonikas M. C., Collins S. R., Denic V., Oh E., Quan E. M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J. S., Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F., Brown E. C., Mak G., Zhuang J., Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bozkurt G., Stjepanovic G., Vilardi F., Amlacher S., Wild K., Bange G., Favaloro V., Rippe K., Hurt E., Dobberstein B., Sinning I. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J., Li J., Qian X., Denic V., Sha B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS ONE. 2009;4:e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mateja A., Szlachcic A., Downing M. E., Dobosz M., Mariappan M., Hegde R. S., Keenan R. J. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suloway C. J., Chartron J. W., Zaslaver M., Clemons W. M., Jr Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. U.S.A. 2009;106:14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamagata A., Mimura H., Sato Y., Yamashita M., Yoshikawa A., Fukai S. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells. 2010;15:29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- 26.Chang Y. W., Chuang Y. C., Ho Y. C., Cheng M. Y., Sun Y. J., Hsiao C. D., Wang C. Crystal structure of Get4/Get5 complex and its interactions with Sgt2, Get3 and Ydj1. J. Biol. Chem. 2010;285:9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chartron J. W., Suloway C. J., Zaslaver M., Clemons W. M., Jr Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leznicki P., Clancy A., Schwappach B., High S. Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 2010;123:2170–2178. doi: 10.1242/jcs.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favaloro V., Vilardi F., Schlecht R., Mayer M. P., Dobberstein B. Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J. Cell Sci. 2010;123:1522–1530. doi: 10.1242/jcs.055970. [DOI] [PubMed] [Google Scholar]
- 30.Ceppi P., Colombo S., Francolini M., Raimondo F., Borgese N., Masserini M. Two tail-anchored protein variants, differing in transmembrane domain length and intracellular sorting, interact differently with lipids. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16269–16274. doi: 10.1073/pnas.0508157102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson M. P., Billen L. P., Kim P. K., Andrews D. W. Cell-free analysis of tail-anchor protein targeting to membranes. Methods. 2007;41:427–438. doi: 10.1016/j.ymeth.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Borgese N., Righi M. Remote origins of tail-anchored proteins. Traffic. 2010;11:877–885. doi: 10.1111/j.1600-0854.2010.01068.x. [DOI] [PubMed] [Google Scholar]
- 33.Brambillasca S., Yabal M., Soffientini P., Stefanovic S., Makarow M., Hegde R. S., Borgese N. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005;24:2533–2542. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim P. K., Janiak-Spens F., Trimble W. S., Leber B., Andrews D. W. Evidence for multiple mechanisms for membrane binding and integration via carboxyl-terminal insertion sequences. Biochemistry. 1997;36:8873–8882. doi: 10.1021/bi970090t. [DOI] [PubMed] [Google Scholar]
- 35.Le Gall S., Neuhof A., Rapoport T. The endoplasmic reticulum membrane is permeable to small molecules. Mol. Biol. Cell. 2004;15:447–455. doi: 10.1091/mbc.E03-05-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mariappan M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R. J., Hegde R. S. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liou S. T., Wang C. Small glutamine-rich tetratricopeptide repeat-containing protein is composed of three structural units with distinct functions. Arch. Biochem. Biophys. 2005;435:253–263. doi: 10.1016/j.abb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 39.Janda C. Y., Li J., Oubridge C., Hernandez H., Robinson C. V., Nagai K. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 41.Wimley W. C., Creamer T. P., White S. H. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry. 1996;35:5109–5124. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- 42.Vilardi F., Lorenz H., Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J Cell Sci. 2011;124:1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochsenbauer-Jambor C., Miller D. C., Roberts C. R., Rhee S. S., Hunter E. Palmitoylation of the Rous sarcoma virus transmembrane glycoprotein is required for protein stability and virus infectivity. J. Virol. 2001;75:11544–11554. doi: 10.1128/JVI.75.23.11544-11554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel K., Cabaniols J. P., Roche P. A. Targeting of SNAP-25 to membranes is mediated by its association with the target SNARE syntaxin. J. Biol. Chem. 2000;275:2959–2965. doi: 10.1074/jbc.275.4.2959. [DOI] [PubMed] [Google Scholar]
- 45.Sato S., Ward C. L., Kopito R. R. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. J. Biol. Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- 46.Auld K. L., Hitchcock A. L., Doherty H. K., Frietze S., Huang L. S., Silver P. A. The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae. Genetics. 2006;174:215–227. doi: 10.1534/genetics.106.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heller H., Schaefer M., Schulten K. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal phase. J. Phys. Chem. 1993;97:8343–8360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.