Abstract
Anopheles stephensi is one of the major vectors of malaria in the Middle East and Indo-Pakistan subcontinent. Understanding the population genetic structure of malaria mosquitoes is important for developing adequate and successful vector control strategies. Commonly used markers for inferring anopheline taxonomic and population status include microsatellites and chromosomal inversions. Knowledge about chromosomal locations of microsatellite markers with respect to polymorphic inversions could be useful for better understanding a genetic structure of natural populations. However, fragments with microsatellites used in population genetic studies are usually too short for successful labeling and hybridization with chromosomes. We designed new primers for amplification of microsatellite loci identified in the A. stephensi genome sequenced with next-generation technologies. Twelve microsatellites were mapped to polytene chromosomes from ovarian nurse cells of A. stephensi using fluorescent in situ hybridization. All microsatellites hybridized to unique locations on autosomes, and 7 of them localized to the largest arm 2R. Ten microsatellites were mapped inside the previously described polymorphic chromosomal inversions, including 4 loci located inside the widespread inversion 2Rb. We analyzed microsatellite-based population genetic data available for A. stephensi in light of our mapping results. This study demonstrates that the chromosomal position of microsatellites may affect estimates of population genetic parameters and highlights the importance of developing physical maps for nonmodel organisms.
Keywords: genome sequence, malaria vector, polymorphic inversion, polytene chromosome, population structure
Anopheles stephensi Liston (Diptera: Culicidae) is an important malaria vector in the Persian Gulf and South Iran (Manouchehri et al. 1976; Vatandoost et al. 2006), in urban areas of the Indian subcontinent (Pant et al. 1981; Hati 1997), as well as in rural areas of North Pakistan and East Afghanistan (Rowland et al. 2002). This species is also an outstanding laboratory model system for malaria parasite transmission studies (Abraham et al. 2004; Baton and Ranford-Cartwright 2007; Bass et al. 2008). Three morpho-ecological variants have been identified within A. stephensi populations—“type, intermediate, and mysorensis”—which can be identified by the number of ridges on the egg (Sweet and Rao 1937; Rao et al. 1938; Subbarao et al. 1987). The type form is mainly an urban mosquito that breeds in temporary water pools, whereas the mysorensis and intermediate variants occupy predominantly rural areas (Nagpal et al. 2003). Although mysorensis is considered mostly zoophilic and is a poor vector of malaria in India (Subbarao et al. 1987), it was found to be a major malaria vector in the southern part of Iran (Manouchehri et al. 1976). In spite of the importance of bionomic differentiation for efficient malaria transmission, the genetic structure of the biological forms and populations of A. stephensi remains unclear, as tools are insufficient to precisely characterize the genetic variation. A study of PCR-based RFLP (PCR-RFLP) of 1512 bp of mitochondrial DNA cytochrome oxidase subunit I and II (COI-COII) and partial sequences of COI and COII genes has found extensive gene flow among the different forms of A. stephensi in Iran (Oshaghi et al. 2006). Analysis of the ribosomal DNA internal transcribed spacer 2 (rDNA-ITS2) and random amplification of polymorphic DNA loci in different populations of A. stephensi has demonstrated very little genetic variation among different populations suggesting that A. stephensi in Iran is a single species with different ecological forms in different zoogeographical zones (Djadid et al. 2006). Sequencing of the rDNA-ITS2 and domain-3 (D3) of rDNA loci of the A. stephensi type and mysorensis in India did not find any intraspecies sequence variation (Alam et al. 2008).
Microsatellites are informative markers for inferring population and taxonomic status of various organisms (Bruford and Wayne 1993). These markers have high levels of polymorphism and tend to evolve neutrally. They have been successfully used to study gene flow in natural populations of A. gambiae, A. funestus, and A. nili (Lehmann et al. 1996; Cohuet et al. 2005; Ndo et al. 2010). A set of 16 microsatellite markers has been developed for A. stephensi (Verardi et al. 2002). A study using 7 of these microsatellite markers has revealed high levels of genetic diversity within populations but not among geographically isolated populations in Pakistan. Deviation from Hardy–Weinberg expectations has been observed for 2 microsatellite loci in 21 tests (Ali et al. 2007). Another study of genetic variation at 8 of the 16 microsatellite loci observed a significant differentiation and a low level of gene flow among 3 ecological variants of A. stephensi in India (Vipin and Gakhar 2010). The study demonstrated that some microsatellites were in significant linkage disequilibrium, whereas other loci had a heterozygote deficit. Reduced recombination and selection can influence loci within polymorphic chromosomal inversions or near inversion breakpoints, resulting in estimates of gene flow that may depart significantly from those based on loci elsewhere in the genome (Lanzaro et al. 1998; Tripet et al. 2005). However, locations of the microsatellites on the chromosomes of A. stephensi were unknown, and interpretation of these differences among the loci was difficult.
Geographical and seasonal changes in chromosomal inversion frequencies have been interpreted as a signature of natural selection (Hoffmann et al. 2004; Balanya et al. 2006; Hoffmann and Rieseberg 2008). Therefore, polymorphic inversions are useful markers for studying ecological adaptations of various species, including malaria mosquitoes (Ayala et al. 2011; Sharakhova et al. 2011). An informative approach to determine the role of inversions in a population structure and species environmental adaptation is to study the genetic variation of molecular markers inside and outside of chromosomal rearrangements (Navarro and Barton 2003). In Diptera, contrasting patterns of polymorphism at microsatellite loci and inversions have been revealed. For example, some microsatellite loci located within known inversions did not display clinal profiles even if inversions had clinal profiles (Onyabe and Conn 2001; Kennington et al. 2003; Cohuet et al. 2005; Michel et al. 2005; Ayala et al. 2011). The number of genes under selection within inversions and the inversion age can be responsible for the observed patterns.
Polymorphic inversions have been employed to investigate the population structure and gene flow between ecological forms of A. stephensi. Breakpoints of at least 24 paracentric inversions have been mapped to polytene chromosomes of A. stephensi (Mahmood and Sakai 1984; Gayathri Devi and Shetty 1992). Seven inversions are located on 2R, 4 inversions on 2L, 3 inversions on 3R, and 10 inversions on 3L. No polymorphic inversions have been identified on the X chromosome. The 2Rb inversion was found to be highly polymorphic and widespread in natural populations of A. stephensi. Population studies provide support for the variation in inversion polymorphism according to geographic distribution of A. stephensi especially with respect to the 2Rb inversion (Coluzzi et al. 1973a; Mahmood and Sakai 1984; Gayathri Devi and Shetty 1992). For example, a chromosomal study revealed striking differences in the kinds and frequencies of paracentric inversions between the urban population in Karachi city and rural populations in Lahore and Kasur districts in Pakistan (Mahmood and Sakai 1984). The type and mysorensis forms showed significant differences in frequencies of inversion 2Rb (Coluzzi et al. 1973b). However, another study did not find any correlation between the inversion and the number of ridges on the egg (Suguna 1981).
In this study, we determined the locations of 12 microsatellite markers on a cytogenetic map of A. stephensi. All microsatellites hybridized to unique locations on autosomes both inside and outside polymorphic inversions. The chromosomal map of microsatellite markers and inversion breakpoints helps to understand better the genetic variation and differentiation in natural populations of A. stephensi.
Materials and Methods
Mosquito Strain and Chromosome Preparation
The Indian wild-type laboratory strain of A. stephensi was used in this study. To obtain the polytene chromosomes, ovaries were taken from half-gravid females and placed in Carnoy’s fixative solution (3 parts of ethanol: 1 part of glacial acetic acid by volume). Ovaries were kept at room temperature overnight before being stored at −20 °C. To obtain chromosomal slides, follicles of ovaries were separated in 50% propionic acid. Then, a cover slip was used to squash the follicles. The quality of slides and the banding pattern of polytene chromosomes were analyzed using an Olympus CX-41 phase contrast microscope (Olympus America Inc., Melville, NY). Slides then were dipped into liquid nitrogen, cover slips were removed, and slides were dehydrated in 50%, 70%, 90%, and 100% ethanol. Slides were air dried and used for further experiments.
Probe Preparation
Three approaches were utilized for the microsatellite probe preparation. First, microsatellites were directly amplified from the genomic DNA using previously designed primers (Verardi et al. 2002). Genomic DNA of A. stephensi was prepared using the Qiagen DNeasy Blood and Tissue Kit (Qiagen Science, Germantown, MD). Approximately 80–200 bp long fragments were amplified. Second, 4 microsatellites were ligated and cloned as a cluster of 2 or 3 repeats in the head-to-tail orientation in the plasmid pBluescript SK(+) (Agilent Technologies, Inc., Santa Clara, CA). Amplicons prepared in the first approach were treated with T4 DNA polymerase (SibEnzyme Ltd., Novosibirsk, Russia) for blunting the ends. Resulting fragments were treated with T4 DNA ligase (SibEnzyme Ltd.) at 14 °C for 1–2 h. The mixture was added to the dephosphorylated plasmid digested with restriction enzyme EcoRV (SibEnzyme Ltd.). Ligation was performed overnight. A ligation mixture was used to transform the XL1-Blue Escherichia coli strain (Agilent Technologies, Inc.). The blue-and-white screening revealed colonies containing insertions. The PCR screening revealed colonies containing a cluster of several repeats. Plasmids containing such clusters were sequenced. In this approach, 500–850 bp long fragments were amplified from the plasmid DNA using standard T7 and T6 primers (Fermentas, Inc., Glen Burnie, MD) and were labeled for in situ hybridization. Third, PCR products were obtained for 12 microsatellites using primers designed with the Primer3 program (Rozen and Skaletsky 2000) based on sequences identified by BLASTN in the genome assembly of A. stephensi (accession numbers: HQ328840–HQ328851). The genome assembly for A. stephensi was obtained using ×16 coverage of 454 shotgun and pair-end sequences at the Core Laboratory Facility of the Virginia Bioinformatics Institute of Virginia Tech. The size of these fragments was from 493 to 584 bp. PCR conditions were as follows: 94 °C for 5 min; 45 cycles of 94 °C for 45 s and of 50 °C for 45 s; 72 °C for 30 s; and 72 °C for 5 min. DNA was purified using the GE healthcare illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare UK Ltd., Buckinghamshire, UK). Probes were labeled using Cy3-AP3-dUTP or Cy5-AP3-dUTP (GE Healthcare UK Ltd.) fluorophores by a Random Primer DNA Labeling System (Invitrogen Corporation, Carlsbad, CA).
Fluorescence In Situ Hybridization and Mapping
Labeled probes were hybridized at 42 °C to A. stephensi polytene chromosome slides overnight. Then, slides were washed in 0.2 × Saline Sodium citrate (SSC, 0.03 M sodium chloride, and 0.03 M sodium citrate) at 42 °C and room temperature. Chromosomes were stained using YOYO-1 (Invitrogen Corporation), and slides were mounted in 1,4-Diazabicyclo[2.2.2]octane (DABCO) antifade solution. A Zeiss LSM 510 Laser Scanning Microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY) was used to detect fluorescent signals. Microscopic images were taken with a digital camera, and the locations of signals were determined using a standard cytogenetic photomap of A. stephensi (Sharakhova et al. 2006).
Results
In the current study, we used 3 approaches to map the set of microsatellites to the polytene chromosomes from ovarian nurse cells of A. stephensi. In the first approach, microsatellites were amplified from genomic DNA using specific primers, which were developed before (Verardi et al. 2002). All microsatellites were successfully amplified from the genomic DNA. However, because of the small size of the products (between 82 and 200 bp), the probes failed to get labeled for successful in situ hybridization. In the second approach, the PCR products of 4 microsatellites (A1, A10, B1, and C1) were cloned in a plasmid as a cluster of 2 or 3 repeats in the head-to-tail orientation. In this approach, 500–850 bp long fragments were amplified from the plasmid DNA using standard T7 and T6 primers and were labeled for in situ hybridization with chromosomes. Labeling and fluorescence in situ hybridization (FISH) of these microsatellite clones were successful. However, the probes hybridized either to multiple sites on chromosomes or to heterochromatic regions, probably because of the increased size of repetitive microsatellite motifs. In the third approach, we used recently obtained genomic sequence assembly of A. stephensi to design primers for 493–584 bp long PCR products that contain microsatellites of interest (Table 1). We attempted to find sequences homologous to the previously described 16 microsatellite loci (Verardi et al. 2002) in the A. stephensi genome by BLASTN. The BLASTN search did not yield either positive or unique hits for microsatellites E7*, A1, and D8T. Primers were designed but no PCR product was obtained for microsatellite H1. Therefore, PCR products were obtained for 12 of 16 microsatellites using primers designed based on the A. stephensi genome sequences (accession numbers: HQ328840–HQ328851). We successfully hybridized these probes to the polytene chromosomes from ovarian nurse cells of A. stephensi by FISH (Figure 1, Table 2).
Table 1.
Primers designed for the microsatellite loci using the Anopheles stephensi genome sequence
| Locus | Forward primer | Reverse primer | Size of the PCR product (bp) |
| E12 | GCGAGAGCGAGAGAGTGAGT | GGCGTTCAGTTCTGTGTGAA | 503 |
| B1 | GCATGGGTATGAGCCAAGTT | ATGAGTGTCGTCGTCCGTTT | 510 |
| D11 | ACCAGGGGTTCACAAATTCA | TGTACACAGGAGATAACGTGCAT | 493 |
| C1 | CACAGGAGATAACGTGCATTT | ACGCTCACACCACAAAACC | 505 |
| E7T | ACGAAGGAGCTGTCCGAGTA | GTCGTGTGGGGATAGTTGCT | 584 |
| B2 | CAGGAAAAGCGAGTGAAAGG | TGACAGCTGTGGAAGATTCG | 531 |
| H2ii | TCGATTCGAGGCATCTTTTC | CCTTCAATGTCCGTCACCTT | 564 |
| F10 | ATCCCACTACTGCACCCACT | CATCGCATGCTGATTGTTCT | 580 |
| A7 | CAGTTTTGCGCAGTAGTTGG | TTTCGCCTTTCATTCCTACG | 549 |
| A10 | CACGCAAGTAGGCTTTGACA | TTGAAATCGCTTCACACGAC | 513 |
| G1 | CAAGCGATTTTGGGGTAGAA | TACACCACCCACCCATAACC | 514 |
| B2N | CGCTCGTAGCTATTACGGATG | CAGGGGAAAATTGCTTTCAA | 570 |
Figure 1.
FISH performed on the chromosomes of Anopheles stephensi. Chromosomes counterstained with the fluorophore YOYO-1 and hybridized with fluorescently labeled probes Cy5 (blue) and Cy3 (red) are shown.
Table 2.
Locations of the Anopheles stephensi microsatellite markers on polytene chromosomes
| Microsatellite locus | Accessions for cloned sequences (with length in bp) | Accessions for assembled sequences (with length in bp) | Location; inside/outside inversions | |
| 1. | E12 | AF418586 (229) | HQ328840 (794) | 2R, B7; outside |
| 2. | B1 | AF418596 (150) | HQ328841 (847) | 2R, C8; inside |
| 3. | D11 | AF418581 (82) | HQ328843 (630) | 2R, 13C; inside |
| 4. | C1 | AF418583 (114) | HQ328844 (539) | 2R, 13C; inside |
| 5. | E7T | AF418593 (144) | HQ328842 (980) | 2R, 14C; inside |
| 6. | B2 | AF418588 (210) | HQ328845 (739) | 2R, 16C; inside |
| 7. | H2ii | AF418595 (189) | HQ328846 (619) | 2R, 17A; outside |
| 8. | F10 | AF418589 (175) | HQ328847 (700) | 2L, 24B; inside |
| 9. | A7 | AF418592 (289) | HQ328848 (985) | 2L, 22B; inside |
| 10. | A10 | AF418591 (186) | HQ328849 (666) | 3R, 30A; outside |
| 11. | G1 | AF412812 (154) | HQ328850 (702) | 3R, 35A; inside |
| 12. | B2N | AF418590 (134) | HQ328851 (814) | 3L, 40C; inside |
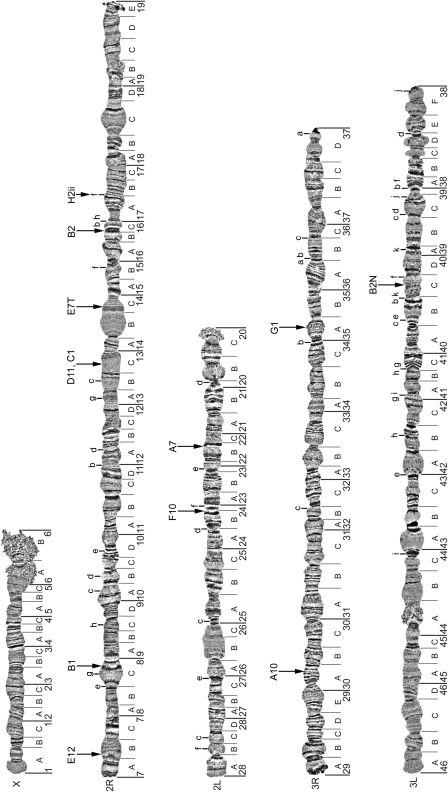
The A. stephensi chromosomal complement in ovarian nurse cells consists of 5 chromosomal arms: X, 2R, 3R, 3L. All 12 microsatellites hybridized to unique locations on autosomes; no hybridization to the X chromosome was detected. We have mapped these microsatellites to the polytene chromosomes of A. stephensi (Figure 2). Seven of 12 microsatellites hybridized to the largest arm 2R; 2 microsatellites localized to each of the 2L and 3R arms, and only 1 microsatellite hybridized to the 3L arm. At least 24 paracentric inversions have been described for A. stephensi (Mahmood and Sakai 1984; Gayathri Devi and Shetty 1992). We placed breakpoints of these inversions on the most recently developed polytene chromosome map of A. stephensi (Sharakhova et al. 2006) (Figure 2). Ten microsatellites were mapped inside of the previously described polymorphic inversions. Four of them were found inside the large 2Rb inversion, which is polymorphic in the Indian wild-type laboratory strain of A. stephensi (Figure 3). The locations of 2 microsatellites, D11 and C1, were cytogenetically indistinguishable; they hybridized to the same band in region 13C on the 2R arm.
Figure 2.
Physical map of 12 microsatellite markers on the Anopheles stephensi polytene chromosomes. Positions of inversion breakpoints are shown with lower-case letters.
Figure 3.
A photomicrograph of the polymorphic inversion 2Rb in a heterozygote state from the Indian wild-type laboratory colony of Anopheles stephensi.
Discussion
Among the 3 approaches to map microsatellite markers to chromosomes, the use of the genome sequence assembly of A. stephensi to amplify and hybridize microsatellites by FISH was successful (Figure 1). This success resulted because the genome sequence allowed us to design primers that would amplify 493–584 bp fragments, which are suitable for effective labeling by the random primer method. The developed microsatellite map (Figure 2) can greatly improve the understanding a population genetic structure of A. stephensi. Three ecological forms of A. stephensi—type, intermediate, and mysorensis, which differ in their habitat preferences and malaria transmission, have been described (Sweet and Rao 1937; Rao et al. 1938; Subbarao et al. 1987). Population analysis using rDNA-ITS2 and mitochondrial DNA loci have demonstrated a low level of gene flow among the 3 variants (Djadid et al. 2006; Oshaghi et al. 2006). Genetic variation at 7 microsatellite loci (F10, H2ii, E12, B1, A7, C1, and A10) has been studied in 153 individuals belonging to 3 populations in Pakistan (Ali et al. 2007). The study has found significant deviation from Hardy–Weinberg equilibrium due to heterozygote deficit in 2 of the 21 tests (loci E12 and B1). Exact tests of linkage disequilibrium showed no significant departure from equilibrium between these or any other locus pairs in any population after Bonferroni correction (Ali et al. 2007). Our study mapped both microsatellite markers, E12 and B1, to the region near the telomere on the 2R arm (Figure 2) suggesting the presence of genes under selection in this chromosomal region.
Another population genetic study utilized 8 microsatellite markers (F10, H2ii, E12, B1, G11, E7T, G1, and A10) to investigate the genetic isolation among the 3 morpho-ecological variants of A. stephensi in India (Vipin and Gakhar 2010). These microsatellites had shown diverse patterns of genetic variation. Locus E7T was found to be highly differentiated between mysorensis and type forms (FST = 0.890), between intermediate and type forms (FST = 0.629), and between mysorensis and intermediate forms (FST = 0.556). Moreover all 3 ecological forms had different nonoverlapping allele sizes for E7T locus (Vipin and Gakhar 2010). These data suggest high genetic differentiation among the forms at the locus E7T. We mapped the microsatellite E7T inside the 2Rb inversion (Figures 2 and 3), which is the most widespread inversion in natural populations. It has been shown that the type and mysorensis forms significantly differ in frequencies of inversion 2Rb (Coluzzi et al. 1973b). Therefore, the presence of this microsatellite inside the 2Rb inversion confirms a possible role of the inversion in differentiating A. stephensi populations. It is possible that the inversion 2Rb acts as a barrier to gene flow among the forms. If this is the case, then E7T should be in linkage disequilibrium with the inversion. Other microsatellites located inside the 2Rb inversion (B2, D11, and C1) have not been included in the population genetics study (Vipin and Gakhar 2010). Significant departure from Hardy–Weinberg equilibrium due to heterozygote deficits was found in intermediate and mysorensis forms across all loci, except for G1 and A10 in all 3 variants and F10 in intermediate (Vipin and Gakhar 2010). Both G1 and A10 are located on 3R, and F10 is located on 2L. Microsatellite G11 has not been mapped in our study. Interestingly, the other 4 microsatellites with significant departure from Hardy–Weinberg equilibrium (H2ii, E12, B1, and E7T) were mapped on 2R arm in our study (Figure 2). We have mapped microsatellites H2ii, B1, and E12 on the 2R arm outside of the 2Rb inversion; E12 and B1 are located close to the telomere. Linkage disequilibrium was found between H2ii and B1, E12 and B1 in mysorensis, and E12 and B1 in intermediate (Vipin and Gakhar 2010). However, the Bonferroni correction for the linkage disequilibrium has not been performed.
The genome sequence assembly of A. stephensi can also be used to discover and develop new microsatellite markers. Future studies of these microsatellites, together with inversion polymorphisms in the natural populations, will provide a better understanding of the population structure of A. stephensi and the effect of inversions on the behavior of microsatellites.
Funding
National Institute of Allergy and Infectious Diseases; National Institutes of Health (grant 1R21AI081023 to I.V.S).
Acknowledgments
We thank Diego Ayala for helpful comments on the manuscript and Melissa Wade for editing the text. Insightful comments from 2 anonymous reviewers helped to improve the manuscript.
References
- Abraham EG, Islam S, Srinivasan P, Ghosh AK, Valenzuela JG, Ribeiro JM, Kafatos FC, Dimopoulos G, Jacobs-Lorena M. Analysis of the Plasmodium and Anopheles transcriptional repertoire during ookinete development and midgut invasion. J Biol Chem. 2004;279:5573–5580. doi: 10.1074/jbc.M307582200. [DOI] [PubMed] [Google Scholar]
- Alam MT, Bora H, Das MK, Sharma YD. The type and mysorensis forms of the Anopheles stephensi (Diptera: Culicidae) in India exhibit identical ribosomal DNA ITS2 and domain-3 sequences. Parasitol Res. 2008;103:75–80. doi: 10.1007/s00436-008-0930-7. [DOI] [PubMed] [Google Scholar]
- Ali N, Hume JC, Dadzie SK, Donnelly MJ. Molecular genetic studies of Anopheles stephensi in Pakistan. Med Vet Entomol. 2007;21:265–269. doi: 10.1111/j.1365-2915.2007.00691.x. [DOI] [PubMed] [Google Scholar]
- Ayala D, Fontaine MC, Cohuet A, Fontenille D, Vitalis R, Simard F. Chromosomal inversions, natural selection and adaptation in the malaria vector Anopheles funestus. Mol Biol Evol. 2011;28:745–758. doi: 10.1093/molbev/msq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanya J, Oller JM, Huey RB, Gilchrist GW, Serra L. Global genetic change tracks global climate warming in Drosophila subobscura. Science. 2006;313:1773–1775. doi: 10.1126/science.1131002. [DOI] [PubMed] [Google Scholar]
- Bass C, Nikou D, Blagborough AM, Vontas J, Sinden RE, Williamson MS, Field LM. PCR-based detection of Plasmodium in Anopheles mosquitoes: a comparison of a new high-throughput assay with existing methods. Malar J. 2008;7:177. doi: 10.1186/1475-2875-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baton LA, Ranford-Cartwright LC. Morphological evidence for proliferative regeneration of the Anopheles stephensi midgut epithelium following Plasmodium falciparum ookinete invasion. J Invertebr Pathol. 2007;96:244–254. doi: 10.1016/j.jip.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Bruford M, Wayne R. Microsatellites and their application to population genetic studies. Curr Opin Gene Dev. 1993;3:939–943. doi: 10.1016/0959-437x(93)90017-j. [DOI] [PubMed] [Google Scholar]
- Cohuet A, Dia I, Simard F, Raymond M, Rousset F, Antonio-Nkondjio C, Awono-Ambene P, Wondji C, Fontenille D. Gene flow between chromosomal forms of the malaria vector Anopheles funestus in Cameroon, Central Africa, and its relevance in malaria fighting. Genetics. 2005;169:301. doi: 10.1534/genetics.103.025031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M, Di Deco M, Cancrini G. Chromosomal inversions in Anopheles stephensi. Parassitologia. 1973a;15:129–136. [PubMed] [Google Scholar]
- Coluzzi M, Di Deco M, Cancrini G. Further observations on the egg length in Anopheles stephensi in relation to chromosomal polymorphism. Parassitologia. 1973b;15:213–215. [PubMed] [Google Scholar]
- Djadid N, Gholizadeh S, Aghajari M, Zehi A, Raeisi A, Zakeri S. Genetic analysis of rDNA-ITS2 and RAPD loci in field populations of the malaria vector, Anopheles stephensi (Diptera: Culicidae): implications for the control program in Iran. Acta Trop. 2006;97:65–74. doi: 10.1016/j.actatropica.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Gayathri Devi K, Shetty J. Chromosomal inversions in Anopheles stephensi Liston—a malaria mosquito. J Cytol Genet. 1992;27:153–161. [Google Scholar]
- Hati AK. Urban malaria vector biology. Indian J Med Res. 1997;106:149–163. [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg L. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation. Annu Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgro CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 2004;19:482–488. doi: 10.1016/j.tree.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Kennington WJ, Gockel J, Partridge L. Testing for asymmetrical gene flow in a Drosophila melanogaster body-size cline. Genetics. 2003;165:667–673. doi: 10.1093/genetics/165.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzaro GC, Toure YT, Carnahan J, Zheng L, Dolo G, Traore S, Petrarca V, Vernick KD, Taylor CE. Complexities in the genetic structure of Anopheles gambiae populations in west Africa as revealed by microsatellite DNA analysis. Proc Natl Acad Sci U S A. 1998;95:14260–14265. doi: 10.1073/pnas.95.24.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Hawley W, Kamau L, Fontenille D, Simard F, Collins F. Genetic differentiation of Anopheles gambiae populations from East and West Africa: comparison of microsatellite and allozyme loci. Heredity. 1996;77:192–200. doi: 10.1038/hdy.1996.124. [DOI] [PubMed] [Google Scholar]
- Mahmood F, Sakai RK. Inversion polymorphisms in natural populations of Anopheles stephensi. Can J Genet Cytol. 1984;26:538–546. doi: 10.1139/g84-086. [DOI] [PubMed] [Google Scholar]
- Manouchehri A, Javadian E, Eshighy N, Motabar M. Ecology of Anopheles stephensi Liston in southern Iran. Trop Geogr Med. 1976;28:228–232. [PubMed] [Google Scholar]
- Michel AP, Guelbeogo WM, Grushko O, Schemerhorn BJ, Kern M, Willard MB, Sagnon N, Costantini C, Besansky NJ. Molecular differentiation between chromosomally defined incipient species of Anopheles funestus. Insect Mol Biol. 2005;14:375–387. doi: 10.1111/j.1365-2583.2005.00568.x. [DOI] [PubMed] [Google Scholar]
- Nagpal BN, Srivastava A, Kalra NL, Subbarao SK. Spiracular indices in Anopheles stephensi: a taxonomic tool to identify ecological variants. J Med Entomol. 2003;40:747–749. doi: 10.1603/0022-2585-40.6.747. [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton NH. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution. 2003;57:447–459. doi: 10.1111/j.0014-3820.2003.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Ndo C, Antonio-Nkondjio C, Cohuet A, Ayala D, Kengne P, Morlais I, Awono-Ambene PH, Couret D, Ngassam P, Fontenille D, et al. Population genetic structure of the malaria vector Anopheles nili in sub-Saharan Africa. Malar J. 2010;9:161. doi: 10.1186/1475-2875-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyabe DY, Conn JE. Genetic differentiation of the malaria vector Anopheles gambiae across Nigeria suggests that selection limits gene flow. Heredity. 2001;87:647–658. doi: 10.1046/j.1365-2540.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- Oshaghi M, Yaaghoobi F, Abaie M. Pattern of mitochondrial DNA variation between and within Anopheles stephensi (Diptera: Culicidae) biological forms suggests extensive gene flow. Acta Trop. 2006;99:226–233. doi: 10.1016/j.actatropica.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Pant CP, Rishikesh N, Bang YH, Smith A. Progress in malaria vector control. Bull World Health Organ. 1981;59:325–333. [PMC free article] [PubMed] [Google Scholar]
- Rao BA, Sweet WC, Subbarao AM. Ova measurements of A. stephensi type and A. stephensi var. mysorensis. J Malar Inst India. 1938;1:261–266. [Google Scholar]
- Rowland M, Mohammed N, Rehman H, Hewitt S, Mendis C, Ahmad M, Kamal M, Wirtz R. Anopheline vectors and malaria transmission in eastern Afghanistan. Trans R Soc Trop Med Hyg. 2002;96:620–626. doi: 10.1016/s0035-9203(02)90331-7. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sharakhova MV, Antonio-Nkondjio C, Xia A, Ndo C, Awono-Ambene P, Simard F, Sharakhov IV. Cytogenetic map for Anopheles nili: Application for population genetics and comparative physical mapping. Infect Genet Evol. 2011;11:746–754. doi: 10.1016/j.meegid.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Xia A, McAlister SI, Sharakhov IV. A standard cytogenetic photomap for the mosquito Anopheles stephensi (Diptera: Culicidae): application for physical mapping. J Med Entomol. 2006;43:861–866. doi: 10.1603/0022-2585(2006)43[861:ascpft]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Subbarao S, Vasantha K, Adak T, Sharma V, Curtis C. Egg-float ridge number in Anopheles stephensi: ecological variation and genetic analysis. Med Vet Entomol. 1987;1:265–271. doi: 10.1111/j.1365-2915.1987.tb00353.x. [DOI] [PubMed] [Google Scholar]
- Suguna SG. Inversion(2)R1 in Anopheles stephensi, its distribution and relation to egg size. Indian J Med Res. 1981;73(Suppl):124–128. [PubMed] [Google Scholar]
- Sweet W, Rao B. Races of Anopheles stephensi Liston, 1901. Ind Med Gaz. 1937;72:665–674. [PMC free article] [PubMed] [Google Scholar]
- Tripet F, Dolo G, Lanzaro GC. Multilevel analyses of genetic differentiation in Anopheles gambiae s.s. reveal patterns of gene flow important for malaria-fighting mosquito projects. Genetics. 2005;169:313–324. doi: 10.1534/genetics.104.026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatandoost H, Oshaghi MA, Abaie MR, Shahi M, Yaaghoobi F, Baghaii M, Hanafi-Bojd AA, Zamani G, Townson H. Bionomics of Anopheles stephensi Liston in the malarious area of Hormozgan province, southern Iran, 2002. Acta Trop. 2006;97:196–203. doi: 10.1016/j.actatropica.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Verardi A, Donnelly M, Rowland M, Townson H. Isolation and characterization of microsatellite loci in the mosquito Anopheles stephensi Liston (Diptera: Culicidae) Mol Ecol Notes. 2002;2:488–490. [Google Scholar]
- Vipin M, Gakhar S. Genetic differentiation between three ecological variants (‘type’,‘mysorensis’ and ‘intermediate’) of malaria vector Anopheles stephensi (Diptera: Culicidae) Insect Sci. 17:335–343. 2010 [Google Scholar]