Abstract
Motivation: Alternative splicing (AS) is a pre-mRNA maturation process leading to the expression of multiple mRNA variants from the same primary transcript. More than 90% of human genes are expressed via AS. Therefore, quantifying the inclusion level of every exon is crucial for generating accurate transcriptomic maps and studying the regulation of AS.
Results: Here we introduce SpliceTrap, a method to quantify exon inclusion levels using paired-end RNA-seq data. Unlike other tools, which focus on full-length transcript isoforms, SpliceTrap approaches the expression-level estimation of each exon as an independent Bayesian inference problem. In addition, SpliceTrap can identify major classes of alternative splicing events under a single cellular condition, without requiring a background set of reads to estimate relative splicing changes. We tested SpliceTrap both by simulation and real data analysis, and compared it to state-of-the-art tools for transcript quantification. SpliceTrap demonstrated improved accuracy, robustness and reliability in quantifying exon-inclusion ratios.
Conclusions: SpliceTrap is a useful tool to study alternative splicing regulation, especially for accurate quantification of local exon-inclusion ratios from RNA-seq data.
Availability and Implementation: SpliceTrap can be implemented online through the CSH Galaxy server http://cancan.cshl.edu/splicetrap and is also available for download and installation at http://rulai.cshl.edu/splicetrap/.
Contact: michael.zhang@utdallas.edu
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
In higher eukaryotes, a given transcribed locus can generate several mature mRNA isoforms via the process of alternative splicing (AS). AS is frequently a regulated mechanism, which coordinates the removal of the internal non-coding portions of the transcripts (introns) with the differential joining of the coding and 5′/3′ untranslated portions (exons). As a result, proteins with similar, different or antagonistic activities can be generated from a single genomic locus (Brett et al., 2002; Maniatis and Tasic, 2002). In addition, AS can lead to downregulation of gene expression by diverting some of the mRNA isoforms to the nonsense-mediated mRNA decay pathway (Lewis et al., 2003).
More than 90% of human genes express primary transcripts that undergo AS (Pan et al., 2008; Wang et al., 2008). Owing to the regulatory power of this process, an increasing number of studies are being directed at understanding AS regulation at the single-exon level (Castle et al., 2008; Johnson et al., 2003; Lewis et al., 2003; Wang et al., 2008). In general, researchers in the splicing regulation field have utilized comparative approaches to reveal tissue-specific (Relogio et al., 2005; Ule et al., 2005) or disease-related (Baumer et al., 2009) AS events. However, such methodologies have not been used to generate maps of AS activity within one cellular condition. The completion of such maps would add a higher level of resolution to transcriptome analysis, allowing precise quantification of exon inclusion levels within a population of related isoforms.
Until recently, systematic analysis of AS was done using expressed sequence tags (EST) (Gupta et al., 2004; Sorek et al., 2004; Xie et al., 2002) or specialized microarrays (Castle et al., 2008; Clark et al., 2002; Johnson et al., 2003; Pan et al., 2008). These techniques facilitated the discovery of a large number of alternative transcripts, and the extraction of distinctive features of alternatively spliced exons. Nevertheless, these techniques suffer from several limitations. ESTs are subject to cloning biases—especially towards the 3′ -end of transcripts—low coverage and insufficient robustness to allow reliable quantification. Likewise, the specificity of splicing microarrays is negatively affected by cross-hybridization with related mRNA molecules.
The development of deep-sequencing technologies provided an alternative to ESTs and microarrays for transcriptomic quantification. Two recent studies utilized single-end RNA-seq to analyze a series of human tissues. In Pan et al. (2008), the inclusion level of alternative exons was quantified as the percentage of the number of reads that match the two splice junctions formed by exon inclusion, over the splice junction formed by exon skipping. Wang et al. (2008) also utilized splice-junction reads for quantification of minor isoforms with different frequencies, as a function of the read coverage or RPKM (reads per kilobase of exon per million mapped reads. Although both studies demonstrated improved coverage relative to microarrays and ESTs, they utilized only isoform-specific reads, leaving out the majority of reads, which map to common exons of different isoforms.
An improved version of the deep-sequencing technique utilizes paired-end tags (Fullwood et al., 2009), which allows a significant gain of coverage and a reduction in read ambiguity through the generation of linked tag pairs that span longer stretches of sequenced template. This technology is especially suitable for AS profiling, because many exon-mapped tags are expected to span splice junctions, and these can be exploited to improve AS quantification.
Two recent methods exploit paired-end sequencing information for transcript quantification: Cufflinks (Trapnell et al., 2009), which is based on a previous RNA-seq model for single-end reads (Jiang and Wong, 2009) and Scripture (Guttman et al., 2010). Both can reconstruct transcript structures using directed graphs, and assign FPKM (fragment per kilobase of exon per million mapped reads) or RPKM values to every transcript, without relying on a reference genome. Cufflinks uses a mathematical model to identify alternatively spliced transcripts at each gene locus. Scripture employs a statistical segmentation model to distinguish expressed loci, and filters out experimental noise. Both methods were originally designed to identify and quantify full-transcript expression levels, but in our tests they appear not to be optimal for inferring local exon-inclusion ratios, presumably due to limited transcript coverage and sequencing noise.
Here we introduce SpliceTrap, a method to quantify local exon inclusion levels in paired-end RNA-seq data. SpliceTrap generates alternative splicing profiles for different splicing patterns, such as exon skipping, alternative 5′ or 3′ splice sites, and intron retention. It utilizes a comprehensive human exon database called TXdb (see Section 2) to estimate the expression level of every exon as an independent Bayesian inference problem. Unlike microarray-based methods, SpliceTrap relies on RNA-seq, and therefore it can determine the inclusion level of every exon within a single cellular condition, without requiring a background set of reads.
We tested SpliceTrap both by simulation and real data analysis. Compared to Cufflinks and Scripture, it demonstrated improved accuracy, robustness and reliability in quantifying a large fraction of AS activity. In particular, SpliceTrap is suitable for studying changes at the single-exon levels and it can facilitate the discovery of nearby cis-regulatory elements in diverse applications. SpliceTrap can be implemented online through the CSH Galaxy (Goecks et al., 2010) server http://cancan.cshl.edu/splicetrap and is also available for download and installation at http://rulai.cshl.edu/splicetrap/.
2 METHODS
2.1 Database construction
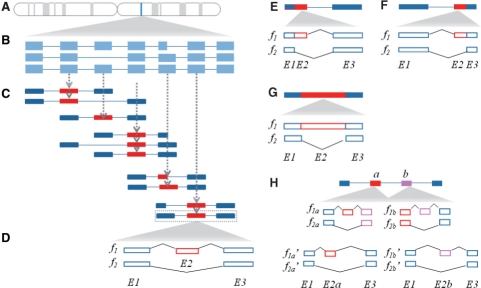
To quantify exon-inclusion levels, we designed an exon-trio database called TXdb. First, we captured all known transcripts encoded by every human gene (Fig. 1A and B), using annotations from RefSeq (Pruitt et al., 2007) (downloaded from the UCSC genome browser, hg18) and the EST-based AS database dbCASE (Zhang et al., 2007). Second, to account for every possible exon-skipping event, we subdivided each transcript set (i.e. encoded by the same gene) into exon trios, by sliding a 3-exon window along the transcript (Fig. 1C). In particular cases in which an exon was flanked by more than one assembly of flanking exons, every possible combination was represented in TXdb as a separate case. About 20% of the exons in TXdb are represented by more than one assembly of flanking exons (i.e. trios or duos). The pie charts in Supplementary Figure S1 show the types and numbers of exons represented by one or multiple assemblies.
Fig. 1.
TXdb assembly. (A) From a given gene expression locus (blue strip) (B) we extracted all the known transcript isoforms using available transcriptome annotations. (C) Using a 3-exon sliding window, we subdivided the transcript isoform population into exon trios, accounting for all known transcriptomic variability. Every exon trio is then used as an independent mappable unit, wherein the middle exon (red block) is queried for alternative splicing activity and the flanking exons (blue blocks) are treated as constitutive exons. (D) Two isoforms are constructed for each trio. Every exon-skipping event is represented by an inclusion isoform (f1) and a skipping isoform (f2) which comprise a pair of flanking exons (E1, E3) and an alternative exon (E2) present in f1 but not in f2. To examine additional types of alternative splicing, such as (E) alternative 5′ splice sites, (F) alternative 3′ splice sites, and (G) intron retention, we generated exon duos to compare extended isoforms to shortened isoforms. (H) SpliceTrap can detect consecutive alternative exons. When the alternative exon a or b is used as a flanking exon in an exon trio (f1a, f2a, f1b and f2b), if it is skipped, the exon trio will not pass the coverage cutoff, and thus will not be considered to be reliable. However, if substitute exon trios are present in TXdb (f1a′, f2a′, f1b′ and f2b′), when f1a′>f2a′ and f1b′<f2b′ or vice-versa (referring to their expression levels), exon a and b are mutually exclusive. Or, if f1a′<f2a′ and f1b′<f2b′, they are skipped together.
Next, we formatted the database to allow quantification of exon skipping (CA: cassette exon). We assumed that the middle exon was a cassette exon (E2) (regardless of whether it is annotated as alternatively or constitutively spliced) and the flanking exons (E1 and E3) are constitutive exons. Accordingly, each exon trio in TXdb was represented by two sequences (Fig. 1D): an inclusion isoform (f1) with all three exons; and a skipping isoform (f2) comprising the flanking exons only. The first and last exons from every transcript were filtered out, because transcriptomic variability in these areas is primarily due to alternative transcription initiation or polyadenylation, rather than to AS per se. By using TXdb as a mapping database, we can approach every exon as an independent case to estimate its AS level.
Based on this concept, we extended TXdb for detecting other types of AS patterns (AA: alternative 3′ splice site, AD: alternative 5′ splice site. IR: intron retention). To analyze AD (Fig. 1E) and AA (Fig. 1F) we compiled exon duos (rather than trios), setting f1 as the extended isoform (spliced via the proximal splice site), and f2 as the shortened isoform (spliced via the distal splice site). In addition, to account for IR (Fig. 1G), we defined f1 as the intron-retaining isoform, and f2 as the spliced isoform.
To estimate the extent to which the trio/duo assemblies can capture local AS variability, we downloaded the compendium of AS events from the AStalavista website (http://genome.crg.es/astalavista/) (Sammeth et al., 2008). For hg18, we counted the numbers of events that could be represented in the format of exon trio/duo. About 74.14% of the events from RefSeq accounted for single-exon AS events, all of which were covered by TXdb with a single exon trio/duo. When ESTs annotations were included, the percentage of covered events was 64.2%. It is important to note that some EST-based annotations might be of poor quality, as a result of noise in EST libraries (Sugnet et al., 2004). In SpliceTrap, ESTs are used only as mapping references, and not as a sole means to predict AS events; in this way, EST-based predictions can be confirmed if they align with a substantial number of RNA-seq reads. In addition, 9.82% of the RefSeq AS events in AStalavista (8.96% with ESTs) could be described by combining two entries in TXdb (e.g. consecutive CAs), and 2.9% (RefSeq only) or 1.97% (ESTs included) corresponded to assemblies of three or more exon trios/duos. The rest corresponded to more complex AS events that could not be handled directly by TXdb.
Complex AS events (i.e. involving two or more exon trios/duos) can be investigated by further comparing SpliceTrap quantifications. For example, Figure 1H illustrates two consecutive exons that are alternatively spliced. If either of them is skipped, the respective exon trios would not pass the coverage cutoff (see Section 2.3 for details). However, if annotations exist, the inclusion ratio may be quantified based on substitute exon trios available in TXdb (Supplementary Fig. S1). By comparing the inclusion ratios of both exons, one may detect if they are mutually exclusive or skipped together. It is important to note that the ability of SpliceTrap to detect complex AS events is limited, and depends on the availability of AS annotations to generate several trios/duos for each examined exon. For this reason, we recognize that some AS events may be overlooked, especially if they involve more than two consecutive alternative exons.
The final assembly of TXdb for hg18 comprises a total of 167 445 CA candidates, of which 11 812 have CA annotation, and the remaining 155 633 are annotated as constitutive exons (CS, to be examined whether they are in fact skipped). In addition, TXdb comprises 8667 AA, 4838 AD, and 1170 IR candidates, based on annotations from dbCASE or RefSeq. All together, SpliceTrap contains 224 995 exon trios (or duos) embodying transcript variability from 182 560 human exons (Supplementary Table S1).
Finally, we wish to bring to the reader's attention that since ~ 6% of the exons in TXdb are uniquely annotated in dbCASE, a slight bias towards the 3′ -end of the transcript may exist, especially for AAs, ADs and IRs, which are generally unique to dbCASE (Supplementary Table S1). TXdb is available online as part of the SpliceTrap package at http://rulai.cshl.edu/splicetrap/.
2.2 A Bayesian model to estimate inclusion ratios
In a paired-end RNA-seq experiment, a fragment is defined as a sequence segment encompassed between the first and last nucleotides of a read-pair. We assume that for each exon trio/duo, the positions of the mapped fragments follow a uniform distribution, and that their sizes follow a nearly normal distribution that depends upon the experimental protocol. Based on these assumptions, a fragment j can be described as a vector rj: (bj, sj), where bj and sj denote the beginning position and size of the fragment, respectively.
Then, for every exon trio (or exon duo), we define the set of all possible isoforms as F={f1, f2}, where f1 is an inclusion (or extended) isoform, and f2 is a skipping (or shortened) isoform (Fig. 1). The lengths and the relative expression levels of these isoforms are L={L1, L2} and E={e1, e2}. Accordingly, the probability of observing an isoform i, given the expression level E, can be written as:
| (1) |
Let m be the number of fragments R={rj, j=1, 2,···, m} that can be mapped to F. Given that for each fragment rj : (bj, sj), bj and sj are independent, the probability of observing rj, given an isoform fi, is:
| (2) |
where li is the effective length of fi (li=Li − sj + 1), and P(sj) is the probability of observing a fragment size sj in the experiment. Note that if only one end can be mapped to fi, then P(sj) is set as 1 to ignore fragment-size information.
For all isoforms in F, we can write P(rj|E) as:
| (3) |
So for the whole data, we can write:
| (4) |
Provided the prior distribution of E (see Section 2.4), a Bayesian posterior function can be written as:
| (5) |
Then, we can maximize P(E|R) to estimate the inclusion ratio e1 for every exon.
Note that throughout the text we refer to P(sj) as FSM (Fragment-Size distribution Model), and to P(E) as IRM (Inclusion-Ratio distribution Model), both of which are prior distributions and will be further described in Section 2.4.
2.3 Pipeline design
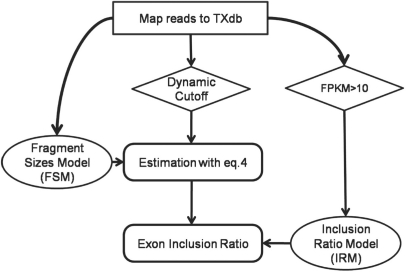
We designed a simple pipeline to run SpliceTrap (Fig. 2). We started by mapping the read-pairs onto TXdb. For this purpose, we independently aligned every read to the inclusion/skipping isoforms in TXdb using Bowtie (Langmead et al., 2009). Then, the fragments unambiguously mapped to single exons were used to build a FSM (Section 2.4).
Fig. 2.
SpliceTrap Pipeline. This chart illustrates the order and interrelation among the different tasks performed by SpliceTrap. Squares represent mapping steps; diamonds are filtering steps; ellipses are prior-information models; rounded-corner rectangles represent steps in the Bayesian model.
To filter out poorly covered exon trios, we applied a dynamic, exon-size-dependent cutoff strategy (Supplementary Fig. S2 and Section 3.1). Basically, we applied different coverage thresholds to every exon, such that the size of the exon and the coverage are inversely correlated. As an extreme example, an exon that is shorter than a read would need to be covered several times to be reliable. However, very long exons may be partially covered and still be reliable. Accordingly, we filtered exon trios with poorly covered flanking exons E1 and E3, but we did not require minimal coverage for exon E2 (i.e. the exon under consideration). This filtering method is intended to reduce noise resulting from rarely expressed transcripts, truncated transcripts, DNA contamination, wrongly mapped reads, etc., while avoiding unnecessary loss of information from exons with good coverage (details in Section 3.1).
Next, we maximized Equation 4 for every exon trio, utilizing all the mapped reads and the FSM to estimate the exon inclusion ratios. Finally, to reduce variability noise, we corrected the results with an inclusion-ratio distribution model (IRM) derived from high-confidence data (see Section 2.4).
2.4 Prior information models (FSM and IRM)
To generate FSMs, we took all the fragments uniquely mapped within the boundaries of constitutive exons (i.e. not spanning across splice junctions) and extracted the fragment sizes according to the positions of the reads. Finally, the occurrence of each fragment size was recorded to generate the distribution. The FSM distribution can be affected by variations in the experimental protocols. In previous studies, it was approximated as a normal distribution (Trapnell et al., 2010); however, to increase prediction accuracy, we chose to derive the FSMs directly from the dataset under study.
We generated IRMs for every type of splicing pattern separately (Supplementary Fig. S3). Essentially, after mapping RNA-seq data onto TXdb, we selected the highest covered exon trios (FPKM > 10) and estimated their inclusion ratios using Equation 4 (Supplementary Fig. S3A–D). As a control, we also generated IRMs with Cufflinks (Supplementary Fig. S3E–H). Notably, the distributions were very similar with both methods. To avoid overfitting, we smoothed the IRMs by fitting beta distributions (Supplementary Fig. S3I–L) to the histograms, which were then used in subsequent correction steps. Note that there is no specific IRM for CS, because every CS is examined as a potential CA with a CA IRM.
2.5 Metrics for accuracy testing
To test the ability of SpliceTrap to discriminate alternative from constitutive exons with inclusion ratios, we designed a series of metrics based on TXdb annotations. The assumption is that exons annotated as CA are enriched within the fraction of exons with inclusion ratio ir<1, and conversely, exons annotated as CS are included at approximately ir=1.
Cassette exon discovery rate (CAD): this metric is analogous to the Positive Predictive Value (PPV). Given an ir<1, all cassette exons above this ir are true positives (CAir denotes the number), whereas all constitutive exons above the same ir are false positives (CSir); then,
| (6) |
Constitutive exon discovery rate (CSD): by analogy to the False Positive Rate, above a certain ir(ir<1), all constitutive exons are false positives (the number of which is denoted by CSir<1), whereas all constitutive exons at ir=1 are true negatives (CSir=1), because these are reported as constitutively spliced, then CSDir can be written as:
| (7) |
Specificity (SP): using the definitions above, we calculate the specificity, which is nearly the converse case of CSD:
| (8) |
3 RESULTS
SpliceTrap is a tool specifically designed to detect local alternative splicing activity and quantify exon-inclusion ratios. Below, we present both simulations and data analysis demonstrating that SpliceTrap is highly accurate, reliable and robust, and we also compare it to state-of-the-art RNAseq analysis tools.
3.1 Simulation of inclusion-ratio quantification
We carried out a simulation in order to test the accuracy of SpliceTrap compared to other methods. A series of exon trios was generated by analogy to TXdb. For every exon trio, the flanking exons (E1 and E3) were fixed to a size of 120 nt (the average exon size in TXdb) whereas the middle exons (E2) varied in size from 9 to 500 nt. For these isoforms, we set expression levels based on the distribution of inclusion ratios. We selected the IRM for CA, which is the most common AS type (Supplementary Fig. S3I).
To simulate an RNA-seq experiment, we randomly fragmented the isoforms into overlapping fragments of sizes following a N(200, 152) distribution (by analogy to typical paired-end datasets), and preserved only the 75 (or 36) nt ends of each fragment as read-pairs. For every exon trio, the number of reads was adjusted to achieve exon coverage between 0 and 10. All together, we ran a total of 5555 simulations with different combinations of middle-exon size and coverage per tested method. For each simulation, 1000 repeats were made, and then the Pearson correlation coefficient (PCC) and the mean absolute error between the predicted and expected inclusion ratios were calculated for accuracy evaluation.
We evaluated five different methods (Table 1): a naïve method based on RPKM counts alone (Wang et al., 2008); Cufflinks (Trapnell et al., 2009); Scripture (Guttman et al., 2010); a maximum likelihood estimation model (MLE) (SpliceTrap using uniform IRM and FSM models); and SpliceTrap. (see Supplementary Method 1.2 for the implementations of Cufflinks and Scripture). Our simulation demonstrated that SpliceTrap can outperform all the other methods, with higher PCCs and lower mean errors (Table 1). We observed that using 36 nt or 75 nt reads, the average PCC of SpliceTrap was the highest (0.83–0.87) compared to RPKM (0.75–0.76), Cufflinks (0.78–0.83) and Scripture (0.61–0.72). In addition, we noticed that by adding prior information (i.e. the full Bayesian model) we obtained better results compared to MLE alone (0.79–0.84) providing evidence for the contribution of the prior-information models to the estimations. A similar pattern can be found in the mean absolute errors, where SpliceTrap attained the lowest errors (0.11–0.13) compared to the rest of the tools (0.11–0.25).
Table 1.
Simulation averages and standard deviations
| Method | Correlation coefficient |
Mean absolute error |
||
|---|---|---|---|---|
| 36 nt | 75 nt | 36 nt | 75 nt | |
| RPKM | 0.76 ± (0.18) | 0.75 ± (0.14) | 0.16 ± (0.11) | 0.17 ± (0.06) |
| Cufflinks | 0.83 ± (0.13) | 0.78 ± (0.12) | 0.11 ± (0.03) | 0.16 ± (0.03) |
| Scripture | 0.72 ± (0.22) | 0.61 ± (0.19) | 0.18 ± (0.10) | 0.25 ± (0.09) |
| MLE | 0.84 ± (0.14) | 0.79 ± (0.12) | 0.10 ± (0.05) | 0.15 ± (0.03) |
| SpliceTrap | 0.87 ± (0.14) | 0.83 ± (0.12) | 0.11 ± (0.05) | 0.13 ± (0.04) |
Next, we carried out simulations for the other three major types of AS patterns (Supplementary Table S3). We kept the same parameters, except for the IRM models, which were adjusted to each splicing type (Supplementary Fig. S3). In all cases, the error means and PCCs obtained were similar to those calculated using a CA IRM with 36 nt and 75 nt reads. (Supplementary Table S3), indicating that SpliceTrap can be used to investigate different splicing patterns.
Notably, these simulations revealed a general association between the prediction accuracy, the size and the coverage of the exons (Supplementary Fig. S4). Specifically, whereas smaller exons required higher coverages, low-coverage but larger exons achieved comparable accuracies. This resulted in a power-law-shaped surface, both for the mean error and the PCC, which was independent of the method used (Supplementary Fig. S4). We took advantage of this observation and designed a dynamic cutoff strategy accordingly. Using the simulation results shown in Supplementary Figure S4A, cutoff curves were derived at different PCCs ranging from 0.1 to 0.9 (Supplementary Fig. S2). For convenience, these dynamic cutoff curves are referred to as 0.1dc to 0.9dc throughout the text. Basically, for every exon in the data, we required a minimum coverage, depending on its size. Smaller exons required higher coverage, and larger exons required lower coverage. In short, this procedure should filter out most of the noise, and yet avoid unnecessary loss of exons that are partially covered, albeit by a sufficient number of reads.
3.2 Running SpliceTrap with RNA-seq data
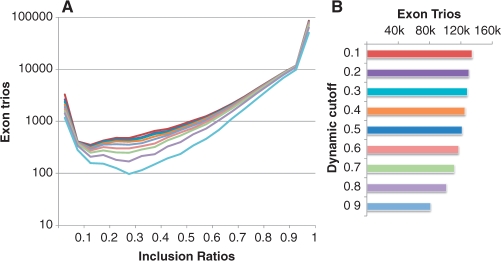
To experimentally test SpliceTrap and compare it to other methods, we generated more than 60 million 36 nt paired-end reads using HeLa cell RNA (see Supplementary Methods 1.1). We applied SpliceTrap to these data, using dynamic cutoffs from 0.1dc to 0.9dc (Fig. 3 and Supplementary Fig. S2). We noticed that in general, the distributions of the inclusion ratios had a ‘U’ shape (Fig. 3A) which was also observed using Cufflinks (Supplementary Fig. S3E–H) and in other studies based on ESTs (Peng et al., 2008). This means that in the sample analyzed, the exons tended to be highly included (i.e. constitutive) or fully skipped from the transcripts. Nevertheless, a substantial proportion of the exons showed intermediate inclusion levels, regardless of the stringency of the dynamic cutoff.
Fig. 3.
Distribution of inclusion ratios (A) and number of detected exon trios (B) based on RNA-seq data from HeLa cells (36 nt paired-end). The colors correspond to dynamic cutoffs from 0.1dc to 0.9dc.
In addition, we noticed that the number of selected exon trios did not vary dramatically within the range of lower dynamic cutoffs (0.1dc–0.6dc) although it dropped considerably above 0.7dc. Based on these observations, we selected low (0.6dc), medium (0.7dc) and high (0.8dc) stringency dynamic cutoffs for further analysis.
We ran Cufflinks and Scripture on the same datasets (provided with TXdb annotations), then we used three different cutoffs: FPKM = 1, FPKM = 2, FPKM = 10 for Cufflinks; and RPKM = 1, RPKM = 2, RPKM = 10 for Scripture. For every AS candidate, Cufflinks and Scripture reported the expression levels of the inclusion and skipping isoforms. We used these numbers to calculate inclusion ratios (See Supplementary Methods 1.2 for details).
3.2.1 Predicting known splicing patterns
We first tested the ability of SpliceTrap and other methods to detect known splicing events. In TXdb, every exon is assigned an annotation based on high-confidence ESTs and/or cDNAs (Supplementary Table S1). Our assumption is that exons annotated as cassette (CA) should be predominantly skipped (ir<1), whereas exons annotated as constitutive (CS) should be highly included at approximately ir=1.
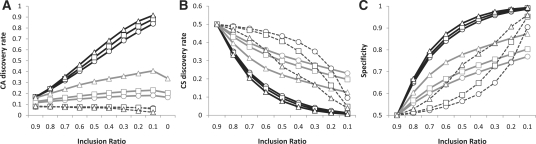
Based on this premise, we extracted all the exons (CA and CS) and their inclusion ratios from the above results. CS exons were examined in SpliceTrap as potential CAs using the CA IRM. Therefore, in this assay, SpliceTrap was ‘blind’ to TXdb annotations (CS or CA). To test the ability of the different methods to discriminate between CA and CS, we calculated the CAD, CSD and SP (see Section 2.5). Notably, for any selected threshold, SpliceTrap performed better at detecting low-included CAs, compared to the other tools (Fig. 4A). Whereas Cufflinks and Scripture detected CAs in any ir range to a similar extent, SpliceTrap was more efficient at identifying known CAs as the ir decreased. For example, at ir = 0.5, the SpliceTrap CAD value was ~ 0.6, and at ir = 0.1, it was almost 1. On the other hand, SpliceTrap detected CSs almost exclusively at high inclusion ratios (Fig. 4B), with CSD values below 0.1 at ir = 0.5 and ~ 0 at ir = 0.1. Accordingly, SpliceTrap exhibited higher specificity than Cufflinks and Scripture (Fig. 4C), achieving levels above 0.5 for ir<0.5.
Fig. 4.
Predicting known splicing patterns using 36 nt paired-end reads from HeLa cells. (A) The CAD, (B) CSD and (C) SP are shown as a function of the inclusion ratios (x-axis) for SpliceTrap (black lines), Cufflinks (gray lines) and Scripture (dashed lines). Each method was applied using low (circles), mid (squares) and high (triangles) cutoffs. (0.6dc, 0.7dc and 0.8dc for SpliceTrap, FPKM = 1, FPKM = 2 and FPKM = 10 for Cufflinks; RPKM = 1, RPKM = 2 and RPKM = 10 for Scripture).
In summary, SpliceTrap quantifications appear to be consistent with previous AS annotations; that is, most annotated CSs are included at around ir=1, whereas CAs are spread through the whole range of inclusion ratios (Fig. 4). Using a U-shaped CA IRM (Supplementary Fig. S3I) as prior information may have contributed to the prediction accuracy of SpliceTrap. Also, wrongly annotated CAs/CSs in TXdb or novel AS events might have affected the accuracy of the metrics.
3.2.2 Robustness and reliability of SpliceTrap
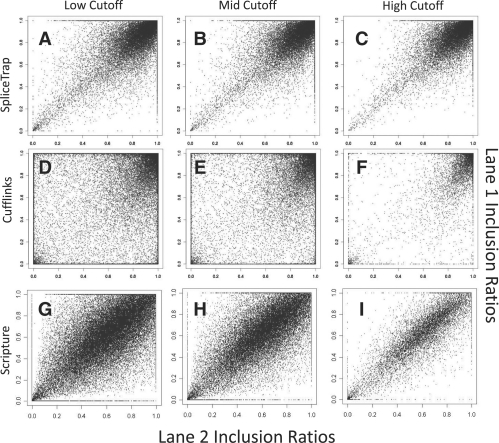
We evaluated the robustness of SpliceTrap estimations to technical variability among different replicates of a same experiment. To this end, we compared the results obtained from two independent RNA-seq lanes (36-nt paired-end) generated under the same conditions. The plots comparing the two lanes, using either SpliceTrap, Cufflinks or Scripture with different thresholds, are shown in Figure 5.
Fig. 5.
Robustness of the inclusion ratio estimations. The charts illustrate the correlations between the inclusion ratios calculated in two independent RNA-seq lanes (36 nt paired-end data from HeLa cells). (A–C) SpliceTrap at 0.6dc, 0.7dc and 0.8dc, (D–F) Cufflinks at FPKM = 1, FPKM = 2 and FPKM = 10, (G–I) Scripture at RPKM = 1, RPKM = 2 and RPKM = 10.
Consistently with Figure 4, SpliceTrap predicted most exons to be constitutively spliced (Fig. 5A–C). Using the stringent cutoff, 78% of the exons were included at ir≥0.9 in both experimental replicates. In contrast, only 30% of the exons for Cufflinks (Fig. 5D–F) and 23% for Scripture (Fig. 5G–I) showed ir≥0.9 in both replicates. Notably, SpliceTrap could reliably reproduce the results (PCC from 0.74 to 0.77) regardless of the threshold used. In contrast, Cufflinks achieved a maximum PCC of 0.7 (Table 2), but only when using the highest threshold (FPKM = 10). In other words, Cufflinks could achieve a reproducibility comparable to that of SpliceTrap, but only at the expense of the number of reported exons. whereas SpliceTrap reported 97 068 exons at PCC = 0.74, Cufflinks reported only 11 606 exons at PCC = 0.7. Scripture performed with high reproducibility (0.83–0.92), however the predicted inclusion ratios were averaged around 0.5. This would signify that most human exons are alternatively spliced (Fig. 5), which is not in agreement with previous transcript annotations.
Table 2.
Comparison of two replicates (36 nt paired-end reads)
| Method | Cutoff | Exons | PCC |
|---|---|---|---|
| Cufflinks | FPKM = 1 | 52 243 | 0.41 |
| Cufflinks | FPKM = 2 | 38 140 | 0.49 |
| Cufflinks | FPKM = 10 | 11 606 | 0.7 |
| SpliceTrap | 0.6dc | 97 068 | 0.74 |
| SpliceTrap | 0.7dc | 90 896 | 0.75 |
| SpliceTrap | 0.8dc | 80 052 | 0.77 |
| Scripture | RPKM = 1 | 70 466 | 0.83 |
| Scripture | RPKM = 2 | 49 816 | 0.87 |
| Scripture | RPKM = 10 | 14 022 | 0.92 |
Of note, Cufflinks achieved a high PCC in reproducing the expression levels of the inclusion (0.91) and skipping (0.81) isoforms (Supplementary Fig. S7), suggesting that Cufflinks has a higher robustness in detecting full transcript expression than AS.
Finally, we wanted to rule out dependencies between the net expression levels and the inclusion ratios detected by SpliceTrap and the other tools. To this end, we ranked all calculated inclusion ratios from one lane in Figure 5(A, D and G) according to the expression levels of the full-length exon trios reported by Cufflinks.
We observed that the inclusion ratios calculated with SpliceTrap were independent of the expression levels, with a constant average rate of ~ 0.95 (Supplementary Fig. S5). In contrast, the inclusion ratios calculated with Cufflinks decreased proportionally to the expression levels. The inclusion ratios calculated with Scripture were also constant; however, as in Figure 5, they averaged around 0.5, meaning that most exons in the data are viewed as alternatively spliced.
In conclusion, SpliceTrap can detect AS events in a more reliable and reproducible way, compared to Cufflinks and Scripture, which can be used to quantify local AS events, although with lower accuracy.
4 DISCUSSION
SpliceTrap is a computational tool fully dedicated to quantify major classes of AS activity based on paired-end RNA-seq data. Unlike other available tools, SpliceTrap focuses on quantifying local exon-inclusion ratios, instead of full-transcript expression levels. Rather than reporting background-based read densities, SpliceTrap utilizes Bayesian statistics to summarize exon-inclusion probabilities derived from every single read-pair. For this reason, SpliceTrap is also insensitive to transcript expression levels.
SpliceTrap was specifically designed to accurately quantify alternative splicing at the single exon level. To achieve this goal, we started by describing the problem with a statistical model based on exon trios/duos, instead of full transcripts. To reduce the number of false positives and yet minimize the loss of information, we applied dynamic cutoffs derived from simulation, rather than using fixed cutoffs. Finally, we adjusted the results using specific inclusion-ratio models for different AS patterns. In theory, full-transcript quantification tools like Cufflinks and Scripture can also be used to calculate inclusion ratios with TXdb annotations (Supplementary Method 1.2), However, these tools were originally designed and optimized for transcript-level estimation, and our analysis indicates that they are less accurate than SpliceTrap for the specific problem of calculating exon inclusion ratios.
We have shown that it is possible to approach every exon as a separate problem, and yet quantify its inclusion ratio without knowledge of the full transcript structure. Given that our quantitative units are the exons, we can disregard information from distant exons (hence reducing complexity and noise), though it is certainly important for transcript-level quantification.
SpliceTrap can detect different splicing patterns. Even though the original algorithm was developed to detect single cassette exons, we have adapted it to other types of AS, such as alternative 3′/5′ splice sites and intron retention. In combination, these patterns account for 75% of all known human AS events in RefSeq (Sammeth et al., 2008). Additionally, some of the complex AS patterns involving multiple exon trios/duos were also detected by SpliceTrap (Supplementary Fig. S6). However, SpliceTrap's ability to detect very complex AS patterns may be limited, depending on the annotations present in TXdb.
At this stage, SpliceTrap does not offer an option for ab initio exon prediction. We chose to focus on a set of ~200 000 well characterized exons, and designed a transcript database (TXdb) as a mapping reference. In this way, we sought to reduce ambiguities generated by rarely expressed isoforms, especially during the mapping procedure. Because TXdb is a collection of exon trios/duos, in the future it can be expanded by adding newly discovered or predicted splice juctions, such as those derived by splice-junction mappers like TopHat, or novel exons predicted by gene-prediction tools, e.g. GENSCAN (Burge and Karlin, 1997). In this way, the depth and sensitivity of SpliceTrap can be enhanced.
SpliceTrap is based on the assumption that the reads are uniformly distributed within the exon trio/duo. Although the uniformity in a small region is presumably a better assumption than in a full transcript, this factor will still bias the results and should be considered in future versions of the model.
SpliceTrap's running time depends on the number of mapped reads. For instance, using a 2 GHz AMD CPU with 8 GB of memory, the running time was ~3 h for one lane (20×2 million 36 nt paired-end reads), and ~12 h for three lanes. Less than 500 MB memory and 10-30 GB of hard disk space were needed. SpliceTrap is easy to operate and requires a small number of input parameters, reducing the user's setup time. SpliceTrap also supports SUN Grid engine (SGE) qsub for parallel computing. It can be implemented online through the CSH Galaxy server (http://cancan.cshl.edu/splicetrap) or downloaded at http://rulai.cshl.edu/splicetrap/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Assaf Gordon for providing access and support to the CSH Galaxy server and Chaolin Zhang for providing dbCASE. Thanks also to Chaolin Zhang and Justin Kinney for helpful discussions.
Funding: National Institutes of Health (GM74688 to M.Q.Z.).
Conflict of Interest: none declared.
REFERENCES
- Baumer D, et al. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett D, et al. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Castle JC, et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, et al. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, et al. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, et al. Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics. 2004;20:2579–2585. doi: 10.1093/bioinformatics/bth288. [DOI] [PubMed] [Google Scholar]
- Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, et al. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Pan Q, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Peng T, et al. Functional importance of different patterns of correlation between adjacent cassette exons in human and mouse. BMC Genomics. 2008;9:191. doi: 10.1186/1471-2164-9-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, et al. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relogio A, et al. Alternative splicing microarrays reveal functional expression of neuron-specific regulators in Hodgkin lymphoma cells. J. Biol. Chem. 2005;280:4779–4784. doi: 10.1074/jbc.M411976200. [DOI] [PubMed] [Google Scholar]
- Sammeth M, et al. A general definition and nomenclature for alternative splicing events. PLoS Comput. Biol. 2008;4:e1000147. doi: 10.1371/journal.pcbi.1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, et al. A non-EST-based method for exon-skipping prediction. Genome Res. 2004;14:1617–1623. doi: 10.1101/gr.2572604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, et al. Transcriptome and genome conservation of alternative splicing events in humans and mice. Pac. Symp. Biocomput. 2004:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, et al. Computational analysis of alternative splicing using EST tissue information. Genomics. 2002;80:326–330. doi: 10.1006/geno.2002.6841. [DOI] [PubMed] [Google Scholar]
- Zhang C, et al. Dual-specificity splice sites function alternatively as 5′and 3′splice sites. Proc. Natl Acad. Sci. US A. 2007;104:15028–15033. doi: 10.1073/pnas.0703773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.