Abstract
Summary: While the R software is becoming a standard for the analysis of genetic data, classical population genetics tools are being challenged by the increasing availability of genomic sequences. Dedicated tools are needed for harnessing the large amount of information generated by next-generation sequencing technologies. We introduce new tools implemented in the adegenet 1.3-1 package for handling and analyzing genome-wide single nucleotide polymorphism (SNP) data. Using a bit-level coding scheme for SNP data and parallelized computation, adegenet enables the analysis of large genome-wide SNPs datasets using standard personal computers.
Availability: adegenet 1.3-1 is available from CRAN: http://cran.r-project.org/web/packages/adegenet/. Information and support including a dedicated forum of discussion can be found on the adegenet website: http://adegenet.r-forge.r-project.org/. adegenet is released with a manual and four tutorials totalling over 300 pages of documentation, and distributed under the GNU General Public Licence (≥2).
Contact: t.jombart@imperial.ac.uk
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The free software R (R Development Core Team, 2011) is becoming a standard for the analysis of genetic data, offering a wealth of packages dedicated to population genetics (Jombart, 2008; Paradis, 2010), phylogenetics (Paradis et al., 2004; Schliep, 2011) or genome-wide association studies (Aulchenko et al., 2007; Clayton and Leung, 2007). Until recently, classical genetic marker data such as microsatellites could be analyzed using standard tools and personal computers. However, the increasing availability of genomic sequence data has challenged both the tools and the ressources needed to carry such analyses. While some specific packages have been developed for human association studies (Aulchenko et al., 2007; Clayton and Leung, 2007), more general tools for the analysis of the genetic structure of biological populations are needed. In this article, we introduce new tools implemented in the R package adegenet (Jombart, 2008) which allow large genomic datasets (e.g. hundreds of individuals typed for hundreds of thousands SNPs) to be analyzed using standard personal computers. As an illustration, we show how a new implementation of the discriminant analysis of principal components (DAPC) (Jombart et al., 2010) can be used to identify structuring alleles from genomic data with minimum computing resources.
2 DESCRIPTION
The sheer size of genomic sequence data often precludes their analysis using standard personal computers. While studies focusing on genetic diversity can reduce the size of the analyzed datasets by considering biallelic SNPs only, the subsequent amount of data often remains considerable and can require prohibitive amounts of random access memory (RAM). To address this issue, we implemented a new data representation which codes each biallelic SNP using a single bit. While such coding is not readily possible in R, the new class genlight internally codes chunks of 8 SNPs using a single byte, resulting in drastic compression of the data. For instance, 50 individuals genotyped for 1 000 000 SNPs classically coded as characters would require ~380 MB of RAM, as opposed to 6 MB using genlight objects. This new coding scheme is also about eight times more compact than other available classes for representing SNP data such as DNAbin (Paradis et al., 2004) or snp.matrix (Clayton and Leung, 2007). A further advantage of genlight is the ability to accomodate any ploidy in the data, even allowing for the ploidy to vary across individuals. The features of the class genlight are fully documented in a tutorial accessible from R by typing vignette("adegenet-genomics").
While the bit-level coding of SNP data is undoubtedly memory efficient, it also makes the internal structure of the objects far more complex. Considerable efforts have been made to simplify the handling and analysis of genlight objects, whose manipulation is very close to matrices of individual allele frequencies. The entire genlight class has been replicated in C, which allowed for optimizing recurrent operations such as conversions from and to integers. Dedicated functions (‘accessors’) facilitate the access and modification of information while preventing the user from interacting directly with the complex internal structure of the objects. As a result, genlight objects act as ‘black boxes’ which resemble matrices of individual allele frequencies, albeit storing the information more efficiently. Basic functions such as mean and variance of SNP frequencies have also been implemented in order to facilitate the development of future dedicated tools.
Beyond the need for efficient data storage, the analysis of genome-wide SNP data also requires significant computing power. Fortunately, most computers now possess processors with multiple cores, which can be used to partition important tasks into several smaller operations executed simultaneously by the different cores. This approach can lead to appreciable reductions in computational time and is most useful for analyzing large datasets. By default, most procedures implemented for genlight objects achieve parallelization using the package multicore (currently available on linux and MacOSX systems), although this can be disabled by the user. For instance, the new implementations of PCA (function glPca) and DAPC (dapc, see example) by default use compiled C code and parallelized computations, while never requiring more than two genomes to be represented as integers at a time. In some cases, this approach turns out to be even faster than other classical implementations of PCA (Supplementary Table S1).
Data interoperability can be a critical issue when large datasets are considered. Therefore, we made sure that genome-wide SNP data could be imported from standard formats into genlight objects as simply as possible. First, genlight objects can be created from lists or matrices of individual allele frequencies. Data can also be imported from the widely used software PLINK (Purcell et al., 2007), which has defined a standard format for storing diploid SNP data. Alternatively, data can also be imported from adegenet's own format (‘.snp’ files), which can accomodate any degree of ploidy and can store any meta-information such as individual group membership or positions of the SNPs. Finally, SNPs can also be directly extracted from aligned DNA sequences stored as FASTA files. Importantly, all these functions allow for processing the data by chunks of a few individuals, which allows for minimizing the RAM required for reading the data in.
3 EXAMPLE
We illustrate how a new implementation of DAPC for genlight objects can be used to identify structuring alleles from genome-wide SNP data. After loading the package, we simulate 1 001 000 SNPs for two groups of 50 individuals using glSim. The first 1 000 000 SNPs have similar distributions for both groups, whereas these distributions differ in the last 1000 SNPs.
> library(adegenet)
> x <- glSim(n.ind=100, n.snp.nonstruc=1e6, n.snp.struc=1e3)
We then apply DAPC to these data, choosing to retain 20 principal components in the prior dimension-reduction step.
> dapc1 <- dapc(x, n.pca=20)
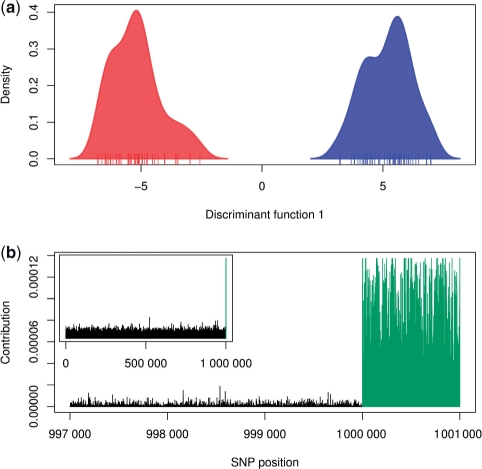
Despite defavourable noise/signal ratio, DAPC discriminates very neatly the two groups of individuals (Fig. 1a). Interestingly, it also clearly identifies the structuring SNPs (Fig. 1b). Despite its simplicity, this example suggests that DAPC could be a useful tool for identifying structuring alleles from genome-wide SNP data.
Fig. 1.
DAPC of simulated data (see text). (a) Density of individual scores on the first discriminant function, with groups represented in red and blue. (b) SNP contribution to the separation of the groups; the last (structured) 1000 SNPs are coloured in green; the last 4000 SNPs are represented in the main plot, while the figure corresponding to all SNPs is shown in inset.
4 CONCLUSION
adegenet 1.3-1 provides new tools for the analysis of genome-wide SNP data using standard personal computers. As the availability of genomic data increases faster than computing resources, efficient data representation and parallel computation represent viable alternatives to the mere increase of raw computing power. As such, we hope that the new class genlight and the associated tools will make a significant contribution to taking population genetics studies into the genomic era and encourage the development of new dedicated methods.
Supplementary Material
ACKNOWLEDGEMENT
We thank David Aanensen, Lucy Weinert, Christophe Knecht and Lee Li-Foh for interesting discussions about genomic data, and two anonymous reviewers for their useful comments.
Funding: ERC Grant (P33585) and NIGMS MIDAS Programme to Neil Ferguson.
Conflict of Interest: none declared.
REFERENCES
- Aulchenko YS, et al. Genabel: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Clayton D, Leung H-T. An R package for analysis of whole-genome association studies. Hum. Hered. 2007;64:45–51. doi: 10.1159/000101422. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, et al. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- Paradis E, et al. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Purcell S, et al. Plink: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.