This paper describes a web environment dedicated to noncoding RNA prediction, annotation, and analysis. Tools for comparison, visualization, editing, and export of noncoding RNA candidates are provided in this environment.
Keywords: bioinformatics, noncoding RNA, prediction, annotation
Abstract
The annotation of noncoding RNA genes remains a major bottleneck in genome sequencing projects. Most genome sequences released today still come with sets of tRNAs and rRNAs as the only annotated RNA elements, ignoring hundreds of other RNA families. We have developed a web environment that is dedicated to noncoding RNA (ncRNA) prediction, annotation, and analysis and allows users to run a variety of tools in an integrated and flexible manner. This environment offers complementary ncRNA gene finders and a set of tools for the comparison, visualization, editing, and export of ncRNA candidates. Predictions can be filtered according to a large set of characteristics. Based on this environment, we created a public website located at http://RNAspace.org. It accepts genomic sequences up to 5 Mb, which permits for an online annotation of a complete bacterial genome or a small eukaryotic chromosome. The project is hosted as a Source Forge project (http://rnaspace.sourceforge.net/).
INTRODUCTION
Noncoding RNAs (ncRNA) are RNAs that are transcribed, but not translated into protein. They include well-characterized transfer RNAs and ribosomal RNAs, snRNAs, snoRNAs, and miRNAs, as well as a plethora of new ncRNAs that have been shown to play major roles in the cellular processes of all living organisms (Amaral et al. 2008; Jacquier 2009; Ponting et al. 2009; Waters and Storz 2009; Liu and Carnilli 2010). Even though a large number of genomes have now been sequenced, the number and the diversity of ncRNAs remain largely unknown. The existence of pervasive conserved elements and the extensive expression of transcripts in the noncoding regions of genomes suggest that an important number of ncRNAs of unknown function and structure remain to be identified (Hüttenhofer et al. 2005). In Eukaryotes, hundreds of thousands of (often short) noncoding transcripts are expressed from the intergenic regions, introns and antisense strands of protein-coding genes (Carninci et al. 2005; van Bakel et al. 2010). Pervasive transcription also occurs in Bacteria (Toledo-Arana et al. 2009) and Archaea (Jäger et al. 2009). Thousands of complete genomes are currently being sequenced, and many more are forthcoming. In such a context of data accumulation, the ability to differentiate automatically known RNA families and the development of additional computational tools dedicated to the improvement of the completeness of the catalog of functional elements are required.
In spite of considerable improvements in the development of ncRNA detection software in the recent years, the prediction and annotation of ncRNAs still remain challenging tasks (Menzel et al. 2009). The most effective ncRNA detection software are distributed as stand-alone applications (Rivas and Eddy 2001; Klein and Eddy 2003; Livny et al. 2005; Washietl et al. 2005), some of them address only specific classes of RNAs (Lowe and Eddy 1997; Laslett and Canbäck 2004; Schattner et al. 2006; Lagesen et al. 2007) or organisms (Klein et al. 2002; Schattner 2002; Upadhyay et al. 2005; Larsson et al. 2008). The integration of these approaches for consistent ncRNA prediction and annotation remains the realm of bioinformatics experts. As a result, ncRNA annotation in genome databases lags behind protein-coding gene annotation. This is also due in large part to the relative difficulty of identifying ncRNA-encoding loci, which requires unique bioinformatic approaches. Integrating the diversity of ncRNA prediction and annotation tools in a unique environment is highly desirable for biologists interested in the prediction, annotation, and basic analysis of known and new ncRNA genes from genomic sequences.
The most ambitious attempt at providing tools for RNA prediction and annotation is that of the RFAM database (Gardner et al. 2009). RFAM 10.0 hosts over 1446 ncRNA families and allows biologists to perform similarity searches against this database. Users can also access precomputed full-genome annotations. The limitations of the RFAM website are the small size limit for online searches and the restriction of matches to existing RNA families, which excludes a large number of ncRNAs for which no alignment is available or submitted to RFAM. Other ncRNA databases exist that focus on a single class of molecules such as miRBase (Griffiths-Jones et al. 2008), on one class for a set of organisms such as the snoRNABase (Lestrade and Weber 2006), or on one genome such as ASRP (http://asrp.cgrb.oregonstate.edu/db/) or that aim at providing an exhaustive catalog of ncRNAs such as NONCODE (Liu et al. 2005) or fRNAdb (Kin et al. 2006). Besides RFAM, several alternative computational methods exist that perform ncRNA prediction in genomic sequences. These can be classified into two types, namely, those that aim at searching for ncRNA homologs using conserved sequence and structure characteristics and those that aim at discovering new ncRNA families. Methods of the former type use sequence/structure alignments in order to identify key conserved motifs involved in the molecule structure. Alignments are then processed and modeled in several ways, for instance, with position weight matrices in ERPIN (Gautheret and Lambert 2001) or covariance models in INFERNAL (Nawrocki et al. 2009). These types of methods also include programs dedicated to one type of ncRNA, such as tRNAScan-SE (Lowe and Eddy 1997) and RNAmmer (Lagesen et al. 2007). Programs such as RNAMOTIF (Macke et al. 2001), PatScan (Dsouza et al. 1997), and DARN! (Zytnicki et al. 2008) provide users with a programming language allowing the description of any ncRNA structure. Methods of the latter type, namely, programs aiming at de novo ncRNA identification, use diverse and often complementary approaches, including sequence composition bias analysis and the identification of significant stable and conserved secondary structure regions in multiple sequence alignments. Successful ncRNA searches using these approaches have been reported in Archaea (Klein et al. 2002; Schattner 2002), Bacteria (Rivas et al. 2001; Livny et al. 2006; Geissmann et al. 2009), and Eukaryotes (Mendes et al. 2009; Li et al. 2010). More recently, the CMfinder tool (Yao et al. 2006) was used to predict large sets of new ncRNA/motifs signatures in Bacteria (Weinberg et al. 2007, 2009).
The absence of a genuine generic ncRNA prediction method and the expertise required to detect ncRNAs based on sequence and structure conservation have led us to develop a web environment, RNAspace.org, that permits users to perform all of these tasks in an integrated fashion. This web environment is dedicated to biologists who are involved in genome annotation projects with a particular interest in known and new ncRNA families. RNAspace.org enables these users to run a variety of ncRNA gene finders in an integrated way and to explore results using dedicated tools for the comparison, visualization, filtering, alignment, and editing of putative ncRNAs. This environment is collaboratively developed and uses open-source software. Thus, computer scientists developing new methods on these topics may be interested in using the web environment to integrate their methods.
RESULTS
The ncRNA prediction and annotation activities are organized into three main steps corresponding to a typical annotation process. In the first step, users upload the sequence(s) to be analyzed, along with related information. In the second step, users select ncRNA gene finders. It will be possible to return to this step later on for selecting other gene finders or changing their parameters. The third step includes a set of functionalities to assess predicted ncRNA genes with regards to their context, annotation, conservation, and secondary structure. Results obtained from an analysis can be saved in various standard formats and are kept 14 d.
Data upload step
The sequences to be analyzed are entered as a fasta or multi-fasta file. One or more sequence files can be uploaded, provided that the total length is <5 Mb. As some software require knowledge of the phylogenetic domain (Archae, Bacteria, Eukarya) of the organism under analysis, this information must be entered. Other optional information can be entered. An e-mail address is required in order to receive the link giving access to the results of the prediction step.
ncRNA prediction step
After sequence uploading, users select ncRNA gene finders. These gene finders were chosen so that a variety of complementary approaches can be used, modeling different characteristics of ncRNAs. Programs are organized into three main groups: homology search, comparative analysis, and ab initio prediction. The first class of tools identifies genes that belong to known families of ncRNAs, whereas the two latter classes aim at finding new families. The list of tools currently available is presented in Table 1.
TABLE 1.
List of software available in the predict step
Homology search tools rely on the knowledge of a signature for a given ncRNA family. This signature contains information at the sequence and/or secondary structure level. For sequence level searches, RNAspace compares user sequences to several databases using the sequence alignment programs BLASTN (Altschul et al. 1990) and YASS (Noé and Kucherov 2005). Both programs implement a heuristic approach for local similarity search. BLASTN is based on contiguous k-mers, while YASS is based on spaced-seeds that are known to achieve higher sensitivity. However, BLASTN is faster than YASS. When aligning the query sequence against all sequences of a given ncRNA database with BLASTN or YASS, it appears frequently that the query sequence matches against many sequences from the same family in the database, giving rise to a high number of alignments. Of course, all these alignments correspond to a single RNA gene on the query sequence. So all alignments referring to the same region on the same strand for a same family are merged in a single prediction provided that the positions between the alignments are consistent. We say that two local alignments A and A′ are consistent if |(a − x) − (a′ − y)| < 10, where a is the start position of the query sequence in A, a′ is the start position of the query sequence in A′, x is the start position of the ncRNA sequence of the family in A, and y is the start position of the ncRNA sequence of the family in A′. Alignments are merged in a greedy fashion, starting from the first alignment on the query sequence, from 5′ to 3′. Among all mutually consistent alignments, only the best alignments (with lower P-value) are stored and can be presented in a visual format to the user. The number of stored alignments is parameterizable. The start (respectively, end) position of the predicted ncRNA is obtained as the lower (respectively, upper) bound of start (respectively, end) positions of alignments on the query sequence. Installed search databases are currently RFAM 9.1 (1371 families) and RFAM 10.0 (1446 families less the SSU_rRNA_5 family) (Gardner et al. 2009), fRNAdb 3.0 (Kin et al. 2006), and miRBase 13.0 (Griffiths-Jones et al. 2008). For secondary structure and sequence-based homology searches, users may run ERPIN (Gautheret and Lambert 2001), INFERNAL (Nawrocki et al. 2009), and DARN! (Zytnicki et al. 2008). These programs use RNA secondary structure profiles. INFERNAL uses covariation models, which are very sensitive profiles, and offers all RFAM 10.0 families. It is possible to launch it on all families simultaneously or to select one family at a time. Almost all RFAM 9.1 families are currently modeled in ERPIN, and the underlying search algorithm is faster than INFERNAL. DARN! provides specific patterns for C/D box sRNA, some riboswitches, and tRNA. Note that these programs are significantly slower than similarity sequence search programs but are able to identify homologous ncRNA with a lower sequence similarity to known ncRNA. Finally, RNAspace users may run specific programs for rRNA and tRNA gene prediction, respectively, RNAmmer (Lagesen et al. 2007) and tRNAscan-SE (Lowe and Eddy 1997), which are both recognized for their high prediction accuracy.
RNAspace users may also identify new ncRNAs with no homology with the known ncRNAs, using comparative sequence analysis. The underlying principle in this approach is that functional ncRNAs are under a positive selection pressure, hence their sequence should be better conserved than random noncoding regions. Furthermore, mutations observed between homologous structured RNA sequences should be consistent with the formation of a conserved secondary structure core. RNAspace includes the basic components required for running a comparative analysis pipeline. For bacterial genomes, the predictions obtained with our comparative pipeline will differ somewhat from those made by the Livny group (Livny et al. 2008), who used the presence of a transcription terminator, and not secondary structure conservation, as the main filtering criteria. Our comparative analysis results should resemble more those obtained in recent studies combining conservation analysis and the RNAz filter, such as that by Sonnleitner et al. (2008). To run the comparative prediction tool, users select a set of species for comparisons (at most four species). Due to the relatively long computer runtimes involved, we limited the comparative analysis to Bacteria and Archaea. In the first component of the pipeline, the query sequence is compared against all intergenic regions of the selected organisms by selecting BLASTN or YASS. The expected result is a set of similar sequences across species. The second component, CG-seq (http://bioinfo.lifl.fr/CGseq/), performs the combination of all pairwise alignments into clusters of significantly conserved regions. Finally users may select caRNAc (Touzet and Perriquet 2004) or RNAz (Washietl et al. 2005) as the third pipeline component, in charge of inferring and scoring the conserved consensus secondary structure. RNAz requires pre-aligned sequences (a ClustalW [Larkin et al. 2007] alignment is first performed by RNAspace) and is known to be well suited to process clusters of highly similar sequences. caRNAc is a simultaneous align/fold algorithm that shows a better specificity when sequences are difficult to align accurately (Gardner and Giegerich 2004).
The third group of tools performs ab initio prediction by seeking intrinsic signals, which aid in distinguishing ncRNA from other elements in the genome. In the past, several attempts were made to characterize ncRNA using folding measures alone (Freyhult et al. 2005). However, none provided a reliable ncRNA prediction at the genome scale. We chose instead to implement a tool that exploits the compositional bias between ncRNA and the rest of the genome. This approach succeeded at predicting ncRNA from (GC)-atypical regions in hyperthermophile archaea (Klein et al. 2002; Schattner 2002). We implemented a method for identifying (GC)-atypical regions in the program AtypicalGC. This program searches for rich (default parameter) atypical GC regions by using a sliding window. Only regions having a GC value distant of 2 SDs from the mean computed on the whole-genomic sequence are considered as atypical.
Users may select a selection of prediction programs simultaneously to obtain a ncRNA catalog that is as complete as possible, given the query sequence. All programs can be run with default parameter values. For most of them, however, parameters can be easily varied through a dedicated interface, according to known biological constraints or user expertise. The analysis performed with a given tool selection is called a run. The set of putative ncRNA genes obtained in a run may contain redundant overlapping predictions, thus representing accumulated evidence from different ncRNA gene finders. These redundant predictions can be partially merged, given they are consistent in location, strand, length, and functional annotation. Redundancy exists when at least two ncRNA gene finders produce candidates that overlap on the same strand and share the same functional annotation. Overlapping is considered if putative RNAs share at least 10 nucleotides. Functional annotation is based on the family name. For that purpose, a table of synonymous names has been built by hand. It gives an equivalence between name families assigned by different software. Predictions share the same functional annotation if they possess identical or synonymous names according to this equivalence table. Redundancy is managed using an algorithm that merges regions by considering positions, functional annotation, and the nature of the priority assigned to gene finders. Because accuracy of some family-dedicated gene finders is now well established, redundancy can be removed by giving them higher priority for assigning begin/end positions. In RNAspace, tRNAScan-SE and RNAmmer were selected as such tools. Finally, for merging ncRNAs, the algorithm first considers the priority assigned to selected gene finders and then the positions to be merged. This rule does not hold for ncRNAs, which are added by the user, and for ncRNAs annotated as unknown (such as those provided by the comparative analysis pipeline or with ab initio approach). In practice, this merging strategy provides a valuable way to deal with redundant predictions and simplifies the analysis of predictions. Note, however, that in some cases the algorithm is not always a satisfying solution. The latter case is illustrated by a simple example when two tRNAs distant of a few nucleotides are predicted by tRNAscan-SE as two separated regions and are also predicted by BLASTN or YASS as a single one. In this case, no merging is realized by the current merging functionality.
Explore step
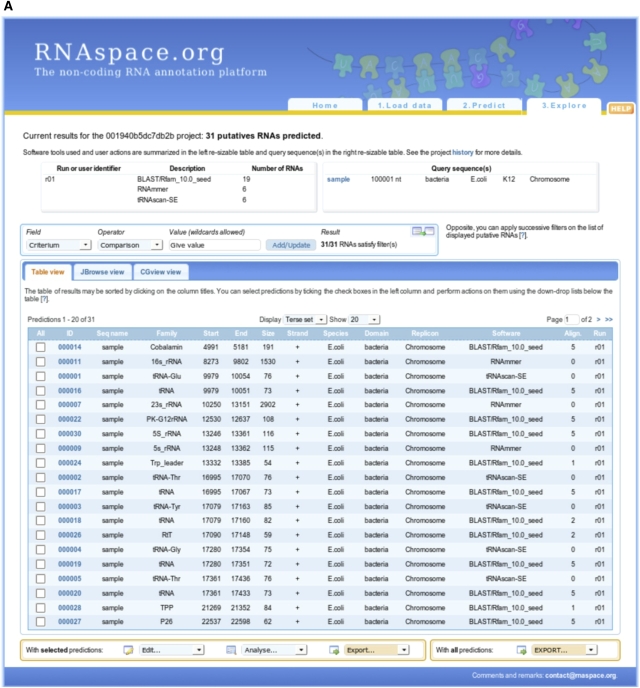
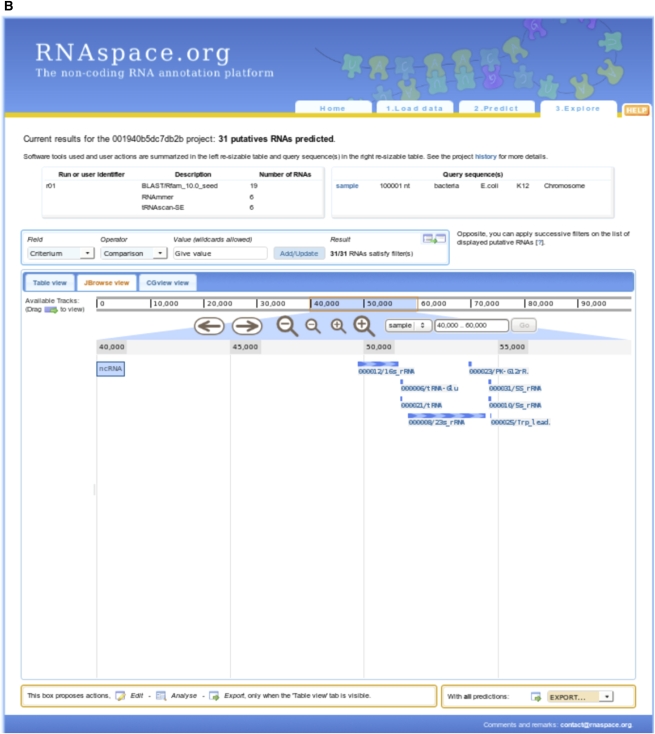
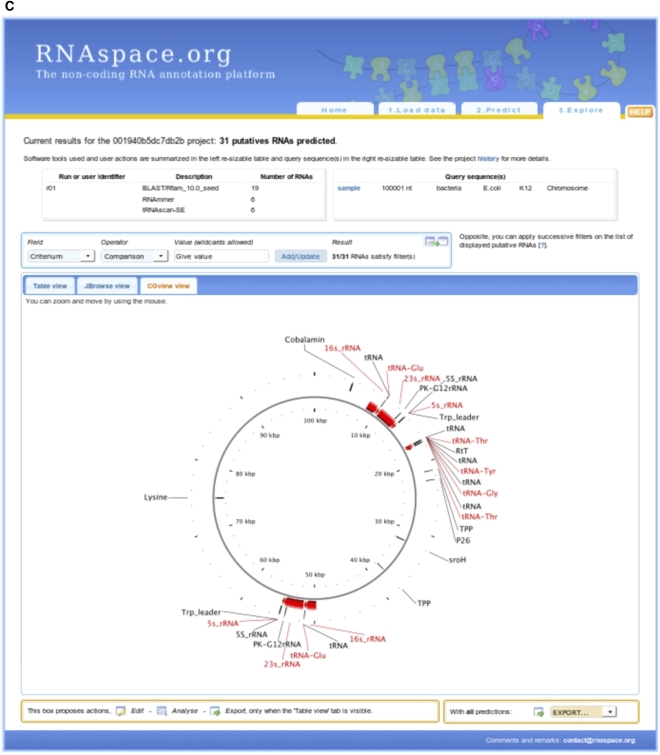
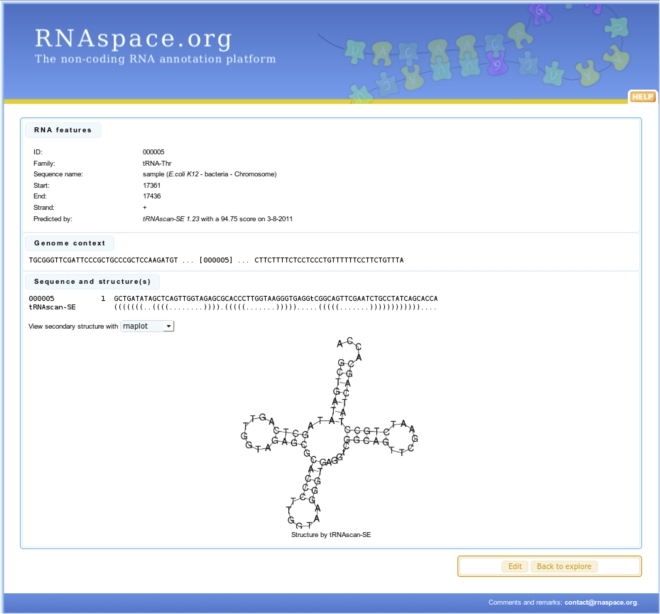
Different gene finders will typically differ in the precise boundaries and annotations of a putative ncRNA. In this case, RNAspace aims to display as much information as possible to help users decide on a suitable selection of evidence and prediction. This explore step provides users with a set of information, functionalities, and programs dedicated to the detailed analysis of predictions and to data export. Prediction results can be visualized as a summary table showing the main characteristics of each hit, or using one of the Jbrowse (Skinner et al. 2009) or CGview (Stothard and Wishart 2005) graphical visualization tools (Fig. 1). Under the table view, users can dynamically sort and explore ncRNA predictions. Users may also apply filtering criteria to any data field by comparing its value to a regular expression. Only predictions matching all user-defined filters are displayed. By default, results are ranked according to their physical location, but any other data field (name, software, etc.) can be used as a sort criterion. Each line of the table shows a ncRNA prediction and offers access to a set of functionalities and data, such as the sequence context of the prediction, existing secondary structures, and alignments in which the sequence is involved (Fig. 2). Other information available for each prediction includes the name of the user sequence, the RNA family (when applicable), the start and end positions on the user sequence, the strand on which the prediction was made (when possible), the domain of life, species and a replicon of the user sequence, the program or set of programs that produced the prediction, the number of alignments in which the prediction is involved, and the identification of the run in which the prediction was performed. Alignments come from two sources. They can be the result of prediction by sequence similarity search. In this case, they are the best pairwise alignments computed by BLASTN and YASS. Otherwise, they consist of multiple alignments computed online from a (multiple) selection of predictions.
FIGURE 1.
Visualization of predictions in the table view (A), with JBrowse (B), and with CGview (C).
FIGURE 2.
Visualization of the characteristics of a predicted ncRNA.
Users may edit most of the information associated to a prediction, produce and save alternative secondary structures computed with one of the RNAfold (Gruber et al. 2008), UNAfold (Markham and Zuker 2008), or RNAshapes (Steffen et al. 2006) programs, or provide their own secondary structure in the form of balanced parentheses. The structures are generated with RNAplot (Gruber et al. 2008) or VARNA (Darty et al. 2009). Users may also select one or more predictions in the table and apply editing functionalities to them. Thus, users can modify the RNA family name, create a new prediction from the selection, delete the predictions, or split the predictions into two new families. Users may also analyze selected predictions. A functionality allows for the alignment of a selection of predictions on the basis of their sequence and structure conservation. The alignment is built with ClustalW (Larkin et al. 2007), and the common structure is derived with RNAz (Washietl et al. 2005). It is also possible to map and visualize a selection of predictions in ApolloRNA (http://carlit.toulouse.inra.fr/ApolloRNA), an extension of the annotation environment Apollo (Lewis et al. 2002) dedicated to RNA analysis. Last, selected predictions can be exported under the formats multifasta, gff, RNAML, and CSV.
DISCUSSION
The annotation of ncRNA remains a major bottleneck on the way toward understanding genome function and evolution. In this context, RNAspace.org provides to biologists a user-friendly web environment dedicated to ncRNA prediction, annotation, and analysis, allowing users to run a variety of complementary tools in an easy and integrated way. Biologists involved in the annotation of ncRNAs in any newly sequenced genome can predict ncRNAs from known families and explore potential novel ncRNAs using different tools. In archaeal or bacterial genomes, users may also discover new ncRNA families using comparative genomics. RNAspace.org does not yet offer access to all the programs and functionalities that may be desirable for ncRNA annotation. We first integrated those tools and databases that partner laboratories had contributed to develop or were used to working with.
In the future, we plan to develop the environment along several lines. First, it will be necessary to extend the scope of ncRNA predictions by including, for example, dedicated CRISPR and miRNA gene finders. Second, as observed in our tests, the combination of gene finders may produce large prediction sets, making it difficult for users to focus on the most significant predictions. A functionality is already implemented to help users reduce redundancy by merging overlapping regions. This functionality will be improved in the future. For instance, the automatic merging of overlapping predictions should benefit strongly from a continuous updating of the dictionary of synonymous RNA gene names. Other functionalities are implemented to help users edit structural and functional annotations and merge or split annotated RNA families. These functionalities will be further developed and improved. For example, we plan to allow for the visualization of predictions in the context of other genome annotation (such as protein-coding gene data). This information will improve ncRNA prediction through knowledge of neighboring gene functions and of the intronic, coding, or intergenic locations of candidate ncRNA. We will also consider the integration of existing tools for editing multiple alignments to enable expert users to improve the accuracy of multiple alignments for consensus structure identification.
With the huge quantity of high-throughput sequencing data obtained by transcriptome studies, it is also highly desirable to consider RNAseq and sRNAseq data. In the future, we will consider handling large sets of RNAseq and sRNAseq with respect to a reference sequence. Such analyses will permit both the annotation and quantification of noncoding transcripts and the search for potential targets of regulatory RNA acting through RNA–RNA interactions.
RNAspace is a collaborative project, and our goal is to make this environment available to the largest community. By making the code open-source, we hope that useful features of RNAspace will be adapted and extended by other developers to serve the needs of particular user groups.
MATERIALS AND METHODS
Implementation and architecture
RNAspace is developed in Python. The web framework CherryPy and the template engine Cheetah were chosen for their simplicity and proximity with Python. A three-tiers client-server architecture is implemented with a presentation tier that displays information, an application tier that controls application's functionality by performing processing, and a data tier (data access) that keeps data independent from the application tier. The Presentation tier relies on a Model-View-Controller (MVC) design pattern. The model is related to domain-specific representation of the information on which the application operates (here, process and access data). The view renders the model into a form suitable for interaction, typically a user interface element (here, template). The controller processes and responds to events, typically user actions, and may invoke changes on the model (here, view as web request entry). The source code is developed under the GPL open-source license. The project is hosted as a Source Forge project (http://sourceforge.net/projects/rnaspace). More information on the architecture and development is available in the RNAspace development documentation.
Data storage
RNAML (Waugh et al. 2002) is used to store and export information on putative ncRNA. RNAML is a standard XML syntax for RNA information. This format is used as an input or output format in a variety of applications (Ruan et al. 2004; Jossinet and Westhof 2005; Noirot et al. 2008; Darty et al. 2009). We choose RNAML to store data produced all along an analysis because it provides a single format for representing information specific to RNA molecules and it proposes a standard syntax to easily express information on ncRNA, thus offering a way to represent a large amount of information in an unambiguous and reusable syntax.
ACKNOWLEDGMENTS
This work was supported by the French “Groupement d'Intérêt Scientifique Infrastructure, Biologie, Santé Agronomie.”
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2844911.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Merce TR, Mattick JS 2008. The Eukaryotic genome as an RNA machine. Science 319: 1787–1789 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y 2009. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25: 1974–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dsouza M, Larsen N, Overbeek R 1997. Searching for patterns in genomic data. Trends Genet 13: 497–498 [DOI] [PubMed] [Google Scholar]
- Freyhult E, Gardner PP, Moulton V 2005. A comparison of RNA folding measures. BMC Bioinformatics 6: 241 doi: 10.1186/1471-2105-6-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P, Giegerich R 2004. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics 5: 140 doi: 10.1186/1471-2105-5-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret D, Lambert A 2001. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J Mol Biol 313: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, François P, Vandenesch F, Gaspin C, et al. 2009. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res 37: 7239–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL 2008. The Vienna RNA Websuite. Nucleic Acids Res 36: W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Schattner P, Polacek N 2005. Non-coding RNAs: hope or hype? Trends Genet 21: 289–297 [DOI] [PubMed] [Google Scholar]
- Jacquier A 2009. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844 [DOI] [PubMed] [Google Scholar]
- Jäger D, Sharma CM, Thomsen J, Ehlers C, Vogel J, Schmitz RA 2009. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci 106: 21878–21882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossinet F, Westhof E 2005. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics 21: 3320–3321 [DOI] [PubMed] [Google Scholar]
- Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y, Kojima A, Kimura Y, Komori T, Asai K 2006. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res 35: D145–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Eddy SR 2003. RSEARCH: finding homologs of single structured RNA sequences. BMC Bioinformatics 4: 44 doi: 10.1186/1471-2105-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Misulovin Z, Eddy SR 2002. Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc Natl Acad Sci 99: 7542–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 3497–3500 [DOI] [PubMed] [Google Scholar]
- Larsson P, Hinas A, Ardell DH, Kirsebom LA, Virtanen A, Söderbom F 2008. De novo search for non-coding RNA genes in the AT-rich genome of Dictyostelium discoideum: performance of Markov-dependent genome feature scoring. Genome Res 18: 888–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 1: D158–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SE, Searle SM, Harris N, Gibson M, Lyer V, Richter J, Wiel C, Bayraktaroglir L, Birney E, Crosby MA, et al. 2002. Apollo: a sequence annotation editor. Genome Biol 3: research0082-0082.14 doi: 10.1186/gb-2002-3-12-research0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xu J, Yang D, Tan X, Wang H 2010. Computational approaches for microRNA studies: a review. Mamm Genome 21: 1–12 [DOI] [PubMed] [Google Scholar]
- Liu J, Carnilli A 2010. A broadening world of bacterial small RNAs. Curr Opin Microbiol 13: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Bai B, Skogerbø G, Cai L, Deng W, Zhang Y, Bu D, Zhao Y, Chen R 2005. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res 33: D112–D115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Fogel MA, Davis BM, Waldor MK 2005. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Res 33: 4096–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Brencic A, Lory S, Waldor MK 2006. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res 34: 3484–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Teonadi H, Livny M, Waldor MK 2008. High-throughput, kingdom-wide prediction and annotation of bacterial noncoding RNAs. PLoS ONE 3: e3197 doi: 10.1371/journal.pone.0003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res 29: 4724–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M 2008. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol 453: 3–31 [DOI] [PubMed] [Google Scholar]
- Mendes ND, Freitas AT, Sagot MF 2009. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res 37: 2419–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel P, Gorodkin J, Stadler PF 2009. The tedious task of finding homologous noncoding RNA genes. RNA 15: 2075–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noé L, Kucherov G 2005. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 33: W540–W543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot C, Gaspin C, Schiex T, Gouzy J 2008. LeARN: a platform for detecting, clustering and annotating non-coding RNAs. BMC Bioinformatics 9: 21 doi: 10.1371/journal.pone.0003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Rivas E, Eddy SR 2001. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics 2: 8 doi: 10.1186/1471-2105-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Klein RJ, Jones TA, Eddy SR 2001. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol 11: 1369–1373 [DOI] [PubMed] [Google Scholar]
- Ruan J, Stormo GD, Zhang W 2004. ILM: a web server for predicting RNA secondary structures with pseudoknots. Nucleic Acids Res 32: W146–W149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P 2002. Searching for RNA genes using base-composition statistics. Nucleic Acids Res 30: 2076–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Barberan-Soler S, Lowe TM 2006. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA 12: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH 2009. JBrowse: a next-generation genome browser. Genome Res 19: 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Sorger-Domenigg T, Madej MJ, Findeiss S, Hackermüller J, Hüttenhofer A, Stadler PF, Bläsi U, Moll I 2008. Detection of small RNAs in Pseudomonas aeruginosa by RNomics and structure-based bioinformatic tools. Microbiology 154: 3175–3187 [DOI] [PubMed] [Google Scholar]
- Steffen P, Voß B, Rehmsmeier M, Reeder J, Giegerich R 2006. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics 22: 500–503 [DOI] [PubMed] [Google Scholar]
- Stothard P, Wishart D 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21: 537–539 [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459: 950–956 [DOI] [PubMed] [Google Scholar]
- Touzet H, Perriquet O 2004. CARNAC: folding families of related RNAs. Nucleic Acids Res 142: W142–W145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay R, Bawankar P, Malhotra D, Patankar S 2005. A screen for conserved sequences with biased base composition identifies noncoding RNAs in the AT-rich genome of Plasmodium falciparum. Mol Biochem Parasitol 144: 149–158 [DOI] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371 doi: 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Stadler PF 2005. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci 102: 2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G 2009. Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh A, Gendron P, Altman R, Brown JW, Case D, Gautheret D, Harvey SC, Leontis N, Westbrook J, Westhof E, et al. 2002. RNAML: a standard syntax for exchanging RNA information. RNA 8: 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35: 4809–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Perreault J, Meyer MM, Breaker RR 2009. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462: 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Weinberg Z, Ruzzo W 2006. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22: 445–452 [DOI] [PubMed] [Google Scholar]
- Zytnicki M, Gaspin C, Schiex T 2008. DARN! A weighted constraint solver for RNA motif localization. Constraints 13: 91–109 [Google Scholar]