This paper shows that THOC5, a member of the THO complex involved in transport of a subset of mRNA, is inactivated upon DNA damage such that it cannot bind to THOC5-dependent mRNAs. Evidence is provided that activation of ATM kinase is involved in suppressing THOC5 RNA-binding activity.
Keywords: FMIP/THOC5, THO complex, mRNA export complex, DNA damage, ATM kinase
Abstract
In response to DNA damage, transcription is blocked by inhibition of RNA polymerase II activity. The regulation of a preexisting pool of mRNAs, therefore, plays a key role in DNA repair, cell cycle arrest, or inhibition of differentiation. THOC5 is a member of the THO complex and plays a role in the export of a subset of mRNA, which plays an important role in hematopoiesis and maintaining primitive cells. Since three serine residues in the PEST domain of THOC5 have been shown to be directly phosphorylated by ataxia-telangiectasia-mutated (ATM) kinase, we examined the THOC5-dependent mRNA export under DNA damage. We show here that DNA damage drastically decreased the cytoplasmic pool of a set of THOC5-dependent mRNAs and impaired the THOC5/mRNA complex formation. The mRNP complex formed with nonphosphorylation mutant (S307/312/314A) THOC5, but not with a C-terminal deletion mutant after DNA damage, suggesting that the C-terminal domain of THOC5, but not its phosphorylation in the PEST domain, is necessary for the regulation of the mRNA-binding potency of THOC5. The cytoplasmic THOC5-dependent mRNAs were recovered by treatment with ATM kinase-specific or p53-specific siRNA, as well as by treatment with ATM kinase inhibitor, KU55933, under DNA damage conditions, suggesting that the ATM–kinase–p53 pathway is involved in this response to the DNA damage. Furthermore, the treatment with KU55933 blocked DNA damage-induced THOC5mRNP complex dissociation, indicating that activation of ATM kinase suppresses the ability of THOC5 to bind to its target mRNAs.
INTRODUCTION
DNA damage response (DDR) involves multiple highly conserved checkpoint pathways that become activated on genotoxic stress. Checkpoint activation triggers a cascade of events that ultimately lead to cell cycle arrest or apoptosis. Key components of this response are the ataxia-telangiectasia-mutated (ATM) and the ATM- and RAD3-related (ATR) kinases that typically function as sensors of DNA damage. In metazoans, the integrity of these pathways is critical to protect cells from agents that cause uncontrolled cellular proliferation and differentiation (Motoyama and Naka 2004). A consequence of ATM kinase loss is a variety of effects, including extinction of hematopoietic stem cells (Ito et al. 2004). Recent analysis of substrates for ATM and ATR kinases revealed that 23 substrates out of 700 belong to a group responsible for “pre-mRNA processing,” and this included members of TREX (transcript/export), which is a protein-complex coupling transcription and nuclear mRNA export. These include THOC4/Aly and FMIP/THOC5/chr22orf19 (Matsuoka et al. 2007), which play roles in mRNA export, suggesting a potential connection between DNA damage and RNA export machinery. In the yeast system, it has been previously shown that Sub2–Yra1 and Thp1–Sac3, the two main mRNA export complexes, are required for efficient transcription-coupled repair (Gaillard et al. 2007). However, the exact role of the TREX complex in mammalian systems under DNA damage conditions is not clear. The THO complex, which is a submember of TREX, was originally identified in Saccharomyces cerevisiae as a 4-protein complex (THO2p, Hpr1p, Mft1p, and Thp2p) (Chavez and Aguilera 1997; Piruat and Aguilera 1998; Chavez et al. 2001; Jimeno et al. 2002; Strasser et al. 2002; Reed and Cheng 2005) that plays a role in transcriptional elongation, nuclear RNA export, and genome stability. In higher eukaryotes such as Drosophila melanogaster (Rehwinkel et al. 2004) or humans (Masuda et al. 2005), three proteins (THOC1/hHpr1/p84, THOC2/hRlr1, and THOC3) and three additional unique proteins were identified, namely, THOC5/Fms-interacting protein (FMIP) (Tamura et al. 1999), THOC6, and THOC7, as members of the THO complex. We have previously shown that depletion of THOC5 by siRNA or ectopic expression causes abnormal hematopoiesis and abnormal muscle differentiation in mouse myeloid progenitor or mesenchymal progenitor cell lines, indicating that the THO complex is essential for the differentiation process in mouse (Tamura et al. 1999; Mancini et al. 2007; Carney et al. 2009). Furthermore, we generated interferon-inducible THOC5 knockout mice to demonstrate that THOC5 is essential for maintaining primitive cells, such as embryonic or hematopoietic stem cells, but not terminally differentiated cells (Mancini et al. 2010), similar to ATM kinase (Ito et al. 2004). THOC5 is phosphorylated by several tyrosine kinases (Tamura et al. 1999; Pierce et al. 2008), protein kinase C (Mancini et al. 2004), and ATM kinase (Matsuoka et al. 2007), suggesting that extracellular stimulation regulates the function of THOC5. To obtain further insight into the role of THOC5 at the molecular level, we performed a microarray analysis using mouse embryonic fibroblasts (MEF) in the presence or absence of THOC5. Surprisingly, only 71 functionally known genes were down-regulated more than threefold by depletion of THOC5/FMIP. Moreover, >43% of these genes are involved in differentiation and development, such as HoxB3 or polycomb-CBX2. (Guria et al. 2011), confirming that the THO complex plays a key role in the differentiation and development system. In this work, we have studied the mechanism of THO complex response to DNA damage signaling. We show that THOC5-dependent mRNAs were spliced, but not detected in the cytoplasm after DNA damage, suggesting that THOC5 is a mediator between DNA damage and the mRNA export of a subset of genes. Since approximately half of the identified THOC5-dependent genes are involved in the differentiation process (Guria et al. 2011), THOC5 may play a role in protecting cells from agents that cause uncontrolled differentiation.
RESULTS
DNA damage drastically decreased the cytoplasmic pool of a set of THOC5-dependent mRNAs
Three THOC5 serine residues, 307, 312, and 314, within the PEST-like domain, were previously identified as phosphorylation sites by ATM kinase using irradiated HEK293 cell extracts (Matsuoka et al. 2007). We confirmed THOC5 phosphorylation after DNA damage by ATM kinase using an ATM kinase substrate antibody. HEK293 cells were treated with etoposide (0.2, 2.0, or 20 μM) for 2 h, then cells were lysed and anti-THOC5 antibody used for immunoprecipitation, followed by ATM substrate (pS/T-Q)-specific immunoblot. At each concentration of etoposide used, THOC5 was detected by ATM substrate-specific antibody. To further confirm whether ATM kinase directly phosphorylates THOC5 in vitro, we performed in vitro kinase assay using flag-tagged ATM kinase cDNA (Kim et al. 1999) and GST-THOC5 (289–378) as substrate. GST-THOC5 (289–378) was phosphorylated by ATM kinase and the mutation of the three serine residues abolished phosphorylation (Supplemental Fig. 1). Thus, after confirming phosphorylation of THOC5 upon etoposide treatment, we then considered the effects of DNA damage and ATM kinase on THOC5-mediated mRNA export. We assessed the protein level of several members of TREX, such as THOC1, THOC5, THOC7, or Aly after etoposide treatment (2 h) in mouse embryonic fibroblasts (MEF). As shown in Figure 1A, the level of these proteins did not change. Furthermore, phosphorylation of p53 was observed only after treatment with etoposide, indicating that a DNA damage signal is induced under this condition in MEF cells (Fig. 1A). In addition, the level of THOC5 mRNA was not changed, suggesting that etoposide treatment did not influence this or THOC5 protein. In addition, we examined the ability of THOC5 to interact with other components of the TREX complex, such as THOC1 or Aly after DNA damage; we did not observe any differences in the THOC1/THOC5 interaction or the THOC5/Aly interaction before and after etoposide treatment (data not shown). We then examined the mRNA export function of THOC5 after etoposide treatment. We had previously generated a MEF cell line from THOC5 flox/flox mice (Mancini et al. 2010). Infection of THOC5/FMIP flox/flox MEF with Adenovirus carrying cre-recombinase and GFP genes (Ad-GFP-Cre) resulted in depletion of THOC5/FMIP expression in these cells within 4 d after infection (Mancini et al. 2010; Guria et al. 2011). Using this system, we previously identified 10 THOC5-dependent genes, such as Grb10 interacting GYF protein 2 (Gigyf2 or Tnrc15), protein kinase C binding protein 1 (Prkcbp1), twisted gastrulation homolog 1 (Twsg1), enhancer of yellow 2 homolog (Eny2), solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 (Slc11a2), DHHC domain containing 2 (Zdhhc2) zinc finger, inhibitor of DNA binding 2 (ID2), or gap junction membrane channel protein α 1 (Gja1) (Guria et al. 2011).
FIGURE 1.
DNA damage drastically decreased the cytoplasmic pool of a set of THOC5-dependent mRNAs. (A) MEF cells were treated with DMSO alone (DMSO) or with 10 μM of etoposide (etoposide) for 2 h, and cell extracts were then subjected to THOC5, THOC1, THOC7, Aly, pp53, p53, and GAPDH-specific immunoblot (immunoblot). In addition, RNA was isolated from the sister culture of A and supplied for THOC5 and actin-specific RT–PCR (RT–PCR). (B) A total of 5000 MEF cells were treated as described above, and then nuclear (Nuc) and cytoplasmic (Cyt) RNAs were isolated and applied for Zdhhc2, Prkcbp1, Gigyf2, Twsg1, Pdrg1, Gadd45a, Myc, and actin-specific RT–PCR using primers as shown in Table 1. We performed three to four independent experiments, and we show one example of representative data. As control for fractionation, aliquots of protein extracts from each sample were supplied for THOC5, Histone H3 (nuclear fraction), and GAPDH (cytoplasmic fraction)-specific immunoblot (immunoblot). (C) Signal intensity from Zdhhc2, Prkcbp1, Gigyf2, Gadd45a, Myc, and actin-specific RT–PCR products from cytoplasmic fraction were quantified using TINA 2.0 software. The reduced percent signal intensity from the treated with DMSO alone (etoposide −: as 100%) of the treated with etoposide (etoposide +) were shown. Mean values ± SD from three independent experiments. (D) Samples were prepared as described in B, but cDNA from all samples were standardized by expression levels of actin mRNA in both fractions, then applied for Zdhhc2, Prkcbp1, Slc11a2, Gigyf2, Eny2, Twsg1, Gja1 (THOC5 dependent), Pdrg1, Gadd45a, Myc, Dusp10, and actin (THOC5 independent)-specific RT–PCR. (E) Signal intensity from Zdhhc2, Prkcbp1, Slc11a2, Gigyf2, Twsg1, Gadd45a, Myc, and actin-specific RT–PCR products was quantified using TINA 2.0 software. The percent signal intensity from the nuclear (Nuc) or the cytoplasmic (Cyt) fraction of total intensity (Nuc + Cyt) in MEF treated with DMSO alone (etoposide −) or with etoposide (+) for 2 h were shown. Mean values ± SD from three independent experiments.
To examine whether THOC5-dependent mRNAs are exported into the cytoplasm after DNA damage, we treated MEF cells with etoposide (10 μM), and 2 h later isolated nuclear and cytoplasmic RNA. To validate the quality of the nuclear and cytoplasmic fractions, the marker proteins GAPDH (cytoplasmic fraction), Histone H3 (nuclear fraction) were used, plus THOC5-specific assay immunoblotting. GAPDH was detected primarily in the cytoplasmic fraction, while Histone H3 was detected only in the nuclear fraction (Fig. 1B). In addition, THOC5 were detected only in the nuclear fraction after treatment with DMSO alone or with 10 μM of etoposide (Fig. 1B), indicating that drug treatment does not influence THOC5 nuclear location. RNA samples of both fractions obtained from 5000 cells were subjected to RT–PCR. Critically, only spliced forms of mRNA can be detected, since we chose the primer pair for RT–PCR, which is located on two different exons (Table 1). In all cases, most mRNA were detected in the cytosol, while mRNAs in the nuclear fraction were effectively undetectable, except for actin mRNA (Fig. 1B). Significantly, by quantifying the signal intensity, a reduction of >85% of THOC5-dependent mRNAs, such as Zdhhc2, Prkcbp1, and Gigyf2 were observed in the cytoplasmic fraction after etoposide treatment (Fig. 1C). As a control, the THOC5-independent genes, myc and actin (Guria et al. 2011), were examined. The expression pattern of these genes was not altered by etoposide treatment (Fig. 1B,C; Table 1). In order to further validate these observations, we examined the growth arrest and DNA-damage-inducible, α (Gadd45a) and p53 and DNA damage-regulated gene 1 (PDRG1) mRNAs that play a role in DNA repair and are well known to be stabilized after DNA damage (Luo et al. 2003; Zhang et al. 2006; Zumbrun et al. 2009; Reinhardt et al. 2010). As expected, both mRNAs were exported to the cytosol (Fig. 1B,C).
TABLE 1.
Comparison of THOC5-dependent mRNAs and mRNAs that are down-regulated by DNA damage
We next examined whether the THOC5-dependent mRNAs were transcribed in the nucleus. The amount of RNA was standardized by using equal expression levels of actin mRNA in both fractions (Fig. 1D,E). Eight THOC5-dependent mRNAs were then analyzed by RT–PCR. Seven out of eight THOC5-dependent mRNAs were barely detectable in the cytoplasmic fraction after cells had been treated with etoposide for 2 h, even in the presence of THOC5 (Fig. 1D; Table 1); however, in the nuclear fraction, spliced mRNAs were detected. The effect was not observed with THOC5-independent mRNAs, suggesting that THOC5-dependent mRNA export may be impaired after etoposide treatment. We obtained similar data using hydroxyurea (20 mM)-treated MEF (data not shown), suggesting that DNA damage may cause the loss of function of THOC5. However, one THOC5-dependent mRNA, ID2, was still exported into the cytoplasm after etoposide treatment (data not shown). In addition, we examined the THOC5-independent genes, myc, dual specificity phosphatase 10 (DUSP10), Gadd45a, and PDRG1 (Guria et al. 2011). The level of these mRNAs was not significantly altered (Fig. 1D). We then examined the ratio between nuclear and cytoplasmic spliced mRNA by quantifying the signal intensity. All seven THOC5-dependent mRNAs in the cytoplasmic fraction were drastically reduced (Fig. 1E), while etoposide treatment caused only slight changes of the ratio between cytoplasmic and nuclear THOC5-independent mRNAs. Although the half-lives of THOC5-dependent mRNAs, Zdhhc2, Prkcbp1, and Gigyf2 are longer than 2 h in the presence or absence of etoposide, these mRNAs disappear from the cytoplasmic fraction within 2 h after etoposide treatment (Supplemental Fig. 2), suggesting that the decrease of cytoplasmic THOC5-dependent mRNAs may be in part due to faster cytoplasmic degradation.
THOC5 does not interact with mRNA after DNA damage
To determine whether the response to the DNA damage is directly dependent on THOC5, we analyzed the mRNP complex (containing THOC5) before and after DNA damage. We have previously shown that THOC5-dependent genes, such as Gigyf2 or Prkcbp1, were detected in the mRNP complex using THOC5 as bait (Guria et al. 2011). Two-sister culture of 4 × 106 MEF cells were treated with DMSO alone or with 10 μM of etoposide for 2 h. After centrifugation, cell extracts from 2 × 106 MEF cells were precipitated by THOC5 antibody or rabbit control IgG with protein G Sepharose. RNA were isolated from immunoprecipitates and supplied for RT–PCR using Gigyf2, Prkcbp1, Zdhhc2, and actin-specific primers. The endogenous Gigyf2, Prkcbp1, and Zdhhc2, but not actin mRNA, were detected in the THOC5-specific immunoprecipitates obtained from the DMSO-treated MEF. However, none of these mRNA species bound to THOC5 after etoposide treatment (Fig. 2A). No mRNAs were detected in precipitates with protein G Sepharose using control rabbit IgG or no IgG (Fig. 2A). Similar data were obtained using exogenous TAP-THOC5-Myc. MEF cells were transfected with TAP-THOC5-myc (Fig. 2B), and were then treated with DMSO alone or with 10 μM of etoposide for 2 h. After isolation of the mRNP complex using streptavidin-conjugated beads, RNA were isolated and supplied for RT–PCR using Gigyf2, Prkcbp1, Zdhhc2, and actin specific primers. The endogenous Gigyf2, Prkcbp1, and Zdhhc2 mRNAs, but not actin mRNA, were detected in the TAP-THOC5–Myc complex; however, after DNA damage, neither of the THOC5-dependent mRNAs were detected in the complex (Fig. 2C). Furthermore, a THOC5–mRNAs complex was isolated exclusively from the nuclear fraction, but not from the cytoplasmic fraction (data not shown), and THOC5 stayed in the nucleus after DNA damage (Fig. 1B), strongly suggesting that DNA damage causes dissociation of THOC5 with mRNAs in the nucleus.
FIGURE 2.
Etoposide treatment impairs the mRNA-binding potential of THOC5. (A) Two-sister culture of 4 × 106 MEF cells were treated with DMSO alone (DMSO) or with 10 μM of etoposide (etoposide) for 2 h. The cells were then extracted with lysis buffer. After centrifugation, cell extracts from 2 × 106 MEF cells were incubated with THOC5 antibody (anti THOC5), rabbit control IgG (CrIgG), and no IgG (−) for 1 h and were then precipitated with protein G Sepharose. RNAs were isolated from immunoprecipitates and supplied for RT–PCR using Gigyf2, Prkcbp1, Zdhhc2, and actin-specific primers. As control, immunoprecipitates were analyzed by THOC5-specific immunoblot (THOC5 [endo] Blot). Aliquotes of cell extracts were supplied for RT–PCR (input). (B) Schematic representation of the pNTAP (TAP-Vector, CBP: Calmodulin binding peptide; SBP: Streptavidin binding peptide) and pNTAP carrying myc-tagged THOC5 (TAP-THOC5wt–Myc), and THOC5 mutants (TAP-THOC5 S307/312/314A-Myc and TAP-THOC5 [1–499]-Myc). (PEST) PEST-like domain; (LZ) putative leucine zipper. Numbers represent amino acid number. (C) pNTAP (Cr) or TAP-THOC5wt–Myc were transfected into MEF cells. After 48 h, cells were incubated with DMSO alone (DMSO) or with 10 μM of etoposide (etoposide) for 2 h, and the RNP complex was then isolated using Streptavidin Sepharose. Bound fractions with Streptavidin Sepharose (bound) and cell extracts (input) were analyzed by immunoblotting using THOC5-specific antibody (Blot). Endogenous Prkcbp1, Gigyf2, Zdhhc2, and actin mRNAs in bound and input RNA samples were analyzed (RT–PCR). (D) pNTAP, TAP-THOC5wt-Myc, TAP-THOC5 S307/312/314A-Myc, and TAP-THOC5 (1–499)-Myc (Blot) were cotransfected with flag-tagged Gigyf2 cDNA or β globin gene cDNA into MEF cells. As control, we transfected MEF cells with pNTAP empty vector (pNTAP Cr). After 48 h, cells were treated as described above, and the RNP complex was then isolated using Streptavidin Sepharose. Bound fractions with Streptavidin Sepharose (bound) and cell extracts (input) were analyzed by immunoblotting using THOC5-specific antibody (Blot). Gigyf2 mRNA in bound and input RNA samples were analyzed by Flag and Gigyf2-specific RT–PCR (Forward [flag]: 5′-ATGGATTACAAGGATGACGACGAT-3′; reverse [Gigyf2]: 5′-TTCATGCCGAAGTCTGGCAGC-3′ [product size: 225bp]) or β globin-specific RT–PCR (Forward: 5′-AGCGGCCGCACACTTGCTTTTTGA-3′; reverse: 5′-TCGAAGAACCTCTGGGTCCATGGG-3′ [product size: 183 bp, spliced form]). (Mock) Cell extract from nontransfectants was applied for THOC5-specific immunoblotting and flag- and Gigyf2-specific RT–PCR. We have performed three independent experiments and we show one example of representative data. (E) Signal intensity from Gigyf2-specific RT–PCR products from the bound fraction using the full-length THOC5 and deletion mutant THOC5 (1–489) were quantified using TINA 2.0 software. The ratio of signal intensity before and after treatment with etoposide is presented from three independent experiments.
We next examined whether phosphorylation of the PEST domain residues is involved in the down-regulation of the mRNP complex formation. We utilized the wild-type TAP-THOC5 and a mutant form generated that lacked ATM kinase phosphorylation sites, TAP-THOC5 (S307/312/314A). MEF cells were transfected with TAP-THOC5–myc or TAP-THOC5 (S307/312/314A) (Fig. 2D) and Flag-tagged Gigyf2 cDNA and were then treated with or without etoposide. After isolation of the mRNP complex using streptavidin-conjugated beads, RNA was isolated and subjected to RT–PCR using Flag and Gigyf2-specific primers. As expected, the exogenous flag-tagged Gigyf2 mRNA was detected in the TAP–THOC5–Myc complex; however, after DNA damage, flag-tagged Gigyf2 mRNA was not detected (Fig. 2D). Since the nonphosphorylation mutant also lost its RNA-binding potential after DNA damage (Fig. 2B,D), the phosphorylation itself is not required for the dissociation of Gigyf2 mRNA from THOC5 after etoposide treatment. Given these data, we examined the domain that is essential for the mRNP complex dissociation using two further deletion mutants, THOC5 (1–499) (Fig. 2B) and THOC5 (1–289) (data not shown) (El Bounkari et al. 2009). Both mutants associated with Gigyf2 mRNA before and after DNA damage equally well (Fig. 2D,E). These data suggest that the C-terminal domain (500–671), but not the PEST domain, is necessary for the regulation of THOC5 interaction, with its target mRNAs resulting from DNA damage. As a negative control, we cotransfected TAP-THOC5–Myc with β-globin cDNA, whose mRNA does not bind to THO complex (Merz et al. 2007). As expected, we did not detect β-globin mRNA from TAP-THOC5–Myc precipitates in any of the samples tested (Fig. 2D).
DNA damage impaired THOC5/mRNA complex formation via the ATM kinase pathway
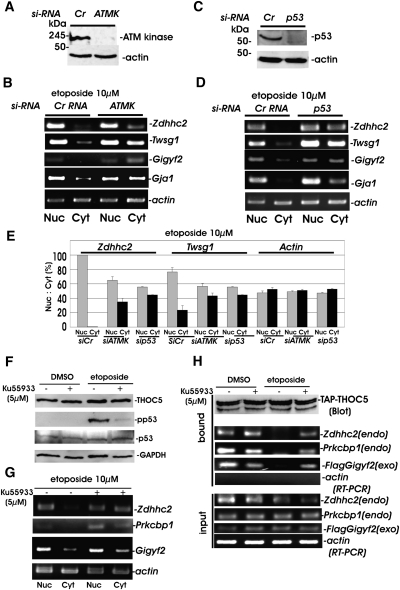
In order to determine whether the effect of etoposide treatment on THOC5-dependent mRNAs is due to ATM-kinase activation, we used the ATM-kinase-specific siRNA, leading to a >10-fold down-regulation of ATM kinase within 48 h after transfection with ATM-kinase-specific siRNA (Fig. 3A). In cells depleted of ATM kinase, Zddhc2, Twsg1, Gigyf2, or Gja1 mRNAs were exported in the cytoplasmic fraction even 2 h after treatment with etoposide (Fig. 3B), suggesting that ATM kinase is required for the inhibition of THOC5-dependent mRNA export after DNA damage. To determine whether p53, a major target substrate of ATM kinase, influences the export of THOC5-dependent mRNAs, we down-modulated p53 using p53-specific siRNA. As shown in Figure 3C, p53 expression was abolished 48 h after siRNA transfection. In these cells, THOC5-dependent mRNA, such as Zdhhc2, Twsg1, Gigyf2, or Gja1 mRNAs were still exported into the cytoplasm 2 h after etoposide treatment (Fig. 3D), indicating that the ATM-kinase/p53 pathway is required for this response to the DNA damage. The intensity of RT–PCR signals was also quantified (Fig. 3E): The export of THOC5-dependent mRNAs, such as Zdhhc2 and Twsg1, were detected in the cytoplasm upon treatment with both ATM-specific (siATMK) and p53-specific (sip53) siRNAs, but not by the treatment with control siRNA (siCr) under DNA damage conditions.
FIGURE 3.
DNA damage impaired the THOC5/mRNA complex formation via the ATM kinase pathway. (A–E) MEF cells were transfected with siRNA against mouse ATM kinase (ATMK) (A,B,E) or p53 (p53) (C–E) (Santa Cruz). (A,C) Forty-eight hours after transfection, cells were lysed and subjected to actin and ATM-kinase- (A) or p53- (C) specific immunoblot. (B,D) Sister cultures from B and D were treated with etoposide (10 μM) for 2 h, then nuclear (Nuc) and cytoplasmic (Cyt) RNAs were isolated, and applied for RT–PCR using Zdhhc2, Twsg1, Gigyf2, and actin-specific primers as described in Table 1. (E) Signal intensity from Zdhhc2, Twsg1, and actin-specific RT–PCR products was quantified using TINA 2.0 software. The percent signal intensity from the nuclear (Nuc) or the cytoplasmic (Cyt) fraction of total intensity (Nuc + Cyt) in MEF transfected with control siRNA (siCr), ATM-kinase-specific siRNA (siATMK), and p53-specific siRNA (sip53) were shown. Mean values ± SD from three independent experiments. (F) MEF cells were incubated with the potent inhibitor of ATM kinase KU55933 (5 μM) 1 h prior to treat with DMSO, or with etoposide for 2 h. Whole-cell extracts were subjected to THOC5, p53, pp53, and GAPDH-specific immunoblot. (G) The nuclear (Nuc) and cytoplasmic (Cyt) RNAs from the sister culture of (F: etoposide) were isolated and applied for RT–PCR using primers as described in Table 1. (H) TAP-THOC5wt-Myc was cotransfected with flag-tagged Gigyf2 (FlagGigyf2) cDNA into MEF cells. After 48 h, cells were treated as described in F, and then RNP complexes were isolated using Streptavidin Sepharose. Bound fractions with Streptavidin Sepharose (bound) were analyzed by immunoblotting using THOC5-specific antibody (Blot) or RT–PCR using Flag-Gigyf2, Prkcbp1, Zdhhc2, and actin-specific primers (RT–PCR). (input) Input RNA samples were analyzed by RT–PCR; (endo) endogenous mRNA; (exo) exogenous mRNA.
To confirm these data, we utilized a potent inhibitor of ATM kinase, KU55933 (Hickson et al. 2004; Adams et al. 2010). MEF cells were incubated with KU55933 1 h prior to the stimulation with etoposide. Under these conditions, the phosphorylation of p53 was drastically reduced (Fig. 3F). RNA was then isolated from the nuclear and cytosolic fractions. In agreement with data using an siRNA approach, the THOC5-dependent mRNAs, Zdhhc2, Prkcbp1, and Gigyf2 were detected in the cytosolic fraction after treatment with ATM-kinase inhibitor (Fig. 3G). We next examined whether DNA damage impaired the THOC5/mRNA complex formation via the ATM-kinase pathway. TAP-THOC5wt–Myc was cotransfected with flag-tagged Gigyf2 cDNA into MEF cells. After 24 h, cells were incubated with or without ATM-kinase inhibitor, KU55933, 1 h prior to the stimulation with DMSO alone or with etoposide for 2 h, and then RNP complexes were isolated using Streptavidin Sepharose. Bound fractions were analyzed by immunoblotting using THOC5-specific antibody. As shown in Figure 3H, the treatment with KU55933 did not change the level of TAP-THOC5–Myc. The FlagGigyf2, Prkcbp1, Zdhhc2, and actin mRNAs-bound fraction were then analyzed by RT–PCR (Fig. 3H), and treatment with ATM-kinase inhibitor recovered the ability of THOC5 to bind to its target mRNAs, such as FlagGigyf2, Prkcbp1, and Zdhhc2, indicating that ATM kinase is required for DNA damage-induced dissociation of THOC5 with mRNA. The THOC5-independent mRNA, actin mRNA, was not detected in the TAP-THOC5–Myc complex under any conditions shown.
In summary, DNA damage impaired the complex formation of THOC5 with its target mRNAs via the ATM-kinase/p53 pathway in the nucleus.
DISCUSSION
DNA damage inhibits the activity of RNA polymerase II at a time when DDR proteins and DNA repair proteins are needed. One of the main post-transcriptional events after DNA damage is the altered stability of mRNAs. Because transcription is reduced after DNA damage, there is an increased need to regulate the production of proteins from a pre-existing pool of mRNAs (Mitchell and Tollervey 2000; Orphanides and Reinberg 2002). On the other hand, the production of proteins that play a role in proliferation or differentiation likely should be blocked, because these may cause uncontrolled cellular proliferation and inappropriate differentiation. mRNAs from a pre-existing pool are potently influenced by RNA-binding proteins and noncoding RNAs, such as microRNA (Keene 2007; Bartel 2009). However, little is known about mRNA export under conditions that damage the DNA template and RNA itself. We have previously shown that depletion of the THOC5 gene in MEF cells causes the down-regulation (more than threefold) of 198 genes (Guria et al. 2011). Forty-three percent of these genes are involved in development and cell differentiation, suggesting that THOC5 plays a role in the export of only a subset of genes, but plays an important role in cell differentiation. Our data obtained from THOC5 knockout mice revealed that primitive hematopoietic cells in vivo not only stopped growing, but became apoptotic after THOC5 gene depletion (Mancini et al. 2010). In addition, conventional THOC1 or THOC5 knockout mice are embryonic lethal (Li et al. 2005; Mancini et al. 2010). On the other hand, we show in the MEF system that THOC5-deficient differentiated fibroblasts just cease growing by THOC5 gene depletion (Guria et al. 2011). We show here that seven out of eight THOC5-dependent mRNAs that were examined were spliced, but were not detected in the cytoplasm within 2 h after DNA damage. Thus, the inhibition of mRNA-binding potential of THOC5 may be essential to block uncontrolled differentiation. Interestingly, it has been reported that the treatment of HeLa cells with DNA damage reagents, such as adriamycin or etoposide, suppressed Glut3 (Slc2a3) mRNA expression (Watanabe et al. 2010). We have previously shown that knockout of THOC5 down-regulated Glut3 mRNA more than sevenfold (Guria et al. 2011), suggesting that the down-regulation of Glut3 mRNA, which was observed under conditions of DNA damage, may also be due to the loss of mRNA-binding potential of THOC5.
In the yeast system, it has been shown that DNA damage impairs tRNA nuclear export via differential relocalization of the karyopherin Los1, the principal tRNA export receptor, to the cytoplasm (Ghavidel et al. 2007). However, THOC5 does not change its subcellular localization nor the complex formation with another member of TREX such as THOC1 or Aly after DNA damage. Strikingly, THOC5-dependent mRNA did not bind to THOC5 after etoposide treatment. How does THOC5 lose its binding potential to mRNAs? Since an ATM kinase phosphorylation site-defective mutant of THOC5 also lost the mRNA-binding potential just as the wild-type does upon DNA damage, the phosphorylation of THOC5 by ATM kinase cannot play a role in the binding potential. Furthermore, since the N-terminal domain that binds to mRNAs (Katahira et al. 2009) contains several phosphorylation sites (Tamura et al. 1999; Pierce et al. 2008), a potential acetylation site, and a potential SUMOylation site, post-translational modification may play a role in the loss of the mRNA-binding potential of THOC5. Alternatively, THOC5-dependent mRNAs may be recruited to other unknown mRNA-binding proteins specifically after DNA damage, annexing them away from THOC5. It has been shown that DNA damage-inducing agents can activate the RNA-binding activity of specific proteins including nucleolin, nucleophosmin, and MCG10, that favors a double stem–loop RNA structure (Carrier et al. 1994; Zhu and Chen 2000; Yang et al. 2002). Notably, some of RNA-binding proteins, such as MCG10 or nucleolin are regulated by p53 (Zhu and Chen 2000; Daniely et al. 2002). On the other hand, the C-terminal deletion mutant still binds mRNAs after DNA damage, suggesting that DNA damage-induced dissociation of THOC5 with its target mRNAs is via the loss of binding potential of THOC5 itself. It could be that the modification of the C-terminal domain by DNA damage influences the THOC5 structure by binding with unknown proteins that are not members of TREX, or alters protein conformation. Interestingly, interactome analysis using THOC5 as bait in HEK293 cells reveals that one of the interacting partners is p53 (AD Whetton and T Tamura, unpubl.). Furthermore, exogenous expression of THOC5 in cancer cells causes accumulation and colocalization of p53 (S Ramachandran and T Tamura, unpubl.). However, we could not find a direct interaction between p53 and THOC5, or indeed, colocalization of the two proteins in MEF cells before or after DNA damage. We are currently studying the binding partners of the THOC5 C-terminal domain before and after DNA damage by interactome analysis, using the MEF cell system.
It is presently unclear whether DNA damage causes inhibition of the export of THOC5-dependent mRNAs. A THOC5/mRNA complex was isolated exclusively from the nuclear fraction, and THOC5 remained in the nucleus after DNA damage, strongly suggesting that DNA damage causes dissociation of THOC5 with mRNAs in the nucleus. These data also suggest that DNA damage may block THOC5-dependent mRNA export. On the other hand, although half-lives of THOC5-dependent mRNAs that were examined here are longer than 2 h, these mRNAs disappear from the cytoplasmic fraction within 2 h after etoposide treatment, suggesting that THOC5-dependent mRNAs are degraded rapidly in the cytoplasm. It may be that THOC5 or other proteins are bound to the mRNA under normal conditions and stabilize the RNA in the cytoplasm. Upon DNA damage, THOC5 (or another protein) no longer binds to the mRNAs, resulting in a shortened half-life for THOC5-dependent mRNAs. Both the inhibition of mRNA export and the faster cytoplasmic degradation may be involved in this response to the DNA damage.
We have previously shown that approximately half of identified THOC5-dependent genes are involved in the differentiation process (Guria et al. 2011), suggesting that THOC5 may play a role in protecting cells from agents that cause uncontrolled differentiation or alternatively disallow differentiation in inappropriate conditions.
MATERIALS AND METHODS
Cell culture, virus infection, plasmid construction, and DNA transfection
Mouse embryonic fibroblast (MEF) cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) FCS. Myc-tagged wild-type and mutant THOC5 were generated using pNTAP (Stratagene), as described previously (Tamura et al. 1999; El Bounkari et al. 2009). DNA transfection was performed with the Polyfect reagent as described by the manufacturer (Qiagen). Gigyf2 cDNA (IMAGE ID 5357184) was purchased by ImaGene. β-globin cDNA was a kind gift from H. Holtmann. Flag-tagged ATM kinase cDNA (Kim et al. 1999) was a kind gift from Michael Kastan. KU55933 was purchased by TOCRIS bioscience.
Antibodies
Mouse monoclonal antibodies against THOC5/FMIP (F6d) were described previously (Mancini et al. 2007). Rabbit polyclonal antibody against THOC5 was from Bethyl Laboratories, Inc. Mouse monoclonal antibodies against Aly and GAPDH, goat antibody against actin, and rabbit polyclonal against p53 were from Santa Cruz Biotechnology. Rabbit antibodies against phospho-p53 (Ser15) and Histone H3 were from Cell Signaling Technology, mouse monoclonal antibody against THOC1 (p84 N5) was from Gene Tex. Inc. Rabbit antibody against THOC7 was from Abgent.
Western blot procedures
Details of immunoblotting have been described previously (Koch et al. 2008). Results were documented on a LAS4000 imaging system (GE Healthcare Bio-Sciences).
RT–PCR analysis
Cytoplasmic RNA was isolated from MEF using the Qiagen RNeasy kit (Qiagen) according to the manufacturer's recommendations (Guria et al. 2011). Reverse transcription was carried out using oligo dT primers and Omniscript reverse transcriptase kit (Qiagen) following the instructions provided. Primer pairs for each PCR are shown in Table 1.
THOC5–mRNA complex isolation
Two-sister culture of 4 × 106 MEF cells were treated with DMSO alone or with etoposide for 2 h. After washing three times, cells were lysed with lysis buffer (10 mM Tris, 150 mM NaCl, 1 mM PMSF, 0.5% NP40, protease inhibitor cocktail, [Sigma-Aldrich] and RNase inhibitor), and were then frozen and thawed three times. After centrifugation, supernatants were incubated with rabbit polyclonal THOC5 antibody or rabbit control IgG with protein G Sepharose, then immunoprecipitates were washed three times. Alternatively, MEF cells were transfected with pNTAP vector (Stratagene) and the same vector carrying myc-tagged THOC5, and its mutants with or without flag-tagged Gigyf2 cDNA using the Polyfect reagent as described by the manufacturer (Qiagen), and the cells were allowed to grow for 48 h. Cells were then treated as detailed above. After centrifugation, supernatants were incubated with Streptavidin Sepharose (GE Healthcare Bioscience) in Streptavidin-binding buffer (lysis buffer with 10 mM β-mercaptoethanol and 0.5 mM EDTA), then washed three times. All steps were carried out at 4°C. Bound RNAs were used for RT–PCR analysis.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank C. Bruce Boschek for critically reading the manuscript, H. Holtmann (Medizinische Hochschule Hannover, Germany) for providing β-globin cDNA, and Michael Kastan (St. Jude Children's Research Hospital, Memphis, Tennessee) for providing Flag-tagged ATM kinase cDNA. This research was supported by the Sonderforschungsbereich 566 (B2), Leistungsorientierte Mittelvergabe with Frauenfactor, PhD program from MHH and Leukaemia Lymphoma Research, UK.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2820911.
REFERENCES
- Adams BR, Golding SE, Rao RR, Valerie K 2010. Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants. PLoS ONE 5: e10001 doi: 10.1371/journal.pone.0010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney L, Pierce A, Rijnen M, Gonzalez Sanchez MB, Hamzah HG, Zhang L, Tamura T, Whetton AD 2009. THOC5 couples M-CSF receptor signaling to transcription factor expression. Cell Signal 21: 309–316 [DOI] [PubMed] [Google Scholar]
- Carrier F, Gatignol A, Hollander MC, Jeang KT, Fornace AJ Jr 1994. Induction of RNA-binding proteins in mammalian cells by DNA-damaging agents. Proc Natl Acad Sci 91: 1554–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Aguilera A 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev 11: 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, García-Rubio M, Prado F, Aguilera A 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol Cell Biol 21: 7054–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely Y, Dimitrova DD, Borowiec JA 2002. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol Cell Biol 22: 6014–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Bounkari O, Guria A, Klebba-Faerber S, Claußen M, Pieler T, Griffiths JR, Whetton AD, Koch A, Tamura T 2009. Nuclear localization of the pre-mRNA associating protein THOC7 depends upon its direct interaction with Fms tyrosine kinase interacting protein (FMIP). FEBS Lett 583: 13–18 [DOI] [PubMed] [Google Scholar]
- Gaillard H, Wellinger RE, Aguilera A 2007. A new connection of mRNP biogenesis and export with transcription-coupled repair. Nucleic Acids Res 35: 3893–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A 2007. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell 131: 915–926 [DOI] [PubMed] [Google Scholar]
- Guria A, Tran DD, Ramachandran S, Koch A, El Bounkari O, Dutta P, Hauser H, Tamura T 2011. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA 17: 1048–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC 2004. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64: 9152–9159 [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. 2004. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431: 997–1002 [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Inoue H, Hurt E, Yoneda Y 2009. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J 28: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274: 37538–37543 [DOI] [PubMed] [Google Scholar]
- Koch A, Scherr M, Breyer B, Mancini A, Kardinal C, Battmer K, Eder M, Tamura T 2008. Inhibition of Abl tyrosine kinase enhances nerve growth factor-mediated signaling in Bcr–Abl transformed cells via the alteration of signaling complex and the receptor turnover. Oncogene 27: 4678–4689 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang X, Zhang X, Goodrich DW 2005. Human hHpr1/p84/Thoc1 regulates transcriptional elongation and physically links RNA polymerase II and RNA processing factors. Mol Cell Biol 25: 4023–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Huang Y, Sheikh MS 2003. Cloning and characterization of a novel gene PDRG that is differentially regulated by p53 and ultraviolet radiation. Oncogene 22: 7247–7257 [DOI] [PubMed] [Google Scholar]
- Mancini A, Koch A, Whetton AD, Tamura T 2004. The M-CSF receptor substrate and interacting protein FMIP is governed in its subcellular localization by protein kinase C-mediated phosphorylation, and thereby potentiates M-CSF-mediated differentiation. Oncogene 23: 6581–6589 [DOI] [PubMed] [Google Scholar]
- Mancini A, El Bounkari O, Norrenbrock AF, Scherr M, Schaefer D, Eder M, Banham AH, Pulford K, Lyne L, Whetton AD, et al. 2007. FMIP controls the adipocyte lineage commitment of C2C12 cells by downmodulation of C/EBPalpha. Oncogene 26: 1020–1027 [DOI] [PubMed] [Google Scholar]
- Mancini A, Niemann-Seyde SC, Pankow R, El Bounkari O, Klebba-Farber S, Koch A, Jaworska E, Spooncer E, Gruber AD, Whetton AD, et al. 2010. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol 8: 1 doi: 10.1186/1741-7007-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Merz C, Urlaub H, Will CL, Luhrmann R 2007. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 13: 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D 2000. mRNA stability in eukaryotes. Curr Opin Genet Dev 10: 193–198 [DOI] [PubMed] [Google Scholar]
- Motoyama N, Naka K 2004. DNA damage tumor suppressor genes and genomic instability. Curr Opin Genet Dev 14: 11–16 [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D 2002. A unified theory of gene expression. Cell 108: 439–451 [DOI] [PubMed] [Google Scholar]
- Pierce A, Carney L, Hamza HG, Griffiths JR, Zhang L, Whetton BA, Gonzalez Sanchez MB, Tamura T, Sternberg D, Whetton AD 2008. THOC5 spliceosome protein: a target for leukaemogenic tyrosine kinases that affects inositol lipid turnover. Br J Haematol 141: 641–650 [DOI] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J 17: 4859–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Cheng H 2005. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol 17: 269–273 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Köcher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11: 558–566 [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Hasskamp P, Schmedding I, Morandell S, van Vugt MA, Wang X, Linding R, Ong SE, Weaver D, Carr SA, et al. 2010. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol Cell 40: 34–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Tamura T, Mancini A, Joos H, Koch A, Hakim C, Dumanski J, Weidner KM, Niemann H 1999. FMIP, a novel Fms-interacting protein, affects granulocyte/macrophage differentiation. Oncogene 18: 6488–6495 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Naraba H, Sakyo T, Kitagawa T 2010. DNA damage–induced modulation of GLUT3 expression is mediated through p53-independent extracellular signal-regulated kinase signaling in HeLa cells. Mol Cancer Res 8: 1547–1557 [DOI] [PubMed] [Google Scholar]
- Yang C, Maiguel DA, Carrier F 2002. Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res 30: 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bhatia D, Xia H, Castranova V, Shi X, Chen F 2006. Nucleolin links to arsenic-induced stabilization of GADD45α mRNA. Nucleic Acids Res 34: 485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen X 2000. MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G2-M. Mol Cell Biol 20: 5602–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrun SD, Hoffman B, Liebermann DA 2009. Distinct mechanisms are utilized to induce stress sensor gadd45b by different stress stimuli. J Cell Biochem 108: 1220–1231 [DOI] [PubMed] [Google Scholar]