Abstract
Fatty acyl-CoA synthetase (fatty acid: CoA ligase, AMP-forming; (EC 6.2.1.3)) catalyzes the formation of fatty acyl-CoA by a two-step process that proceeds through the hydrolysis of pyrophosphate. Fatty acyl-CoA represents bioactive compounds that are involved in protein transport, enzyme activation, protein acylation, cell signaling, and transcriptional control in addition to serving as substrates for beta oxidation and phospholipid biosynthesis. Fatty acyl-CoA synthetase occupies a pivotal role in cellular homeostasis, particularly in lipid metabolism. Our interest in fatty acyl-CoA synthetase stems from the identification of this enzyme, long-chain fatty acyl-CoA ligase (LCFA) by microarray analysis. We found this enzyme to be differentially expressed by Leishmania donovani amastigotes resistant to antimonial treatment. In the present study, we confirm the presence of long-chain fatty acyl-CoA ligase gene in the genome of clinical isolates of Leishmania donovani collected from the disease endemic area in India. We predict a molecular model for this enzyme for in silico docking studies using chemical library available in our institute. On the basis of the data presented in this work, we propose that long-chain fatty acyl-CoA ligase enzyme serves as an important protein and a potential target candidate for development of selective inhibitors against leishmaniasis.
1. Introduction
Leishmaniasis is a disease caused by protozoan parasites of the Leishmania genus. Visceral leishmaniasis (VL), also known as kala-azar, is the most severe form of leishmaniasis (http://www.dndi.org/diseases/vl.html). With no vaccine in sight, treatment for kala-azar relies primarily on chemotherapy [1].
Phylogenetics suggests that Leishmania is relatively early branching eukaryotic cells and their cell organization differs considerably from that of mammalian cells [2, 3]. Hence, the biochemical differences between the host and parasite can be exploited for identification of new targets for rational drug design. It is also imperative that the probability of developing drug resistance should be less with these targets. This can be achieved by targeting an essential cellular process, which has the pressure to remain conserved and cannot be bypassed by using alternative pathway.
One interesting target which emerged from our microarray experiments [4] was long-chain fatty acid-CoA ligase (EC 6.2.1.3) (GenBank Accession No. XM_001681734), a key enzyme involved in the metabolism of fatty acids in all organisms [5–9]. Fatty acyl-CoA has multiple roles involved in protein transport [10, 11], enzyme activation [12], protein acylation [13], cell signaling [14], transcriptional regulation [15], and particularly β-oxidation and phospholipid biosynthesis. Especially in Leishmania, long-chain fatty acids are predominant precursors of total lipid composition (the combination of phospholipids, sphingolipids, and ergosterol). Long-chain fatty acyl-CoA ligase is critical enzyme processing long-chain fatty acid acylation which is essential for lysophosphatidylinositol (lyso-PI) incorporation into glycosyl phosphatidylinositols (GPIs) [16, 17]. These GPIs-anchors are the major surface virulent factors in Leishmania and have received considerable attention [18]. De novo sphingolipid biosynthesis starts with the condensation of serine and the product of long-chain fatty acyl-CoA ligase. L. major preferentially incorporates myristoyl-CoA (C14) over palmitoyl-CoA (C16) into their long-chain base [19, 20]. This selection of specific long-chain fatty acyl-CoA reflects the presence of myristoyl-specific long-chain fatty acyl-CoA ligase in Leishmania [21].
Gaining new knowledge on fatty acid metabolism will not only provide fundamental insight into the molecular bases of Leishmania pathogenesis but also reveal new targets for selective drugs. Enzymes involved in fatty acid and sterol metabolism have been shown to be important pharmaceutical targets in Leishmania and other kinetoplastida [22]. Triacsin C, a specific inhibitor of long-chain fatty acyl-CoA synthetase, was shown to have an inhibitory effect on the growth of Cryptosporidium parvum in vitro [23].
Four fatty acyl-CoA synthetases have been described previously in Trypanosoma brucei, displaying different chain-length specificities [24, 25]. The whole genome sequence of three Leishmania spp. (L. major, L. infantum, and L. braziliensis) has been sequenced, and the availability of putative long-chain fatty acyl-CoA ligase genes was present in all three Leishmania spp. at chromosome 13, which would be required for initiation of β-oxidation and fatty acid metabolism.
In the present study we confirm the presence of long-chain fatty acyl-CoA ligase gene in Leishmania donovani clinical isolate collected from, the state of Bihar India [26–29], which alone accounts for 50% of the total burden of visceral leishmaniasis worldwide [30]. Further progress in the understanding of this enzyme is likely to be achieved through the whole genome sequence (WGS) project of these clinically important isolates [26–29], underway in our laboratory (http://www.leishmaniaresearchsociety.org/).
2. Material and Methods
2.1. Collection of Clinical Isolates
The clinical isolates of L. donovani were collected from two kala-azar patients selected from Muzaffarpur, Bihar. The criterion for visceral leishmaniasis diagnosis was the presence of Leishman-Donovan (LD) bodies in splenic aspirations performed, which was graded to standard criteria [30]. Response to sodium antimony gluconate (SAG) treatment was evaluated by repeating splenic aspiration at day 30 of treatment. Patients were designated as antimonial responsive (L. donovani isolate 2001) based on the absence of fever, clinical improvement with reduction in spleen size, and absence of parasites in splenic aspirate while patients who showed presence of parasites in splenic aspiration were considered to be antimonial unresponsive (L. donovani isolate 39) [26–29].
2.2. Sample Collection and Nuclear DNA Isolation
L. donovani isolates 2001 (SAG-sensitive) and 39 (SAG-resistant) used in the present study, were maintained in culture as described previously in [26–29]. For nuclear DNA isolation 10–15 mL log-phase culture was taken and centrifuged at 5,000 rpm for 8 min at 4°C. The supernatant was decanted; cell pellet was resuspended in 3–6 mL NET buffer and centrifuged at 5,000 rpm for 8 min at 4°C. The supernatant was discarded, and the pellet was redissolved in 750 μL NET buffer, 7.5 μL proteinase K (10 mg/mL stock) (MBI, Fermentas, Cat No. EO0491), and 50 μL of 15% sarkosyl. Sample was incubated at 37°C overnight for proteinase K activity. The cell lysate was centrifuged at 18,000 rpm for 1 hr at 4°C. The supernatant containing nuclear DNA was transferred to a fresh tube and given RNase treatment (20 μg/mL) (MBI, Fermentas, Cat No. EN0531) at 37°C for 30 min. DNA was extracted first with one volume phenol/chloroform/isoamyl alcohol (25 : 24 : 1) and finally with chloroform. Nuclear DNA was precipitated with 2.5 volumes of prechilled absolute ethanol, dissolved in nuclease-free water and stored at 4°C for future use.
2.3. Primer Design, PCR Amplification, and Sequencing of Long-Chain Fatty Acyl-CoA Ligase Gene
PCR amplification was carried out using Pfu DNA polymerase (MBI, Fermentas, Cat No. EP0501). Reactions were carried out in a Perkin Elmer GeneAmp PCR system with 2001 nuclear DNA (10–50 ng) as template. The following oligonucleotide primers were designed on the basis of available gene sequence of L. major (GenBank Accession No. XM_001681734): forward primer: 5′GGGCCATATGCTGCAGCG 3′ (18 mer) and reverse primer: 5′GGCCTCGAGCTAAAACAAATCATCG3′ (25 mer). The amplification conditions were initial denaturation at 95°C for 10 min, denaturation at 95°C for 30 sec, annealing at 65°C for 1 min, extension at 72°C for 2 min, and final extension at 72°C for 10 min; 30 cycles. The PCR product was purified from agarose gel using MBI Fermentas DNA Extraction kit (MBI, Fermentas, Cat No. K0513) and further for DNA sequencing by Bangalore Genei, India.
2.4. Characterization of Long-Chain Fatty Acyl-CoA Ligase Gene
L. donovani nuclear DNA (16 μg for each reaction) of two different clinical isolates, drug (SAG) sensitive 2001 and drug (SAG) resistant 39, were digested with 40-unit three different restriction enzymes (PvuII, BamHI, and XhoI), which were cut overnight and separated on 0.8% agarose gel by electrophoresis at 50 V. In order to improve transfer efficacy, DNA in agarose gel was treated with 0.25 N HCl for 15 min (partial depurination), rinsed with autoclaved water 3x, and treated with 0.4 N NaOH (breaking backbone at depurinated region) for 30 min. DNA was transferred to nylon membrane by conventional downward capillary transfer method for 5 h using 3 mm Whatman paper wick [8]. The efficiency of transfer was assessed by visualizing DNA by methylene blue staining. After transfer on nylon membrane the DNA was neutralized in 0.5 M Tris (pH 7.4), 1.5 M NaCl, 2x for 5 min at room temperature. The membrane was then washed in 2X SSC, 2x for 15 min. Nylon membrane was incubated with 2.5 mL of prehybridization buffer (0.6 M NaCl, 0.5 M Tris-HCl (pH 7.5), 0.008 M EDTA, 1% sodium pyrophosphate, 0.2% SDS, and 50 μg/mL heparin) and incubated in a hybridization oven at 65°C for 2 h. Radioactive probe was prepared by labeling 25 ng of the DNAs with [α-32P] dCTP by random priming method (BRIT/BARC, India) and purified using a desalting column (sephadex G-50). The radioactivity was checked with a Geiger Muller Counter (dosimeter) and stored at −20°C. The probe was added to the prehybridization buffer and incubated at 65°C overnight in hybridization oven. Membrane was washed twice with 2X SSC, 0.1% SDS (15 min each) at 65°C and then washed with 2X SSC, 0.1% SDS for 30 min at 65°C to reduce background signals. Hybridized membrane was layered over a wet Whatman paper sheet to soak extra solution and covered with Saran Wrap (cellophane paper) and exposed to X-ray film. After 4–18 h exposure in an exposure cassette at −70°C, X-ray film was developed for analysis.
2.5. Phylogenetic Analysis
The amino acid sequence of Leishmania long-chain fatty acyl-CoA ligase, obtained from our microarray experiments [4], was compared with sequences available in GeneDB ORTHOMCL4080 database (http://www.genedb.org/) to identify the nearest ortholog of this sequence in kinetoplastida. Multiple sequence alignments were performed using Clustal W version 1.8 (http://www.ebi.ac.uk/clustalw) and T-cofee [31]. To calculate evolutionary distances of kinetoplastida long-chain fatty acyl-CoA ligases with human acyl CoA synthetases (ACSs) [32], phylogenetic dendrograms were constructed by neighbor-joining method and tree topologies were evaluated by performing bootstrap analysis of 1000 data sets using MEGA 3.1 (Molecular Evolutionary Genetics Analysis) [33]. All 26 human ACSs amino acid sequences were selected [32], along with their transcript variants which are aligned with different long-chain fatty acyl-CoA ligase ortholog present in kinetoplastida family, to define the clade difference with Trypanosome and Leishmania long-chain fatty acyl-CoA ligase, and human acyl-CoA synthetases.
2.6. Homology Modeling of Leishmania Long-Chain Fatty Acyl-CoA Ligase
The amino acid sequence of Leishmania long-chain fatty acyl-CoA ligase was retrieved from the NCBI database (GenBank Accession No. XM_001681734). It was ascertained that the 3D structure of Leishmania long-chain fatty acyl-CoA ligase protein was not available in Protein Data Bank (PDB); hence, the present exercise of developing the 3D model of this protein was undertaken. cBLAST (http://www.ncbi.nlm.nih.gov/Structure/cblast/cblast.cgi) and PSI-BLAST search was performed against PDB with the default parameter to find suitable templates for homology modeling. The sequence alignment of Leishmania long-chain fatty acyl-CoA ligase and respective templates was carried out using the CLUSTALW (http://www.ebi.ac.uk/clustalw) and MODELLER9V8 programs [34, 35]. The sequences that showed the maximum identity with high score and lower e-value were used as a reference structure to build a 3D model.
The retrieved sequences of Thermus thermophilus (PDB Accession Code: 1ULT, 1V25, 1V26) [36] and Archaeoglobus fulgidus (PDB Accession Code: 3G7S) long-chain fatty acyl-CoA ligases served as template for homology modeling based on its maximum sequence similarity to Leishmania long-chain fatty acyl-CoA ligase. The alignment was manually refined at some loops region of the templates. The resulting alignment was used as an input for the automated comparative homology modeling for generating 3D model structure of Leishmania long-chain fatty acyl-CoA ligase. The academic version of MODELLER9V8 was used for model building. The backbones of core region of the protein were transferred directly from the corresponding coordinates of templates. Side chain conformation for backbone was generated automatically. Out of 50 models generated by MODELLER, the one with the best DOPE score, minimum MOF (Modeller Objective Function), and best VARIFY 3D profile was subjected to energy minimization. In order to assess the stereochemical qualities of 3D model, PROCHECK analysis [37] was performed and Ramachandran plot was drawn.
3. Results
3.1. Metabolism of Long-Chain Fatty Acyl-CoA Ligase Enzyme
Three types of fatty acyl CoA ligase have been defined with respect to the length of the aliphatic chain of the substrate: short (SC-EC 6.2.1.1), medium (MC-EC 6.2.1.2), and long-chain (LC-EC 6.2.1.3) fatty acyl-CoA ligase. These utilize C2-C4, C4-C12, and C12-C22 fatty acids as substrates, respectively [9]. Fatty acid activation step involves the linking of the carboxyl group of the fatty acid through an acyl bond to the phosphoryl group of AMP. Subsequently, a transfer of the fatty acyl group to the sulfhydryl group of CoA occurs, releasing AMP [38–40]. This magnesium-dependent two-step acylation of fatty acid by fatty acyl CoA synthetases was defined as unidirectional Bi Uni Uni Bi Ping-Pong mechanism [36, 39].
Genome analysis suggests that L. major oxidizes fatty acids via β-oxidation in two separate cellular compartments: the glycosome and mitochondria [41]. An argument for the involvement of glycosome in lipid metabolism is the fact that in each of three trypanosomatid genomes three genes called half ABC transporters (GATI 1-3) have been found identical with peroxisomal transporters involved in fatty acid transport. In T. brucei, it was conformed that these transporters are associated with glycosomal membrane [42]. These transporters might be coupled with fatty acyl-CoA ligase in glycosome, which can provide activated form of fatty acids to these transporters like oleoyl-CoA, and also other acylated fatty acids.
In T. brucei, little β-oxidation was observed in mitochondria. However, T. brucei contains at least two enzymes involved in β-oxidation of fatty acid (2-enoyl-CoA hydratase and hydroxyacyl-dehydrogenase, encompassed in a single protein) with glycosome localization [43]. The presence of a PTS (Peroxisomal Targeting Sequence) on T. brucei and T. cruzi carnitine acetyl transferase, catalysing the last peroxisomal step in fatty acid oxidation, suggests that the major β-oxidation processes are situated in glycosomes [44]. In L. donovani, one of the β-oxidation enzyme 3-hydroxyacyl-CoA dehydrogenase has been localized to glycosomes [45]. The hypothetical localization of Leishmania long-chain fatty acyl-CoA ligase was predicted in mitochondria or glycosome but, with the reference of other organisms, the specialized localization of specific long-chain fatty acyl-CoA ligase family protein needs to be taken into account in future.
As mentioned in a previous study β-oxidation has been found to be unregulated in Leishmania's amastigotes then in promastigote stage [46–48]. This specialized increase was described so that, in infectious stage, energy requirement was supplemented to utilize fatty acid as carbon and energy source rather than glucose [47]. Long-chain fatty acyl-CoA ligase is the key enzyme involved in β-oxidation of fatty acids, and its compartmentation in glycosome supports a strong evidence of the involvement of this enzyme in cellular biogenesis and its importance at particular stage of Leishmania life cycle. In the same way upregulation of long-chain fatty acyl-CoA ligase with combination of other enzymes involved in fatty acid catabolism might play a crucial role in cell survival at infectious stage of Leishmania, and these analyses must be supplemented with experimental biology.
3.2. Characterization of Leishmania Long-Chain Fatty Acyl-CoA Ligase Gene
The presence of L. donovani long-chain fatty acyl-CoA ligase gene in the clinical isolates was ascertained by PCR amplification. The putative long-chain fatty acid-CoA ligase gene of L. major is present in the Leishmania Genome Databank (http://www.genedb.org/) on chromosome 13 (Figure 1). Specific 2010 bp size amplified product was obtained, showing the presence of long-chain fatty acyl-CoA ligase gene in the L. donovani clinical isolate (Figure 2(D)). The amplified product was sequenced and confirmed to be long-chain fatty acid-CoA ligase gene by performing NCBI-BLAST identity with L. major gene. NCBI-BLAST result showed 96% sequence similarity and 1% gaps with L. major long-chain fatty acyl-CoA ligase gene (GenBank Accession No. XM_001681734). The starting 18 nucleotides and 19 nucleotides from the end sequence were missed due to direct amplified product sequencing. These nucleotides were collected from its maximum similar L. major long-chain fatty acyl-CoA ligase sequence (GenBank Accession No. XM_001681734).
Figure 1.
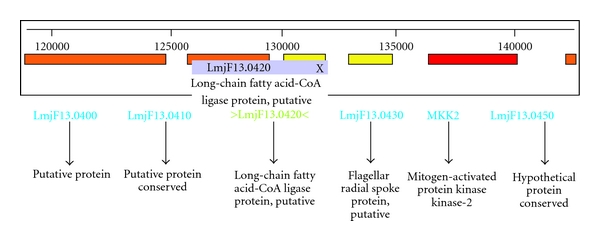
Graphical representation of long-chain fatty acyl-CoA ligase (LCFA) gene (in Artemis) on chromosome 13 of Leishmania major.
Figure 2.
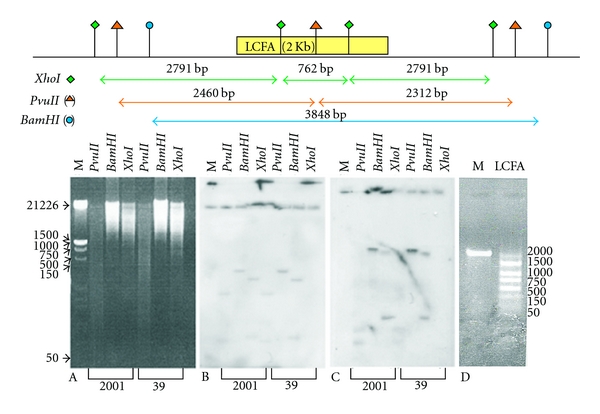
Determination of long-chain fatty acyl-CoA ligase gene copy number. The nuclear DNA of L. donovani 2001 and 39 promastigotes was isolated and 16 μg was digested with different restriction enzymes. (A) Nuclear DNA digest stained with ethidium bromide (B) Southern blot of “A” with α-tubulin gene probe. (C) Southern blot of “A” with long-chain fatty acyl-CoA ligase probe. (D) PCR amplification of long-chain fatty acyl-CoA ligase gene (M: Marker, LCFA: 2010 bp of long-chain fatty acyl-CoA ligase gene).
For the determination of long-chain fatty acid-CoA ligase gene copy number, nuclear DNA from the L. donovani clinical isolates (2001, 39) was digested with various restriction enzymes. The restriction map was designed from the complete putative long-chain fatty acyl-CoA ligase gene and the flanking region present in chromosome 13 of L. major (Figure 2). Southern hybridization was performed using the 2010 bp long-chain fatty acid-CoA ligase gene PCR product as probe (Figure 2(C)). The same blot was also probed with alpha tubulin gene probe as an internal control, showing equal loading (Figure 2(B)). Complete digestion resulted in a single copy within the L. donovani genome, as BamHI enzyme showed only one band of approximately 3848 bp, except PvuII which was cut once into the gene sequence and XhoI which was cut twice into the gene sequence, which exhibited two and three hybridizing bands, respectively. The results showed that long-chain fatty acid-CoA ligase is present as a single copy gene in the L. donovani genome. The restriction pattern also verifying the restriction pattern of L. donovani and L. major long-chain fatty acyl CoA ligase coding region is almost the same.
3.3. Identification of Conserved Domains and Structure-Function Correlation in Leishmania Long-Chain Fatty Acyl-CoA Ligase
Leishmania long-chain fatty acyl-CoA ligase gene was translated from full length ORF on the basis of its nucleic acid sequence. Long-chain fatty acyl-CoA synthetase from T. thermophilus, yeast, and E. coli and all 26 distinct human acyl-CoA synthetases were subjected to phylogenetic analysis to facilitate the evaluation of conserved motif with relationship of reference Leishmania long-chain fatty acyl-CoA ligase amino acid sequence. The amino acid sequence of Leishmania long-chain fatty acyl-CoA ligase (LdLCFA) was aligned with LC-FACS from T. thermophilus (TtLC-FACS,1ultA), human (LCFA_HUMAN, P41215), yeast (LCF1_YEAST, P30624), and E. coli (LCFA_ECOLI, P29212) on the basis of PSI-BLAST (Figure 3). However the overall similarity of Leishmania long-chain fatty acyl-CoA ligase (LdLCFA) with other fatty acyl-CoA synthetases family proteins is low, about 17% with TtLC-FACS, 15% with LCFA_HUMAN, 14% with LCF1_YEAST, and 13% with LCFA_ECOLI. Based on the crystal structure of TtLC-FACS and alignment with other long-chain fatty acyl-CoA synthetases [36], the amino acid sequence of Leishmania long-chain fatty acyl-CoA ligase shows conserve region corresponding to the linker (L), adenine (A), and gate (G) motifs as well as the P-loop, the phosphate-binding site. Previous studies [32, 36] put forward different motifs which can give insight to enhance our understanding of predicted structure-function relationships in Leishmania. P-loop is the Motif I which is also known as AMP-binding domain found in a close proximity to the adenosine moiety and helps to maintain the substrate in the proper orientation. The consensus sequence of Motif I, [Y,F]TSG[T,S]TGXPK shows high level of conservation with respect to Leishmania long-chain fatty acyl-CoA ligase, that is, 237-FTAGTTGPPK-246. Motif II contains the L-motif (432-DRLKDL-437) that acts as a linker between the large N-terminal domain and the smaller C-terminal domain in TtLC-FACS. The linker region is thought to be critical for catalysis function as it facilitates a conformational change upon ATP binding that permits subsequent binding of the fatty acyl and/or CoA substrates. In Leishmania long-chain fatty acyl-CoA ligase, this linker region (517-GNKDVL-522) is less similar compared with other organisms and is likely to be critical in enzyme activity. Motif III was found to be in all acyl CoA synthetases and a part of A-motif (adenine motif). This region has been described as an ATP/AMP-binding domain in other acyl-CoA synthetases [49–51]. The conserved consensus sequence of A-motif is YGXTE, a highly conserved motif with respect to Leishmania long-chain fatty acyl-CoA ligase region, that is, YGFME. From the crystal structure of TtLC-FACS, it was proposed that Y324 was an adenine-binding residue [42] and also conserved throughout all organisms including Leishmania. The crystal structure of S. enterica acetyl-CoA synthetase revealed that the glutamate residue of A-motif is positioned near oxygen O1 of the AMP phosphate [52]. This region was predicted to be involved in substrate binding or stabilization, conserved in Leishmania long-chain fatty acyl-CoA ligase also. Motif IV comprises the first five residues of the nine-amino acid G-(or gate) motif (226-VPMFHVNAW-234) of ttLC-FACS (36), showing less sequence similarity with Leishmania long-chain fatty acyl-CoA ligase (281-CSWCVAGAL-289). From the crystal structure of TtLC-FACS, it was proposed that the indole ring of W234 acts as a gate and blocks the entry of fatty acids into its substrate binding tunnel unless ATP is first bound, resulting in a conformational change that swings the gate open (36). However, a tryptophan residue corresponding to W234 was not found in any Leishmania, human, yeast, and E. coli fatty acyl-CoA synthetase sequences. In contrast, although no highly conserved sequences were identified, a corresponding gate residue may be located elsewhere in the structure of Leishmania long-chain fatty acyl-CoA ligase.
Figure 3.
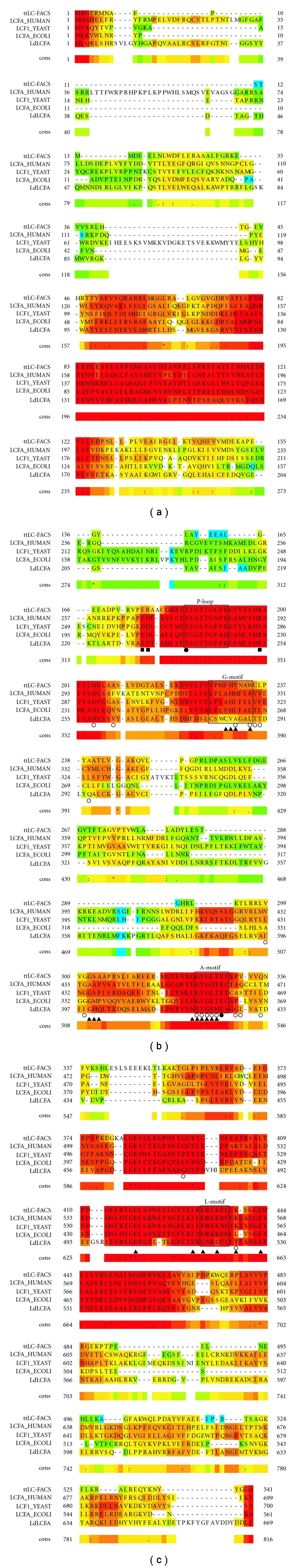
Amino acid sequence alignments of long-chain fatty acyl-CoA synthetases. The amino acid sequence of Leishmania LCFA (LdLCFA) was aligned with LC-FACS from T. thermophilus (ttLC-FACS, Q6L8FO), human (LCFA_HUMAN, P41215), yeast (LCF1_YEAST, P30624), and E. coli (LCFA_ECOLI, P29212). The boxed areas denoted with bold letters correspond to conserved motifs of long-chain fatty acyl-CoA ligase: G, A, and L motifs as well as the P-loop. Filled squares, open circles, filled circles, and filled triangles indicate residues believed to be involved in dimer formation, fatty acid binding, magnesium ion binding, and adenylate binding, respectively.
The fatty acyl-CoA synthetases are part of a large family of proteins referred to as the ATP-AMP-binding proteins. A common feature of enzymes in this family is that they all form an adenylated intermediate as part of their catalytic cycle. This group of enzymes is diverse in catalyzing the activation of a wide variety of carboxyl-containing substrates, including amino acids, fatty acids, and luciferin. Sequence comparison of members of the ATP-AMP-binding protein family has identified two highly conserved sequence elements, [53] Y[T]S[GTTG]X[PKGV]⋯G[YG]XT[E] (the bracket shows the conserved sequence in Leishmania long-chain fatty acyl-CoA ligase), which encompass the ATP-AMP signature motif (Figure 4).
Figure 4.
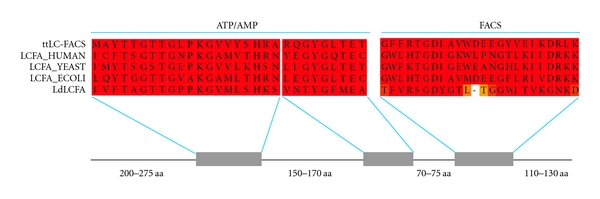
Domain organization of amino acid sequence alignments of ATP-AMP and fatty acyl CoA synthetase (FACS) motif from T. thermophilus (ttLC-FACS), human (LCFA_HUMAN), yeast (LCF1_YEAST), E. coli (LCFA_ECOLI), and Leishmania (L LCFA).
In fatty acyl-CoA synthetases family proteins, there was a third sequence element defined as FACS signature motif that was less conserved and partially overlaps the FACS signature motif, which is involved in both catalysis and specificity of the fatty acid substrate [54]. There are a number of notable features within the FACS signature motif: (i) this region contains two invariant glycine residues (at positions 2 and 7) and a highly conserved glycine at position 16, Leishmania long-chain fatty acyl-CoA ligase shares glycine residue with other FACSs at position 7 and 16 but Tyr instead of Gly was found in position 2. (ii) This region contains additional six residues that are invariant in the fatty acyl-CoA synthetases: W[3], T[6], D[8], D[22], R[23], and K[25], but in Leishmania long-chain fatty acyl-CoA ligase these residues are F[3], S[6], D[8], G[22], N[23], and D[25]. (iii) The residue in the fourth position is hydrophobic and is a leucine, a methionine, or phenylalanine. However, in Leishmania long-chain fatty acyl-CoA ligase hydrophobic residue valine was situated in position 4. (iv) This region of enzyme contains hydrophobic residues (leucine, isoleucine, or valine) at positions 4, 9, 18, 20, and 21. These residues, in addition to tryptophan or phenylalanine residues at position 3, may comprise part of a fatty-acid-binding pocket. All of these five conserved regions from FACS signature motif are having similarity among them except Leishmania long-chain fatty acyl-CoA ligase, with some variable regions. These less conserved regions in Leishmania long-chain fatty acyl-CoA ligase-FACS signature motif were predicted to adopt inconsistent specificity and catalytic activities of the fatty acid substrate compared to other fatty acyl CoA synthetases.
3.4. Phylogenetic Analysis of Leishmania Long-Chain Fatty Acyl-CoA Ligase and Human Acyl-CoA Synthetases Sequences
We performed phylogenetic analysis to infer evolutionary relationships of all available sequences from kinetoplastida long-chain fatty acyl-CoA ligases (Table 1) and human (host) ACSs family sequences. This experiment was performed to validate that the parasite enzyme is unquestionably different from the human enzyme, and this aspect merits further study to validate this enzyme as a drug target. We obtained comparable results using the neighbor-joining distance-based algorithm as well as maximum parsimony. We found 9 clades, including kinetoplastida clade (one set of six kinetoplastida long-chain fatty acyl-CoA ligase protein family) forming a clade with high bootstrap support (Figure 5). kinetoplastida clade was highly dissimilar and distinct from all ancestral nodes with other human ACSs family proteins and showing distinctiveness of kinetoplastida long-chain fatty acyl-CoA ligases, including Leishmania long-chain fatty acyl-CoA ligase. This divergence of Leishmania long-chain fatty acyl-CoA ligase with respect to the homologous human enzymes may be an important protein as a potential target candidate for chemotherapeutic antileishmanial drugs.
Table 1.
Selected ortholog for Leishmania donovani long-chain fatty acyl-CoA ligase gene in kinetoplastida: ORTHOMCL4080 (http://www.genedb.org/).
| Systematic ids | Organism | Product |
|---|---|---|
| LbrM13_V2.0240 | L. braziliensis MHOM/BR/75/M2904 | Fatty acid thiokinase (long chain), putative; acyl-CoA synthetase, putative; long-chain-fatty acid-CoA ligase protein, putative |
|
| ||
| LinJ13_V3.0300 | L. infantum JPCM5 | Fatty acid thiokinase (long chain), putative; long-chain-fatty acid-CoA ligase protein, putative; acyl-CoA synthetase, putative |
|
| ||
| LmjF13.0420 | L. major strain Friedlin | Long-chain fatty acid-CoA ligase protein, putative; acyl-CoA synthetase, putative; fatty acid thiokinase (long chain), putative |
|
| ||
| Tb11.02.2070 | T. brucei 927 | Long-chain fatty acid-CoA ligase protein, putative; fatty acid thiokinase (long chain), putative; acyl-CoA synthetase, putative |
|
| ||
| Tc00.1047053504089.40 | T. cruzi | Long-chain fatty acid-CoA ligase protein, putative; acyl-CoA synthetase, putative; fatty acid thiokinase, long chain, putative |
|
| ||
| TvY486_1104610 | T. vivax | Long-chain fatty acid-CoA ligase protein, putative |
Figure 5.
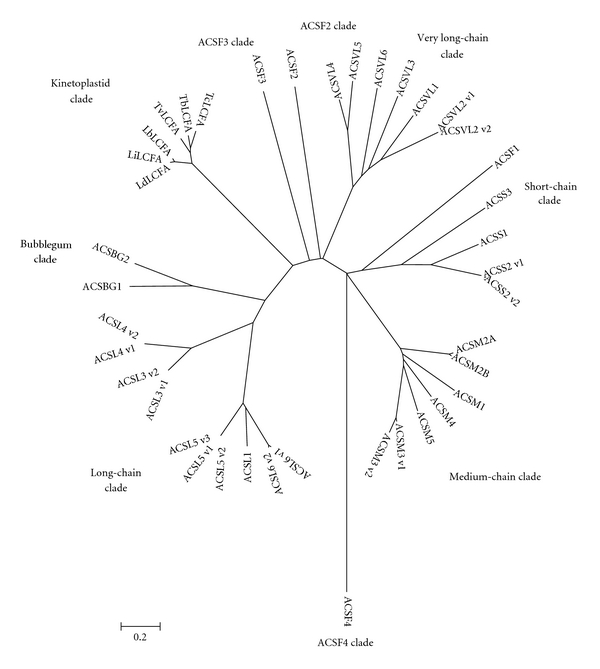
Phylogenetic trees based on human acyl CoA synthetases (ACSs) gene sequences [38] showing the relationship of all Leishmania long-chain fatty acyl-CoA ligase orthologs (Table 1), with their nearest phylogenetic relatives. Phylogenetic trees were constructed by the neighbour-joining method as well as the maximum likelihood method as implemented in MEGA4 software. Numbers at nodes are bootstrap values (ML/NJ; xx represents no bootstrap value in NJ tree where nodes differ in both dendrograms;—represents value <50). The bar represents 0.02 substitutions per alignment position. The bar represents substitutions per alignment position.
3.5. Homology Modeling of Leishmania Long-Chain Fatty Acyl-CoA Ligase Protein
The backbone root-mean-square-deviation (RMSD) values between final model and template crystal structure used are 1.04 Å with Thermus thermophilus (PDB Accession Code: 1ULT, 1V25, 1V26) and 1.40 Å with Archaeoglobus fulgidus (PDB Accession Code: 3G7S) long-chain fatty acyl-CoA ligase. Small RMSD can be interpreted as structures share common structural homology and the generated structure is reasonable for structural similarity analysis (Figure 6). The final modeled structure of Leishmania long-chain fatty acyl-CoA ligase was evaluated for overall quality using available analyses procedures. These analysis compare specific properties of the model with those of known high-quality protein structures using programs like PROCHECK, Verify3D, and WHATIF (Table 2). An important indicator of the stereochemical quality of the model is distribution of the main chain torsion angles phi and psi in Ramachandran plot (Figure 7). The plot clearly shows the vast majority of the amino acids in a phi-psi distribution consistent with right α-helices, and the remaining fall into beta configuration. Only three residues fall outside the allowed regions. Plots comparison shows that the structure is reasonable overall because the space distribution for the homology-modeled structure was similar to the X-ray structure of the Thermus thermophilus long-chain fatty acyl-CoA ligase (PDB Accession Code: 1ULTA). The results showed that our modeled structure was reasonably good at that much less sequence identity.
4. Discussion
Earlier during the course of work, microarray analysis was performed on the same clinical L. donovani isolates (2001 and 39) in order to identify differential gene expression [4]. Out of all genes found differentially expressed, significant upregulation of long-chain fatty acyl-CoA ligase gene in SAG unresponsive clinical isolate [33] was found to be intracellular amastigote specific and has confirmed the involvement of long-chain fatty acyl-CoA ligase in resistance. Similarly, it has been proven before that the rate limiting enzyme, long-chain fatty acyl-CoA ligase of β-oxidation, was found to be upregulated in amastigotes derived from cloned line of L. donovani ISR because, during late stages of differentiation, the parasites shift from glucose to fatty acid oxidation as the main source of energy, and thereby there is increase in enzyme activity associated with β-oxidation capacity [47, 48]. Early in vivo studies showed that enzymatic activities associated with β-oxidation of fatty acids were significantly higher in L. mexicana amastigotes [47]. Additionally microarray experiments with intracellular amastigotes hybridized onto Affymetrix Mouse430_2 GeneChips showed that several genes involved in fatty acid biosynthesis pathway were found to be upregulated [55]. Presently studies are ongoing in our laboratory on microarray analysis using intracellular amastigotes hybridized to Affymetrix GeneChip human genome U133 Plus 2.0 array which will further yield useful information towards the fatty acid/lipid metabolism within this clinical isolate. A very recent study by Yao et al., 2010, on differential expression of plasma membrane proteins in logarithmic versus metacyclic promastigotes of L. chagasi has also identified long-chain fatty acyl-CoA synthetase [56].
As mentioned before, long-chain fatty acid-CoA ligase is present in both prokaryotes and eukaryotes. This divergence of Leishmania long-chain fatty acyl-CoA ligase with respect to the homologous human enzymes may be an important protein as a potential target candidate for chemotherapeutic antileishmanial drugs. Many differences exist between host and parasite pertaining to the structure and arrangement of this enzyme. However, Leishmania has significant divergence and adaptation to specific environmental conditions between its two life stages, in the insect vector and human host. This can affect the parasites metabolic machinery in terms of presence of certain pathways, their subcellular localization and expression at different developmental stages, and the interplay between scavenging and synthesis of key metabolites. It has been argued previously [57] that successful targets for metabolic intervention are most likely to be found among enzymes exerting strong control of flux through metabolic pathways. These control points are likely to be species and development dependent. Even if a unique or highly divergent enzymatic process is found in the parasites, this does not necessarily mean it can be developed as a target for useful inhibitors. On the other hand, enzymes that are present in both the parasites and their animal hosts will often differ sufficiently in their sequence for inhibitors to be specific. Finally, even orthologous enzymes functioning in the same pathway and in the same subcellular compartment of the parasites may have different inhibitor binding properties, leading to variability in the effectiveness and specificity of inhibitors targeting any particular enzyme.
The detection of the long-chain fatty acid-CoA ligase gene in the genome of L. donovani clinical isolate, in the present study, deserves a full exploration with respect to its potential as a drug target. Changes in membrane lipids/deficiency of certain fatty acids and disease association have been documented [34, 58]. Modulation of enzymes involved in lipid synthesis and of others possibly involved in cell wall metabolism may modify access of drug to the plasma membrane. Moreover, our microarray experiment indicated that this enzyme was amastigote specific making it all the more important to study it further and test if it can be exploited as a validated drug target. We have also shown earlier in our laboratory [34] that modification of lipid composition on the plasma membrane of the parasite might have important implications towards generating susceptibility/resistance to antileishmanial drugs. As this enzyme stipulates several important cellular processes in Leishmania like stage-specific expression [47, 48], host-parasite interaction [55], cell membrane composition [17, 18], phospholipid biosynthesis [16, 21], and drug resistance [4], the present study proposed further evaluation of Leishmania long-chain fatty acyl-CoA ligase as a candidate drug target.
Figure 6.
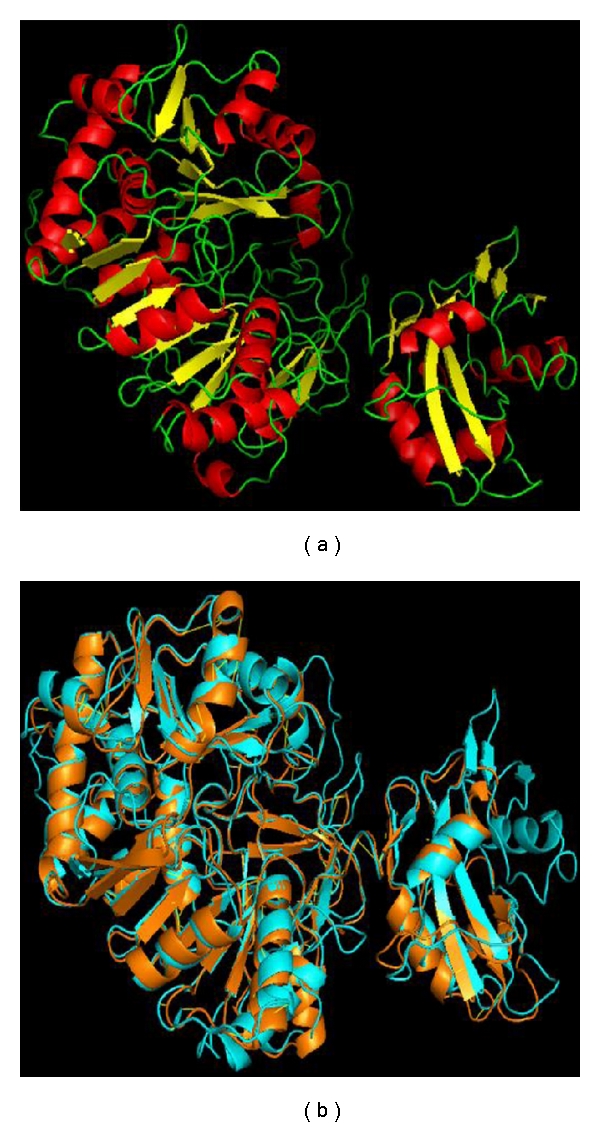
Leishmania long-chin fatty acyl-CoA ligase model. (a) The larger left hand-side domain is the N-terminal domain and the smaller one is the C-terminal domain which is connected by a linker chain. (b) Superposition of the modeled structure of Leishmania long-chain fatty acyl-CoA ligase (Orange) with the crystal structure of the T. thermophilus long-chain fatty acyl-CoA synthetase (PDB code: 1ult A) (Blue).
Figure 7.
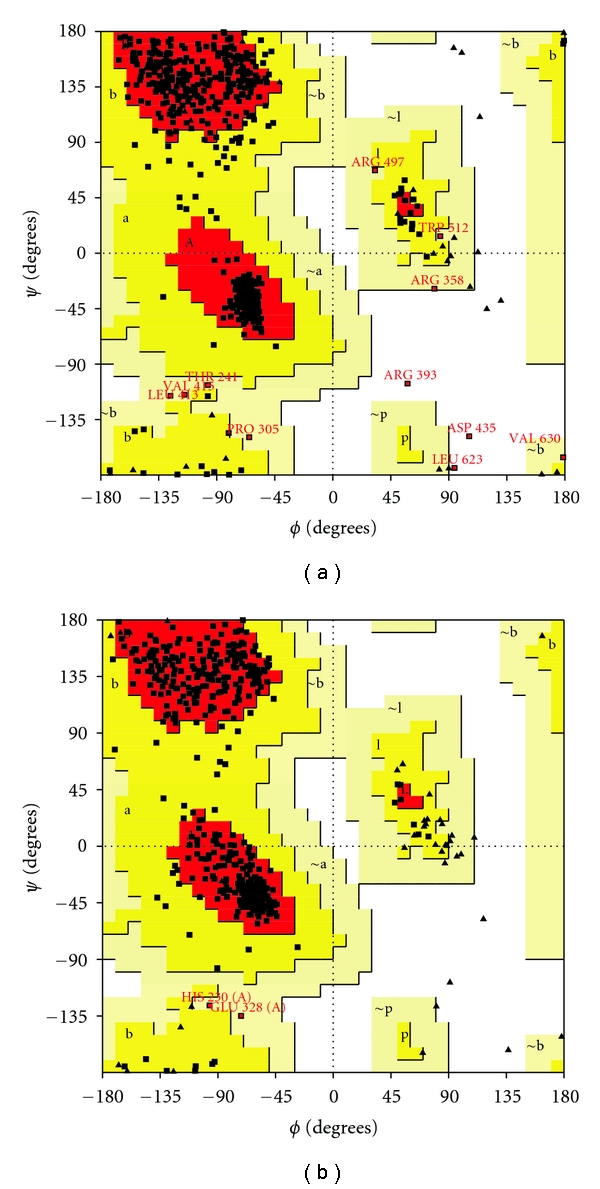
Ramachandran plot of (a) modeled structure Leishmania long-chain fatty acyl-CoA ligase (b) and the crystal structure of the Tt0168 (PDB code: 1ult A).
Table 2.
Results of protein structure by PROCHECK and VERIFY 3D.
| Leishmania long-chain fatty acyl-CoA ligase | T. thermophilus long-chain fatty acyl-CoA synthetase (1ULTA) | |
|---|---|---|
| Residues in most favoured regions | 521 (87.9%) | 405 (90.0%) |
| Residues in additional allowed regions | 62 (10.5%) | 43 (9.6%) |
| Residues in generously allowed regions | 7 (1.2%) | 2 (0.4%) |
| Residues in disallowed regions | 3 (0.5%) | 0 (0.0%) |
| Number of nonglycine and nonproline residues | 593 | 450 |
| Number of end-residues (excl. Gly and Pro) | 2 | 155 |
| Number of glycine residues (shown as triangles) | 48 | 48 |
| Number of proline residues | 26 | 34 |
| Total number of residues | 669 | 687 |
| Residues with Verify 3D Score >0.2 | 52.24% | 96.63% |
| Errat overall quality factor | 44.154 | 89.655 |
Acknowledgments
This work was supported by Department of Biotechnology, New Delhi, India (Grant No. BT/PR5452/BRB/10/430/2004, BT/PR9266/BID/07/221/2007 and BT/PR13384/MED/29/166/2009). J. Kaur and R. Tiwari contributed equally to the present work.
References
- 1.Croft SL, Sundar S, Fairlamb AH. Drug resistance in leishmaniasis. Clinical Microbiology Reviews. 2006;19(1):111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuon FF, Neto VC, Amato VS. Leishmania: origin, evolution and future since the Precambrian. FEMS Immunology and Medical Microbiology. 2008;54(2):158–166. doi: 10.1111/j.1574-695X.2008.00455.x. [DOI] [PubMed] [Google Scholar]
- 3.Kerr SF. Molecular trees of trypanosomes incongruent with fossil records of hosts. Memorias do Instituto Oswaldo Cruz. 2006;101(1):25–30. doi: 10.1590/s0074-02762006000100006. [DOI] [PubMed] [Google Scholar]
- 4.Singh N, Almeida R, Kothari H, et al. Differential gene expression analysis in antimony-unresponsive Indian kala azar (visceral leishmaniasis) clinical isolates by DNA microarray. Parasitology. 2007;134(6):777–787. doi: 10.1017/S0031182007002284. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien WJ, Frerman FE. Evidence for a complex of three beta-oxidation enzymes in Escherichia coli: induction and localization. Journal of Bacteriology. 1977;132(2):532–540. doi: 10.1128/jb.132.2.532-540.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishina M, Kamiryo T, Tashiro S, et al. Subcellular localization of two long-chain-acyl-coenzyme-A synthetase in Candida lipolytica. European Journal of Biochemistry. 1978;89:321–328. doi: 10.1111/j.1432-1033.1978.tb12533.x. [DOI] [PubMed] [Google Scholar]
- 7.Krisans SK, Mortensen RM, Lazarow PB. Acyl-CoA synthetase in rat liver peroxisomes. Computer-assisted analysis of cell fractionation experiments. Journal of Biological Chemistry. 1980;255(20):9599–9607. [PubMed] [Google Scholar]
- 8.Fulda M, Shockey J, Werber M, Wolter FP, Heinz E. Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation. Plant Journal. 2002;32(1):93–103. doi: 10.1046/j.1365-313x.2002.01405.x. [DOI] [PubMed] [Google Scholar]
- 9.Schomburg I, Chang A, Ebeling C, et al. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Research. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glick BS, Rothman JE. Possible role for fatty acyl-coenzyme A in intracellular protein transport. Nature. 1987;326(6110):309–312. doi: 10.1038/326309a0. [DOI] [PubMed] [Google Scholar]
- 11.Pfanner N, Glick BS, Arden SR, Rothman JE. Fatty acylation promotes fusion of transport vesicles with Golgi cisternae. Journal of Cell Biology. 1990;110(4):955–961. doi: 10.1083/jcb.110.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai JCK, Liang BB, Jarvi EJ, Cooper AJL, Lu DR. Differential effects of fatty acyl coenzyme A derivatives on citrate synthase and glutamate dehydrogenase. Research Communications in Chemical Pathology and Pharmacology. 1993;82(3):331–338. [PubMed] [Google Scholar]
- 13.Li ZN, Hongo S, Sugawara K, et al. The sites for fatty acylation, phosphorylation and intermolecular disulphide bond formation of influenza C virus CM2 protein. Journal of General Virology. 2001;82(5):1085–1093. doi: 10.1099/0022-1317-82-5-1085. [DOI] [PubMed] [Google Scholar]
- 14.Murakami K, Ide T, Nakazawa T, Okazaki T, Mochizuki T, Kadowaki T. Fatty-acyl-CoA thioesters inhibit recruitment of steroid receptor co-activator 1 to α and γ isoforms of peroxisome-proliferator-activated receptors by competing with agonists. Biochemical Journal. 2001;353(2):231–238. doi: 10.1042/0264-6021:3530231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Aalten DMF, DiRusso CC, Knudsen J. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO Journal. 2001;20(8):2041–2050. doi: 10.1093/emboj/20.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlandi PA, Turco SJ. Structure of the lipid moiety of the Leishmania donovani lipophosphoglycan. Journal of Biological Chemistry. 1987;262(21):10384–10391. [PubMed] [Google Scholar]
- 17.McConville MJ, Thomas-Oates JE, Ferguson MAJ, Homans SW. Structure of the lipophosphoglycan from Leishmania major . Journal of Biological Chemistry. 1990;265(32):19611–19623. [PubMed] [Google Scholar]
- 18.Schneider P, Ferguson MAJ, McConville MJ, Mehlert A, Homans SW, Bordier C. Structure of the glycosyl-phosphatidylinositol membrane anchor of the Leishmania major promastigote surface protease. Journal of Biological Chemistry. 1990;265(28):16955–16964. [PubMed] [Google Scholar]
- 19.Zhang K, Showalter M, Revollo J, Hsu FF, Turk J, Beverley SM. Sphingolipids are essential for differentiation but not growth in Leishmania. EMBO Journal. 2003;22(22):6016–6026. doi: 10.1093/emboj/cdg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu FF, Turk J, Zhang K, Beverley SM. Characterization of inositol phosphorylceramides from Leishmania major by tandem mass spectrometry with electrospray ionization. Journal of the American Society for Mass Spectrometry. 2007;18(9):1591–1604. doi: 10.1016/j.jasms.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Beverley SM. Phospholipid and sphingolipid metabolism in Leishmania. Molecular and Biochemical Parasitology. 2010;170(2):55–64. doi: 10.1016/j.molbiopara.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts CW, McLeod R, Rice DW, Ginger M, Chance ML, Goad LJ. Fatty acid and sterol metabolism: potential antimicrobial targets in apicomplexan and trypanosomatid parasitic protozoa. Molecular and Biochemical Parasitology. 2003;126(2):129–142. doi: 10.1016/s0166-6851(02)00280-3. [DOI] [PubMed] [Google Scholar]
- 23.Camero L, Shulaw WP, Xiao L. Characterization of a Cryptosporidium parvum gene encoding a protein with homology to long chain fatty acid synthetase. Journal of Eukaryotic Microbiology. 2003;50:534–538. doi: 10.1111/j.1550-7408.2003.tb00621.x. [DOI] [PubMed] [Google Scholar]
- 24.Jiang DW, Ingersoll R, Myler PJ, Englund PT. Trypanosoma brucei: four tandemly linked genes for fatty acyl-CoA synthetases. Experimental Parasitology. 2000;96(1):16–22. doi: 10.1006/expr.2000.4547. [DOI] [PubMed] [Google Scholar]
- 25.Jiang DW, Englund PT. Four Trypanosoma brucei fatty acyl-CoA synthetases: fatty acid specificity of the recombinant proteins. Biochemical Journal. 2001;358(3):757–761. doi: 10.1042/0264-6021:3580757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh N. Is there true Sb (V) resistance in Indian kala-azar field isolates? Current Science. 2002;83:101–102. [Google Scholar]
- 27.Singh N, Singh RT, Sundar S. Novel mechanism of drug resistance in kala azar field isolates. Journal of Infectious Diseases. 2003;188(4):600–607. doi: 10.1086/377133. [DOI] [PubMed] [Google Scholar]
- 28.Singh N, Dube A. Fluorescent Leishmania: application to anti-leishmanial drug testing. American Journal of Tropical Medicine and Hygiene. 2004;71(4):400–402. [PubMed] [Google Scholar]
- 29.Kothari H, Kumar P, Sundar S, Singh N. Possibility of membrane modification as a mechanism of antimony resistance in Leishmania donovani . Parasitology International. 2007;56(1):77–80. doi: 10.1016/j.parint.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Chulay JD, Bryceson ADM. Quantitation of amastigotes of Leishmania donovani in smears of splenic aspirates from patients with visceral leishmaniasis. American Journal of Tropical Medicine and Hygiene. 1983;32(3):475–479. doi: 10.4269/ajtmh.1983.32.475. [DOI] [PubMed] [Google Scholar]
- 31.Notredame C, Higgins DG, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 32.Watkins PA, Maiguel D, Jia Z, Pevsner J. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. Journal of Lipid Research. 2007;48(12):2736–2750. doi: 10.1194/jlr.M700378-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 35.Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Šali A. Comparative protein structure modeling of genes and genomes. Annual Review of Biophysics and Biomolecular Structure. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 36.Hisanaga Y, Ago H, Nakagawa N, et al. Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. Journal of Biological Chemistry. 2004;279(30):31717–31726. doi: 10.1074/jbc.M400100200. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 38.Groot PHE, Scholte HR, Hülsmann WC. Fatty acid activation: specificity, localization, and function. Advances in Lipid Research. 1976;14:75–126. doi: 10.1016/b978-0-12-024914-5.50009-7. [DOI] [PubMed] [Google Scholar]
- 39.Bar-Tana J, Rose G, Shapiro B. The purification and properties of microsomal palmitoyl-Coenzyme A synthetase. Biochemical Journal. 1973;131:199–209. doi: 10.1042/bj1220353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Experimental Biology and Medicine. 2008;233(5):507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myler PJ, Fasel N. Leishmania: after the Genome. Caister Academic Press; 2006. [Google Scholar]
- 42.Igoillo-Esteve M, Mazet M, Deumer G, Wallemacq P, Michels PAM. Glycosomal ABC transporters of Trypanosoma brucei: characterization of their expression, topology and substrate specificity. International Journal for Parasitology. 2011;41(3-4):429–438. doi: 10.1016/j.ijpara.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Wiemer EAC, IJlst L, Van Roy J, Wanders RJA, Opperdoes FR. Identification of 2-enoyl coenzyme A hydratase and NADP-dependent 3-hydroxyacyl-CoA dehydrogenase activity in glycosomes of procyclic Trypanosoma brucei. Molecular and Biochemical Parasitology. 1996;82(1):107–111. doi: 10.1016/0166-6851(96)02710-7. [DOI] [PubMed] [Google Scholar]
- 44.Parsons M. Glycosomes: parasites and the divergence of peroxisomal purpose. Molecular Microbiology. 2004;53(3):717–724. doi: 10.1111/j.1365-2958.2004.04203.x. [DOI] [PubMed] [Google Scholar]
- 45.Hart DT, Opperdoes FR. The occurrence of glycosomes (microbodies) in the promastigote stage of four major Leishmania species . Molecular and Biochemical Parasitology. 1984;13(2):159–172. doi: 10.1016/0166-6851(84)90110-5. [DOI] [PubMed] [Google Scholar]
- 46.Coombs GH, Craft JA, Hart DT. A comparative study of Leishmania mexicana amastigotes and promastigotes, enzyme activities and subcellular locations. Molecular and Biochemical Parasitology. 1982;5(3):199–211. doi: 10.1016/0166-6851(82)90021-4. [DOI] [PubMed] [Google Scholar]
- 47.Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB Journal. 2008;22(2):590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- 48.Saxena A, Lahav T, Holland N, et al. Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Molecular and Biochemical Parasitology. 2007;152(1):53–65. doi: 10.1016/j.molbiopara.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weimar JD, DiRusso CC, Delio R, Black PN. Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids: amino acid residues within the ATP/AMP signature motif of Escherichia coli fadD are required for enzyme activity and fatty acid transport. Journal of Biological Chemistry. 2002;277(33):29369–29376. doi: 10.1074/jbc.M107022200. [DOI] [PubMed] [Google Scholar]
- 50.Horswill AR, Escalante-Semerena JC. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue lys592 is required for propionyl-AMP synthesis. Biochemistry. 2002;41(7):2379–2387. doi: 10.1021/bi015647q. [DOI] [PubMed] [Google Scholar]
- 51.Chang KH, Xiang H, Dunaway-Mariano D. Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. Strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry. 1997;36(50):15650–15659. doi: 10.1021/bi971262p. [DOI] [PubMed] [Google Scholar]
- 52.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. The 1.75 Å crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry. 2003;42(10):2866–2873. doi: 10.1021/bi0271603. [DOI] [PubMed] [Google Scholar]
- 53.Black PN, DiRusso CC. Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiology and Molecular Biology Reviews. 2003;67(3):454–472. doi: 10.1128/MMBR.67.3.454-472.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Black PN, Zhang Q, Weimar JD, DiRusso CC. Mutational analysis of a fatty acyl-coenzyme a synthetase signature motif identifies seven amino acid residues that modulate fatty acid substrate specificity. Journal of Biological Chemistry. 1997;272(8):4896–4903. doi: 10.1074/jbc.272.8.4896. [DOI] [PubMed] [Google Scholar]
- 55.Fortéa JO, de La Llave E, Regnault B, et al. Transcriptional signatures of BALB/c mouse macrophages housing multiplying Leishmania amazonensis amastigotes. BMC Genomics. 2009;10, article 119 doi: 10.1186/1471-2164-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao C, Li Y, Donelson JE, Wilson ME. Proteomic examination of Leishmania chagasi plasma membrane proteins: contrast between avirulent and virulent (metacyclic) parasite forms. Proteomics. 2010;4(1):4–16. doi: 10.1002/prca.200900050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cascante M, Boros LG, Comin-Anduix B, De Atauri P, Centelles JJ, Lee PWN. Metabolic control analysis in drug discovery and disease. Nature Biotechnology. 2002;20(3):243–249. doi: 10.1038/nbt0302-243. [DOI] [PubMed] [Google Scholar]
- 58.Rakotomanga M, Saint-Pierre-Chazalet M, Loiseau PM. Alteration of fatty acid and sterol metabolism in miltefosine-resistant Leishmania donovani promastigotes and consequences for drug-membrane interactions. Antimicrobial Agents and Chemotherapy. 2005;49(7):2677–2686. doi: 10.1128/AAC.49.7.2677-2686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


