Abstract
Carbapenam synthetase (hereafter named CPS) catalyzes the formation of the β-lactam ring in the biosynthetic pathway to (5R)-carbapen-2-em-3-carboxylate, the simplest of the carbapenem antibiotics. Kinetic studies showed remarkable tolerance to substrate stereochemistry on the turnover rate, but did not distinguish between chemistry and a non-chemical step such as product release or conformational change as rate-determining. Also, X-ray structural studies and modest sequence homology to β-lactam synthetase, an enzyme that catalyzes the formation of a monocyclic β-lactam ring in a similar ATP/Mg2+ dependent reaction, implicate K443 as an essential residue for substrate binding and intermediate stabilization. In these experiments, we use pH-rate profiles, deuterium solvent isotope effects and solvent viscosity measurements to examine the rate-limiting step in this complex overall process of substrate adenylation and intramolecular ring formation. Mutagenesis and chemical rescue demonstrate that K443 is the general acid visible in the pH-rate profile of the wild-type CPS catalyzed reaction. On the basis of these results, we propose a mechanism where the rate-limiting step is β-lactam ring formation coupled to a protein conformational change, and underscore the role of K443 throughout the reaction.
Resistance to antibiotics commonly used to combat infectious diseases is rising (1, 2). The β-lactam antibiotics, represented most prominently by penicillins and cephalosporins, constitute the largest portion of the world’s antibiotic market despite inroads from resistant organisms (3). An important part of the continued successful use of penicillins and cephalosporins has been the introduction of clavulanic acid to overcome several widely encountered β-lactamases that confer resistance (4, 5), and the advent of carbapenems that combine potent, broad-spectrum activity with reduced sensitivity to β-lactamases. These advances have extended the clinical usefulness of the β-lactam antibiotics now to more than 50 years, but the inexorable adaptation of disease-causing bacteria motivates continued efforts in semi-synthesis and pathway engineering (6) to yield new structures to counter evolving mechanisms of resistance.
The centrally important β-lactam rings in clavulanic acid and the carbapenems are derived from β-amino acids by coupling their formation to the hydrolysis of ATP—a biosynthetic process wholly different from the oxidative cyclization reactions seen in penicillin and cephalosporin biosynthesis (7). β-Lactam synthetase (β-LS) closes N2-carboxyethyl-L-arginine (CEA) to the monocyclic product, deoxyguanidinoproclavaminate (DGPC), enroute to clavulanic acid (8, 9). In the (5R)-carbapen-2-em-3-carboxylate pathway, CPS, the distant homologue of β-LS (22% identity, 36% similarity) cyclizes (2S,5S)-5-carboxymethylproline [(2S,5S)-CMPr] to (3S,5S)-carbapenam-3-carboxylate (10). While β-LS is specific for the L-configured substrate, CPS can cyclize (2S,5S)-CMPr and the three other possible stereoisomers of this precursor with approximately equal ease (9, 10). The notable stereochemical flexibility of CPS make it an especially attractive target for protein engineering to produce modified carbapenems.
Both CPS and β-LS exhibit Bi-Ter sequential kinetic mechanisms where ATP binds first followed by the modified amino acid substrate (9, 10). Adenylation and 4-membered ring formation occur followed by product release, PPi being the last to leave the active site (9, 10). This kinetic picture agrees well with what is known about the progenitor asparagine synthetases, Class B (AS-B), a highly conserved family of primary metabolic enzymes that convert aspartate to asparagine (11). All three of these proteins proceed through an intermediate acyl-adenylate during the course of their individual reactions (Scheme 1). The chemical and structural similarities among these three proteins suggest an evolutionary link between the β-lactam forming enzymes and AS-B (9, 10, 12, 13). The structural and catalytic machinery of AS-B for ATP binding and for synthesis of a distal amino diacid acyl-adenylate is shared among all three proteins.
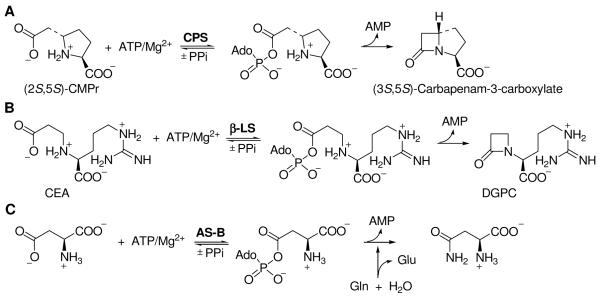
Scheme 1.
Comparison of the reactions catalyzed by CPS, β-LS and AS-B.
(A) CPS catalyzes the formation of the bicyclic β-lactam, (3S,5S)-carbapenam-3-carboxylate, from (2S,5S)-CMPr. (B) β-LS catalyzes the ATP/Mg2+-dependent intramolecular closure of the β-amino acid CEA to the monocyclic β-lactam DGPC. (C) AS-B catalyzes the transfer of the amide nitrogen of glutamine to the side chain of apsartate.
Compared to AS-B, however, evolutionary changes are evident in the enlarged and remodeled active sites of the β-lactam forming enzymes. Analysis of the CPS and β-LS structures suggests that K443 not only interacts with ATP, but could also play a role in stabilizing the oxyanion transition state traversed in the formation of the acyl-AMP intermediate (12-14). To better understand the complex two-stage formation of a β-lactam ring carried out by these enzymes, and as a prelude to engineering these proteins for semi-synthetic purposes, we provide experimental evidence to identify the rate-determining step in the CPS catalytic reaction, and characterize the role of K443. pH dependence, viscosity and solvent kinetic isotope effect studies point to the β-lactam ring-forming step and an enzyme conformational change as both contributing to the overall rate limitation of CPS mediated synthesis. Mutagenesis and chemical rescue experiments identify K443 as the general acid stabilizing the intermediate acyl-adenylate.
EXPERIMENTAL PROCEDURES
Materials and Methods
All buffers, all resins, all coupling enzymes, glycerol, sucrose and PEG 8000 were obtained from Sigma (St. Louis, MO). Plasmid pUC19 was from Invitrogen (Carlsbad, CA) and pCDFDuet-1 was purchased from Novagen (La Jolla, CA). Plasmids pET24a/cps, pET24a/carBC and pCDFDuet-1/cps were generous gifts from Dr. R.-F. Li of this laboratory (15) (and unpublished experiments). Nitrocefin was kindly provided by K. A. Moshos of this laboratory. D2O (99%) was purchased from Cambridge Isotope Laboratories (Andover, MA), while DCl, NaOD, 2,2,2-trifluoroethylamine, propylamine and propargylamine were obtained from Aldrich (Milwaukee, WI). (2S,5S)-5-Carboxymethyl proline was prepared by the method previously described(10).
Plasmid Construction
For mutagenesis purposes, pUC19/cps was obtained by ligation of cps at the HindIII and XbaI sites of pUC19 plasmid. For co-expression of pET24a/carB_carC with the mutant cps genes in the Nitrocefin assay, the mutant cps genes were ligated into the NdeI and XhoI sites of pCDFDuet-1.
Mutagenesis
The singly mutated cps genes corresponding to K443A and K443M were generated using QuikChange Site-Directed Mutagenesis (Stratagene) and pUC19/cps as the template. The presence of the desired mutations in all mutant cps genes as well as the absence of adventitious mutations was confirmed by complete gene sequencing (DNA Sequence Facility, Johns Hopkins University, Baltimore, MD). The mutant cps genes were cloned into pET24a (Novagen, Madison, WI) for subsequent enzyme overproduction.
Overexpression and Purification of Native CPS and Variants
E. coli electrocompetent BL21(DE3) cells were transformed with pET24/cps or mutant gene for enzyme overproduction. Cells were grown at 37 °C in 2xYT medium with 25 μg/mL kanamycin A and induced (OD600 = 0.6) with 1mM IPTG for 4 h at 28 °C. Cells were harvested by centrifugation and the pellet was flash frozen in liquid nitrogen. Usually 10 g of cells for the native CPS enzyme were obtained from 3 L of medium. All purification procedures were carried out on ice or at 4 °C. The frozen cells were resuspended to 0.35 g/mL in lysis buffer (100 mM Tris-HCl, pH 8.0, 1.8 mM EDTA, 1 mM Benzamidine, 1 mM PMSF, 1 mM DTT) and lysed by three passes through a French Press at 18,000 psi. Cell debris was removed by centrifugation and excess DNA was removed by streptomycin sulfate precipitation at a final concentration of 3%. The resulting supernatant was subjected to ammonium sulfate fractionation at 35% and 65% saturation. The final pellet was then resuspended in less than 10 mL of dialysis buffer (50 mM Tris-base, pH 7.5, 0.1 mM EDTA, 1 mM Benzamidine, 1 mM PMSF, 1 mM DTT) and dialyzed against 3.5 L of dialysis buffer with two buffer changes in 6 h. The dialyzed enzyme solution was then loaded onto a Q-Sepharose FF column (pre-equilibrated with 50 mM Tris-base, pH 7.5, 150 mM NaCl, 1 mM DTT) by gravity. After a 250 mL wash of 50 mM Tris-base, pH 7.5, 150 mM NaCl, 1 mM DTT, the enzyme was eluted with a 1.6 L gradient of 150 – 450 mM NaCl in 50 mM Tris-base, pH 7.5, 1 mM DTT. Fractions containing the desired protein were pooled and ammonium sulfate was added to a final concentration of 1 M. This solution was then loaded onto a Butyl-Sepharose FF column (pre-equilibrated with 50 mM Tris-base, pH 7.5, 1 M ammonium sulfate, 1 mM DTT) by gravity. After washing the column with 50 mL of 50 mM Tris-base, pH 7.5, 1 M ammonium sulfate, 1 mM DTT, the enzyme was eluted with a 800 mL gradient of 1 – 0 M ammonium sulfate in 50 mM Tris-base, pH 7.5, 1 mM DTT. The fractions containing CPS or its variants were pooled and dialyzed against 50 mM Tris-base, pH 7.5, 1 mM DTT. The enzyme was concentrated to approximately 10 mg/mL by ultrafiltration in an Amicon stirred cell over a YM10 membrane, and flash frozen dropwise into liquid nitrogen and stored at −80 °C. Protein concentrations were measured spectrophotometrically at 280 nm using the extinction coefficient previously determined for CPS [€280 = 1.82 (± 0.16) mL mg−1 cm−1 (pH 7.0, 25 °C)(10). The relative purity was calculated by SDS-PAGE analysis and was ≥95%.
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectroscopy was performed using a Jasco J-810 Spectropolarimeter with a temperature control unit. Wild-type and variants were prepared in 50 mM Tris-base, pH 7.5, 1 mM DTT, at a final concentration of 20 μM and UV CD spectra (300 to 180 nm) were acquired.
Nitrocefin Assay
E. coli electrocompetent BL21(DE3) cells were co-transformed with pET24a/carBC and pCDFDuet-1/cps or the a mutant cps gene for overexpression. Cells were grown at 37 °C in 2xYT medium supplemented with 25 μg/mL kanamycin A and 25 μg/mL spectinomycin and induced (OD600 = 0.6) with 1mM IPTG for 5 h at 28 °C. 1 mL samples were taken at 0, 3, 5 h, spun down at 14,600g and the pellet was separated from the supernatant. The ability of the overexpression supernatant to catalyze carbapenem synthesis was monitored in a Nitrocefin paper disk assay (16-18). BA2 agar plates (18), inoculated with Bacillus licheniformis (ATCC 14580), were used to detect the presence of the carbapenem antibiotic. Supernatant (250 μL) was applied to the assay plates on 1.25 cm paper discs and incubated at 37 °C for 3 h. After incubation, a freshly prepared 1 mL solution of Nitrocefin (300 ug/mL) in PBS buffer (18) was applied to the assay plates. The appearance of a red zone indicated the induction of β-lactamase. Negative and positive control experiments were conducted with pET24a/carBC and the co-transformation of pET24a/carBC and pCDFDuet-1/cps, respectively. To test the sensitivity of the Nitrocefin assay, a range of ampicillin (2.5 pg – 25 μg) was applied to the assay plates on paper discs and assayed as described above.
CPS Assay
PPi release catalyzed by CPS was detected by coupled enzyme assay described by Van Pelt and Northrop (19) as modified and used previously (10). The progress of the enzymic reaction was monitored at 340 nm (€280 = 6.22 mM−1 cm−1) as the increase in the rate of NADPH production at 25 °C. Assay mixtures of 200 μM UDP-glucose, 200 μM NADP, 0.2 μM glucose-1,6-biphosphate, 1.5 mM ATP, 1 mM DTT, 15 mM MgCl2 were made up in a buffer system of 100 mM HEPES and 80 mM piperazine at an ionic strength of 0.15 kept constant with KCl (20) and at the indicated pH. The concentrations of the coupling enzymes at pH 8.0 used were 2 U/mL phosphoglucomutase, 1 U/mL glucose-6-phosphate dehydrogenase, and 4 U/mL UDP-glucose pyrophosphorylase. Reactions were carried out in a final volume of 500 μL and initiated by the addition of CPS. Controls for each condition (pH, viscosity, D2O…etc), as described below, were conducted to ensure that CPS was the rate-determining enzyme in the assay. The initial velocity conditions of the assays were not limited by the coupling enzymes since a linear dependence of v versus CPS concentration was obtained.
Dependence of CPS Activity on pH
pH studies on the wild-type enzyme were performed to generate a profile of enzyme activity as a function of pH. The initial velocity patterns were measured with the PPi coupled enzyme assay described above where ATP was held constant (1.0 mM) and (2S,5S)-CMPr was varied (0.075 – 2.5 mM). The amount of coupled enzymes, phosphoglucomutase and glucose-6-phosphate dehydrogenase, was increased up to a maximum of 4 U/mL and 3 U/mL, respectively, at the high and low pH values. Initial velocity measurements were determined at several pH values (6.67, 7.33, 8.0, 8.67, 9.33, 9.67, 9.8, 10.0, 10.33). Assay reactions were run in duplicate. The resulting kinetic data were fit to the eq 1 to obtain the first- and second-order kinetic parameters, kcat and kcat/Km, respectively. Profiles were generated by plotting log kcat or log (kcat/Km) as a function of pH and relevant pKa values were identified by a fit of eq 2.
Solvent Viscosity Studies
CPS kinetic assays were performed with the macroviscogen PEG 8000 (6% w/w) and the microviscogen glycerol (0 – 30% w/w). The viscosity of the solutions was determined in quadruplicate at 25 °C using a Brookfield viscometer, relative to the standard CPS assay buffer consisting of 80 mM piperazine, 100 mM HEPES, I.S. 0.15, pH 8.0. PPi coupled enzyme assays were performed as described above where ATP was held constant (1.0 mM) and (2S,5S)-CMPr was varied (0.1 – 1.5 mM). Kinetic parameters were determined from fits to eq 1.
Solvent Kinetic Isotope Effects
Initial velocity patterns were measured with the PPi coupled enzyme assay described above where ATP was held constant (1.0 mM) and (2S,5S)-CMPr was varied (0.075 – 0.75 mM). All divalent metal ions and substrates were exchanged in 99.9% D2O and lyophilized. This process was repeated four times. Under argon, the samples were redissolved in D2O to give the desired concentrations and stored in air-tight flasks. The pD (pD = pH + 0.4) (21) of the HEPES and piperazine buffer was adjusted using DCl and NaOD. Assays mixtures were prepared and covered with Parafilm prior to use. Assays were performed in triplicate. pL-Rate profiles were constructed at various mole fractions of D2O (n = 0.15, 0.3, 0.5, 0.7, 0.85 and 1.0) to determine appropriate pL values to perform proton inventories. Proton inventories were constructed by varying the atom fraction of D2O from n = 0 – 1 at the optimum pL value for each isotopic mixture.
Chemical Rescue of the K443A Mutant by Exogenous Amines
All exogenous amines, propylamine, propargylamine and 2,2,2-trifluoroethylamine, were prepared in 100 mM HEPES and 80 mM piperazine at the desired pH, ionic strength and concentration. The chemical rescue effects of the primary amines were determined by measuring the intitial velocity of the PPi coupled enzyme assay at 25 °C as a function of added exogenous amine. Assay mixtures contained the components of the PPi assay and various concentrations of the rescue amines (5 – 500 mM exogenous amines). Reaction was initiated by the addition of the K443A variant. Rescue parameters (Vmax and Kmamine) for CPS by added primary amines were obtained from fits of initital velocities vs. concentrations of the amines to eq. 1. With primary amines (pKa < 8.2), the kinetic parameters were corrected for the fraction of the protonated amine as described below. Such a correction was not necessary for amines with pKa ≥ 9.0 (ie ≥ 97% acid form at pH 8.0) (22, 23). The apparent rate constant of the reaction catalyzed by the exogenous amine is expressed by the following equation:
where ka is the rate constant of reaction catalyzed by the protonated form of amine; ksol is the rate constant of reaction proceeding independently of the amine; and [amine]protonated is the concentration of the protonated amine1. Plots of kobs versus ([amine]total / (Ka/[H+]+1)) were fit to eq 1 to obtain ka.
Steady-State Kinetic Analysis
All steady-state kinetic data were fitted using the FORTRAN programs of Cleland (24). Initial velocity patterns were fitted to eq 1. pH Profiles of the CPS reaction were fitted using eq 2. Proton inventories were fitted using Kaleidagraph® to perform non-linear least squares regression analysis of eq 3 and eq 4 with the errors propagated according to eq 5 where k corresponds to the standard error.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
RESULTS
Purification and Characterization of Wild-type and K443 Variants
The wild-type, K443A and K443M CPS mutant enzymes were purified to >95% homogeneity, based on SDS-PAGE, from an average of 10 mg of protein / L of culture. The kinetic parameters kcat and kcat/Km for wild-type CPS at pH 8.0 were 0.36 ± 0.01 s−1 and 1.04 ± 0.04 mM−1 s−1, respectively. At pH 8.0, 6.33 and 9.33 the K443A and K443M CPS variants showed no measurable activity with reaction concentrations of 0.9 mg/mL and 0.4 mg/mL, respectively, at substrate concentrations ten times the Km of native CPS. The CD spectra of the variants (data not shown) showed minimal deviations from that of the wild-type CD spectrum. Thus, the observed decrease in activity of the single-site mutants is not due to major structural changes in the protein.
A Nitrocefin assay was used to determine whether the K443 active site mutations formed the carbapenem product of the wild-type CPS reaction. This assay confirmed the results of the PPi coupled enzyme assay, the K443A and K443M variants do not catalyze the formation of (3S,5S)-carbapenam-3-carboxylate to a detectable level. Using ampicillin as a positive control, the sensitivity limit of the assay was determined to be < 2.5 pg. Therefore, if the K443 variants catalyze the formation of the more potent (3S,5S)-carbapenam-3-carboxylate, then it is at a level below 2.5 pg.
pH Effects on Catalysis by Wild-Type CPS
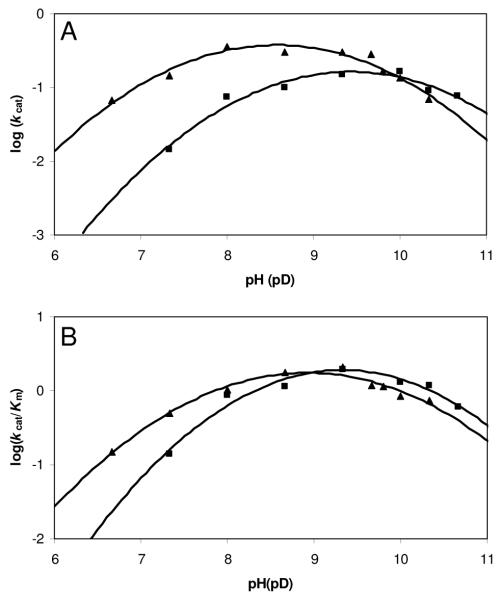
Wild-type CPS pH-rate profiles were constructed to identify ionizable groups that are important for catalysis (20, 25). In an ordered mechanism, kcat/Km for the first substrate normally equals the bimolecular rate constant for combination with the enzyme and shows only binding information, therefore kcat/Km for the second substrate is the one of interest (25). Consequently, the pH-dependence of the parameters kcat and kcat/Km was measured by varying the concentration of (2S,5S)-CMPr at saturating ATP over a pH range of 6.67 – 10.33 (Figure. 1). Log kcat and log (kcat/Km) profiles were similar and bell-shaped with slopes of +1 and −1 in the acidic and basic limbs, respectively, corresponding to single protonation and deprotonation events. The pKa values for kcat and kcat/Km obtained are 7.4 ± 0.1 and 9.7 ± 0.1, and 7.8 ± 0.1 and 10.0 ± 0.1, respectively.
Figure 1.
pH and pD Dependence of kcat and kcat/Km for wild-type CPS. (A) Plot of log (kcat) versus (▲) pH and pD (■) determined the pKa values to be 7.4 ± 0.1 and 9.7 ± 0.1, and 8.3 ± 0.2 and 10.4 ± 0.3, respectively. (B) Plot of log (kcat/Km) versus (▲) pH and pD (■) determined the pKa values to be 7.8 ± 0.1 and 10.0 ± 0.1, and 8.4 ± 0.2 and 10.3 ± 0.2, respectively.
Solvent Viscosity Studies
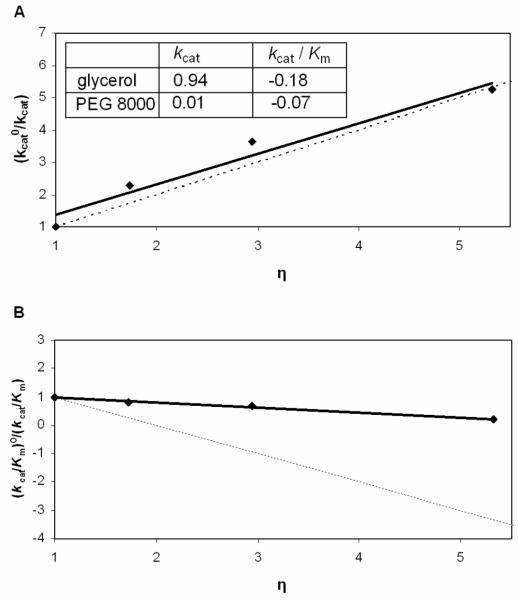
The effect of solvent viscosity on enzyme activity was investigated to assess whether diffusion-controlled events, such as product release or a conformational change, are rate-limiting. It has been reported that the response of kinetic parameters to solution viscosity increase can be significantly affected by the type of viscogenic reagent used (26-28). Therefore, both sucrose and glycerol were initially tested to study the CPS reaction. Unfortunately, sucrose acts as an activator for CPS hydrolysis of ATP to AMP and PPi, in the absence of (2S,5S)-CMPr, at low and high ATP concentrations, but it is unclear how this activation is achieved. Therefore, our studies were carried out only with glycerol as the microviscogen. As shown in Figure 2, there is a significant solvent viscosity effect on kcat (0.94) and only a minor effect on kcat/Km (−0.18). PEG 8000 was employed to ensure that the rate changes observed with glycerol are effects on a diffusion-controlled phenomenon and not the result of changes in global viscosity. Polymeric species, such as PEG 8000, increase the macroviscosity of a solution but do not significantly change the rate at which small molecules diffuse. In general, the addition of macroviscogen agents affect the measured viscosity of the solution but have no effect on the rates of diffusion-controlled reactions (27). Since enzyme activity was not altered by the presence of PEG 8000, the rate changes observed with glycerol are effects on diffusion-controlled phenomena.
Figure 2.
Viscosity effects on the kinetic parameters of CPS catalyzed reactions with glycerol and PEG 8000 as cosolutes. Effects of glycerol are shown on kcat (A) and kcat/Km (B) with variable (2S,5S)-CMPr at constant ATP. The dashed lines have slopes of +1 and −1 in (A) and (B), respectively. Reported in the inset are the slopes of plots from either kcat°/kcat or (kcat/Km)°/(kcat/Km) versus the relative viscosity (η) of the solution.
Solvent Kinetic Isotope Effects
Solvent isotope experiments were carried out to investigate the effects of heavy water on kcat and kcat/Km. Since pKa values change significantly with the isotopic composition of the solvent, resulting in shifts of the pH-profiles in deuterated aqueous environments (21, 29), pL-Rate profiles were constructed at varying mole fractions of D2O (n = 0.15, 0.3, 0.5, 0.7, 0.85 and 1.0) (Supplemental Information and Figure 1). For n = 1.0, the pKa values for kcat and kcat/Km of 8.3 ± 0.2 and 10.4 ± 0.3, and 8.4 ± 0.2 and 10.3 ± 0.2, resulting in a normal ΔpKa ~ 0.5, within experimental error (30, 31) (Table 1). Kinetic assays were performed in either 100% H2O or 95% D2O and solvent isotope effects, Dkcat or D(kcat/Km), were determined at the appropriate pL-optima. A small, but significant solvent isotope effect was observed on the second-order rate constant D(kcat/Km) of 1.4. In contrast, a larger isotope effect was observed on Dkcat of 2.2.
Table 1.
pL Dependence of k cat and k cat/Km for wild-type CPS
| n |
kcat pKa |
pKb |
kcat/Km pKa |
pKb | pL-optimum |
|---|---|---|---|---|---|
| 0 | 7.4 ± 0.1 | 9.7 ± 0.1 | 7.8 ± 0.1 | 10.0 ± 0.1 | 8.75 |
| 0.15 | 7.5 ± 0.2 | 9.9 ± 0.2 | 8.0 ± 0.2 | 9.7 ± 0.2 | 8.8 |
| 0.3 | 7.2 ± 0.1 | 10.0 ± 0.1 | 7.7 ± 0.3 | 9.7 ± 0.3 | 8.9 |
| 0.5 | 7.2 ± 0.2 | 10.3 ± 0.2 | 7.9 ± 0.2 | 10.0 ± 0.3 | 9.0 |
| 0.7 | 7.6 ± 0.2 | 10.3 ± 0.3 | 8.0 ± 0.1 | 10.0 ± 0.1 | 9.15 |
| 0.85 | 7.8 ± 0.1 | 10.2 ± 0.2 | 7.8 ± 0.2 | 10.2 ± 0.3 | 9.3 |
| 1.0 | 8.3 ± 0.2 | 10.4 ± 0.3 | 8.4 ± 0.1 | 10.3 ± 0.2 | 9.4 |
A proton inventory was conducted at the pL-optimum for each isotopic mixture (Table 1). n(kcat/Km) was invariant with respect to increasing mole fraction of D2O, while nkcat (kn/kD) decreased in a non-linear fashion to produce a concave, or “bowl-shaped” plot of nkcat vs. n (Figure 3). The data were fit to the modified Gross-Butler equations 3 and 4 for two- and three-proton inventories, respectively, assuming equivalent transition state fractionation factors for both transfers. The two- and three-proton models both fit well to the modest downward curvature of the plot and were within the limits of the propagated error. In an attempt to distinguish between two and three protons in flight, the square and cubic roots of the relative rate constants versus n were plotted (Figure 3, inset); both displayed a linear dependence on mole fraction of D2O and could not further discriminate between the two- and three-proton models.
Figure 3.
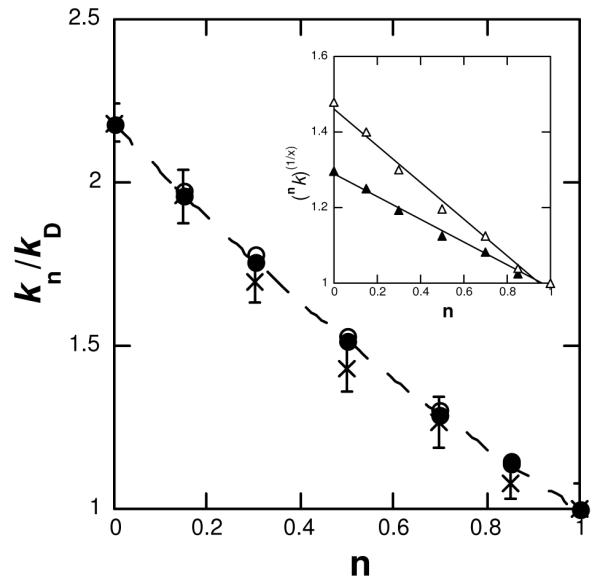
Proton inventory for the CPS catalyzed reaction. (x) nkcat is plotted against mole fraction of D2O (n) and the fit (- - -) to Dkcat [1 – n + n/(Dkcat)1/3]3 is displayed. Using the observed Dkcat 2.2, theoretical curves for (○) two- and (•) three-proton models were generated and plotted against n; it is evident that the propagated errors of the proton inventory encompass both archetypes. (Inset) Plot of the square (Δ) and cubic (▲) root of nkcat versus n with a linear (–) fit for both.
Chemical Rescue of the K443A Variant by Exogenous Amines
The pH-rate profile of wild-type CPS suggested that a general acid and a general base are involved in the catalytic mechanism. Substitution of alanine or methionine for K443 caused the activity of the CPS reaction to plummet to an immeasurably low level, indicating that the amino group of Lys443 is important for catalysis. Non-covalent chemical rescue (32) of inactive K443A was employed to validate the integrity of the mutant protein and gauge the essentiality and catalytic contribution provided by the affected side chain. Rescue efficiency of exogenous amines is dependent on the molecular volume of the rescue agent, as would be anticipated for a finite active site volume (32-34). Substitution of alanine for lysine in the active site of an enzyme creates a molecular volume void of 94.2 Å3 (33), and exogenous amines used for this study were chosen based on their molecular volume. The Vmax of the K443A variant increased to a level equal to 5.7% that of wild-type CPS activity by exogenous addition of 50 mM propylamine (pKa 10.5) (Table 2). 2,2,2-Trifluoroethylamine (pKa 5.7) was more efficient in rescuing the mutant activity at pH 6.33 (Vmax = 0.019 ± 0.002 μmol min−1 mg−1; 28% that of wild-type activity) than at pH 8.0 (Vmax = 0.0078 ± 0.0005 μmol min−1 mg−1; 2.3% that of wild-type activity), indicating that the protonated form of the amine functions as a general acid in catalysis. A full pH-rate profile of K443A with the exogenous amine propargylamine (pKa 8.2 and 82.3 Å3 ) was constructed and the resulting values were pK1 6.8 ± 0.2 and pK2 10.2 ± 0.2. For each of the three rescue agents, the maximum addition of the exogenous amines did not significantly affect the activity of wild-type CPS, arguing against nonspecific activation by amines.
Table 2.
Exogeneous amine rescue of CPS activity of the K443A variant.
| amine | pKa | molecular volume (Å3)a |
Kmamine (mM−1) | Vmax (μmol min−1 mg−1) |
% recovery, Vmax |
|---|---|---|---|---|---|
| propylamine, pH 8.0 | 10.5 | 94.2 | 0.0004 ± 0.0001 | 0.019 ± 0.002 | 5.7 |
| 2,2,2-trifluoroethylamineb, pH 8.0 | 5.7 | 94.2 | 0.35 ± 0.08 | 0.0078 ± 0.0005 | 2.3 |
| 2,2,2-trifluoroethylamineb, pH 6.33 | 5.7 | 94.2 | 39 ± 9 | 0.019 ± 0.002 | 28 |
| propargylamineb, pH 8.0 | 8.2 | 82.3 | 35 ± 9 | 0.021 ± 0.003 | 5.8 |
The molecular volume for amines reported in Zheng et al . (30)
Corrected for the fraction of protonated amine.
DISCUSSION
Rate-limiting Step(s) inWild-type CPS Catalyzed Reaction
The broad substrate specificity of CPS and the lack of stereochemical dependence on kcat (10), suggest that either a step other than chemistry is rate-limiting, or that chemistry is rate-limiting and the in-line attack between the substrate distal carboxylate and the α-phosphoryl bridge oxygen of ATP and subsequent cyclization step are not significantly effected by the various stereochemistries (10). pH, solvent kinetic isotope effects and solution viscosity studies were employed to investigate the identity of the rate-limiting step in the CPS catalyzed reaction.
pH-Rate studies show changes in the protonation state involved in the rate-limiting step of the enzymatic reaction. Log kcat/Km vs. pH profiles show changes in protonation state of those groups involved in the rate-limiting step from the substrate binding step up to the first irreversible step. Log kcat vs. pH profiles describe the ionizations of groups in the rate-limiting step after the formation of the substrate-enzyme complex up to product release. Therefore, groups involved in substrate binding will not be present in log kcat vs. pH profiles. The fact that the pKa and pKb values are very similar for both log kcat/Km and log kcat vs. pH profiles indicates that steps other than substrate binding are rate-limiting.
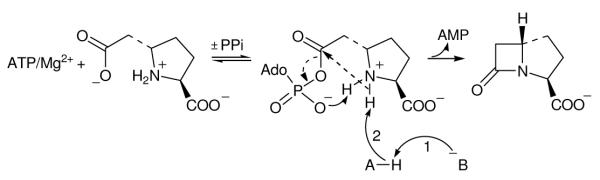
Accordingly, a large solvent kinetic isotope effect (SKIE) on kcat and a moderate SKIE on kcat/Km suggests that the rate-determining step involves kinetically significant proton transfers. Moderate D(kcat/Km) values in the range of 1.5 – 2.1 have been observed in situations where proton transfer was rate-limiting (35-38). Typical SKIE values on kcat for reactions which are subject to general acid and general base catalysis are 2-3 (21). Proton inventories of the wild-type CPS catalyzed reaction indicate that there are at least two protons in flight during the rate-limiting step. We propose that these proton transfers are occurring during rate-limiting β-lactam ring formation (Scheme 2). One of these two protons can be attributed to acid-base chemistry of the proposed catalytic Glu-Tyr dyad (12-14). The second proton can be credited to the deprotonation of the acyl-AMP intermediate to enable ring closure. The highly acidic nature of the developing protonated β-lactam could be visualized to impose no rate-limiting effect in its loss. Alternatively, the departing AMP, for example, could accept this proton to account for a cubic dependence in the proton inventory.
Scheme 2.
Mechanistic proposal to account for the protons in flight observed in the proton inventory.
Proton (1) is due to acid-base chemistry of the proposed Glu380 (B) -Tyr345 (A) dyad. The second proton (2) is removed from the nitrogen of the acyl-adenylate intermediate by Tyr345(A). After β-lactam formation, represented by dashed arrows, the departing AMP could serve to deprotonate the newly formed lactam nitrogen.
On the other hand, viscosity experiments indicate that the rate-limiting step is not solely composed of a chemical transformation. Solvent viscosity studies have been classically used to demonstrate that a diffusion-controlled process, usually substrate binding, is the rate-determining step in an enzymatic reaction (27, 39, 40). Since the catalytic efficiency of CPS (1.04 mM−1 s−1) does not approach the diffusion-controlled limit, substrate binding was not expected to be rate-limiting and is evident by the lack of kcat/Km dependence upon increased viscosity. In contrast, as shown in Figure 2, the kinetic parameter kcat for the reaction catalyzed by wild-type CPS is substantially dependent on viscosity of the medium indicating that a step other than chemistry is rate-limiting. The effects of changing viscosity on kcat have been used to assign product release or a conformational change as the rate-determining step (41-43).
Two theories have been proposed to model the effects of a viscosity dependence on kcat. The first theory, originally proposed by Kramers (44), postulates that in a random field environment, diffusion controls the rate at which the enzyme complex will pass over the activation energy barrier (45). Based upon Kramers’ theory, the relationship between kcat and viscosity should be η−δ (46) if a conformational change associated with product release is limiting. The second theory, attributed to Somogyi et al., postulates that if the rate-limiting step is the dissociation of product from the enzyme active site, without a structural rearrangement, the relationship between kcat and η would obey the function e−γη2 (47). For CPS, a plot of ln(kcat) vs. η2 was nonlinear (data not shown), whereas that of ln(kcat) vs. ln(η) showed a very good linear relationship with a value of 0.98, in accord with the model of Kramers (Figure 4).
Figure 4.
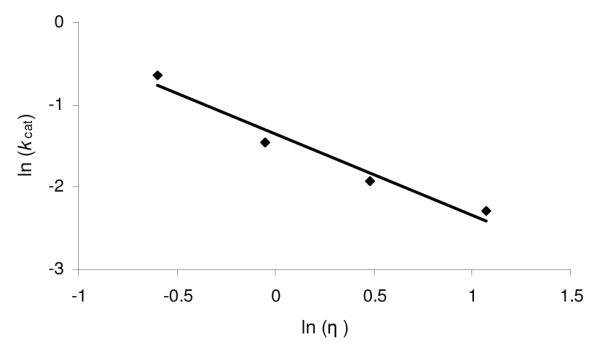
Plot of ln (kcat) vs. ln (η) for the CPS catalyzed reaction with glycerol as a cosolvent. The linear relationship between activity and viscosity resulted in a δ value of 0.98.
It is observed that the reaction rate constant, kcat, shows a negative power relationship to the medium viscosity, η. The reaction rate constant can be presented in the form:
| (3) |
where kT is the product of the Boltzmann constant and the absolute temperature, Ea is the activation energy, and A and δ are the empirical coefficients. At high viscosities, the reaction rate constant, kr, is an explicit function of viscosity:
| (4) |
where ηris a parameter that describes the dissipation of energy (internal friction) in the process of activational transition, i.e. local viscosity in the region of the reaction center (46). The two parameters: η (Eq 3) and ηr (Eq 4) differ by the factor which describes the coupling between the dynamics of protein and that of solvent molecules (46). Since the δ value is high (0.5 < δ < 1), this coupling is significant (46).
Deuterium solvent isotope effects indicate that chemistry is the rate-limiting step in the wild-type CPS catalyzed reaction, and the viscosity dependence of kcat suggests that it is a conformational change. This apparent dichotomy can be explained if the CPS catalyzed reaction is limited by a chemical step coupled to a conformational change of the enzyme. As mentioned before, CPS shows mechanistic and structural similarity with both β-LS and AS-B. All three proteins catalyze the formation of a distal acyl-AMP amino diacid intermediate in the first step of their individual reactions (9, 10, 48). Steady state studies of AS-B show a burst of PPi when no nitrogen source is available, which is indicative that a step after the formation of the intermediate is rate-limiting (11). These results agree with the SKIE and proton inventory results for wild-type CPS that the rate-limiting step occurs after the formation of the acyl-adenylate intermediate. The crystal structure of β-LS (12, 14) indicates that a conformational change occurs once the distal acyl-AMP amino diacid intermediate is formed. In a series of crystallographic snapshots, it was found that a loop comprised of residues 444 – 453 in β-LS becomes highly ordered and blocks the active site after formation of the acyl-adenylate intermediate (14). This loop is also conserved in CPS at residues 377-388 (13). Consistent with the proposed role for this conformational change, the kinetic mechanism for both -LS and CPS show that PPi is the first side-product of the reaction, but the last product to dissociate from the active site.
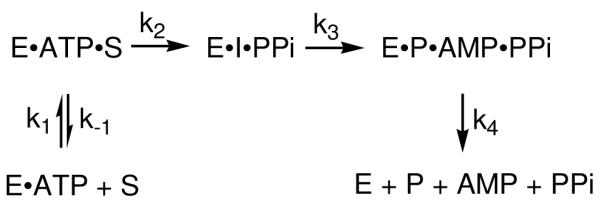
Combining the results of all the wild-type CPS studies, the kinetic mechanism shown in Scheme 3 can be constructed. k1and k−1 are the association and dissociation rate constants for (2S,5S)-CMPr and the E·ATP complex. This reversible step was proven to be independent of solution viscosity and, therefore, not rate-limiting. k2 describes the irreversible formation of the acyl-adenylate intermediate. Although acyl-adenylation reactions are typically reversible, the lack of 32PPi/ATP exchange (10) and SKIE measurements suggest otherwise in the CPS reaction. If β-lactam formation was the first irreversible step, then kcat and (kcat/Km) should exhibit comparable isotope effects and mole fraction D2O dependencies. As discussed previously, kcat displays a proton inventory of 2 – 3 and a much larger isotope effect than the second order rate constant. Therefore, it is reasonable to propose an irreversible step (k2) before the rate-limiting cyclization step to account for these differences. k3 is the rate-limiting unimolecular rate constant that describes coupling of the chemical transformation of the acyl-adenylate intermediate into the carbapenam product to a conformational change. Finally, the dissociation of product, AMP and PPi is combined in k4.
K443 as the general acid in the CPS Reaction
The pH-rate profile of wild-type CPS suggests that a general acid and a general base are involved in the catalytic mechanism. Substituting K443 with alanine or methionine caused a complete loss of activity assessed by coupled enzyme assay and Nitrocefin paper disc assay, indicating it to be essential for catalysis. A general acid function for K443 has been proposed for CPS based upon its structure (13) and sequence similarity to β-LS. CPS crystals were obtained at the relatively low pH of 5.6 and their structure revealed the substrate to be in a non-productive conformation, as a consequence the β-LS structure was taken to be more reliable for active site visualization. In the β-LS active site, K443 is hydrogen bonded to the non-bridging γ-phosphate oxygen of ATP and is proposed to stabilize the oxyanion intermediate (12, 14). The inactive K443A variant was successfully rescued with protonated amines strongly indicating that the €-amino group of K443 functions as a general acid in the CPS catalyzed reaction. This assignment was further confirmed by the pH-rate profile of the rescued variant with propargylamine. This profile showed not only a very similar bell-shaped pH profile as wild-type CPS, but also similar pKa values in the basic limb. The present data argue that K443 is the residue whose pKa of 9.7 is observed in the wild-type CPS pH-rate profile.
Mechanistic Conclusions
pH-dependence, SKIE and solvent viscosity studies on wild-type CPS indicate that the rate of the reaction is limited by a chemical transformation that is coupled to a conformational step. The chemical step is the β-lactam ring closure while the conformational change is proposed to be the movement of a catalytic loop. Scheme 3 shows the proposed mechanism of the CPS catalyzed reaction where ATP/Mg2+ binds first followed by (2S,5S)-CMPr in an ordered, sequential manner (10). The ATP is positioned and readied for attack by the (2S,5S)-CMPr carboxylate by the protonated form of the K443 residue through coordination to the γ-phosphate oxygen. The distal acyl-AMP intermediate and pyrophosphate are then formed, and the random loop comprised of residues 377-388 becomes ordered and covers the active site, sequestering the intermediate and side-product. Stabilization of the intermediate acyl-adenylate is achieved by general acid interactions of K443 with the α-phosphate oxygen atom. The rate-limiting cyclization step is then catalyzed by deprotonation of the substrate amine where two or more protons are in flight, ending with the formation of AMP and (3S,5S)-carbapenam-3-carboxylate and the relaxation of the ordered loop. Next, the products dissociate from the enzyme in an ordered manner with PPi leaving last. The presence and assignment of the rate-limiting step proposed here will be further investigated through the use of pre-steady state experiments. The nature and identity of the acid residue revealed in the wild-type pH-rate profile and its role in the formation of the β-lactam will be addressed in a subsequent publication.
Supplementary Material
The concentration of the protonated amine depends on ionization constant (Ka) of the amine and the proton concentration [H+] based on:
ACKNOWLEGEMENT
We are grateful to Professors A. S. Mildvan, B. Garcia-Moreno and J. P. Roth (Johns Hopkins University) for insightful discussions. We thank Dr. R.-F. Li and K. A. Moshos in our laboratory for pET24a/cps, pET24a/carB_carC and pCDFDuet-1/cps plasmids and for Nitrocefin, respectively.
This research was supported by NIH grant RO1 AI014937.
Footnotes
SUPPORTING INFORMATION AVAILABLE An overlay of the pL-rate profiles for kcat and kcat/Km. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations and Textual Footnotes: CPS, carbapenam synthetase; β-LS, β-lactam synthetase; AS-B, asparagine synthetase, class B; IPTG, isopropyl β-Dthiogalactopyranoside; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; PMSF, phenylmethanesulfonyl fluoride; Tris-HCl, tris(hydroxymethyl)aminomethane hydrochloride; (2S,5S)-CMPr, (2S,5S)-5-carboxymethyl proline; PPi, pyrophosphate; CEA, N2-(carboxyethyl)-L-arginine; DGPC, deoxyguanidinoproclavaminic acid.
REFERENCES
- 1.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 2.Thomson JM, Bonomo RA. The threat of antibiotic resistance in Gram-negative pathogenic bacteria: beta-lactams in peril! Curr Opin Microbiol. 2005;8:518–24. doi: 10.1016/j.mib.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Demain AL, Vaishnav P. Involvement of nitrogen-containing compounds in beta-lactam biosynthesis and its control. Crit Rev Biotechnol. 2006;26:67–82. doi: 10.1080/07388550600671466. [DOI] [PubMed] [Google Scholar]
- 4.Reading C, Cole M. Clavulanic acid: a beta-lactamase-inhiting beta-lactam from Streptomyces clavuligerus. Antimicrob Agents Chemother. 1977;11:852–7. doi: 10.1128/aac.11.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgopapadakou NH. Beta-lactamase inhibitors: evolving compounds for evolving resistance targets. Expert Opin Investig Drugs. 2004;13:1307–18. doi: 10.1517/13543784.13.10.1307. [DOI] [PubMed] [Google Scholar]
- 6.Li R, Townsend CA. Rational strain improvement for enhanced clavulanic acid production by genetic engineering of the glycolytic pathway in Streptomyces clavuligerus. Metab Eng. 2006;8:240–52. doi: 10.1016/j.ymben.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Townsend CA. New reactions in clavulanic acid biosynthesis. Curr Opin Chem Biol. 2002;6:583–9. doi: 10.1016/s1367-5931(02)00392-7. [DOI] [PubMed] [Google Scholar]
- 8.Bachmann BO, Li R, Townsend CA. beta-Lactam synthetase: a new biosynthetic enzyme. Proc Natl Acad Sci U S A. 1998;95:9082–6. doi: 10.1073/pnas.95.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachmann BO, Townsend CA. Kinetic mechanism of the beta- lactam synthetase of Streptomyces clavuligerus. Biochemistry. 2000;39:11187–93. doi: 10.1021/bi000709i. [DOI] [PubMed] [Google Scholar]
- 10.Gerratana B, Stapon A, Townsend CA. Inhibition and alternate substrate studies on the mechanism of carbapenam synthetase from Erwinia carotovora. Biochemistry. 2003;42:7836–47. doi: 10.1021/bi034361d. [DOI] [PubMed] [Google Scholar]
- 11.Boehlein SK, Stewart JD, Walworth ES, Thirumoorthy R, Richards NG, Schuster SM. Kinetic mechanism of Escherichia coli asparagine synthetase B. Biochemistry. 1998;37:13230–8. doi: 10.1021/bi981058h. [DOI] [PubMed] [Google Scholar]
- 12.Miller MT, Bachmann BO, Townsend CA, Rosenzweig AC. Structure of beta-lactam synthetase reveals how to synthesize antibiotics instead of asparagine. Nat Struct Biol. 2001;8:684–9. doi: 10.1038/90394. [DOI] [PubMed] [Google Scholar]
- 13.Miller MT, Gerratana B, Stapon A, Townsend CA, Rosenzweig AC. Crystal structure of carbapenam synthetase (CarA) J Biol Chem. 2003;278:40996–1002. doi: 10.1074/jbc.M307901200. [DOI] [PubMed] [Google Scholar]
- 14.Miller MT, Bachmann BO, Townsend CA, Rosenzweig AC. The catalytic cycle of beta -lactam synthetase observed by x-ray crystallographic snapshots. Proc Natl Acad Sci U S A. 2002;99:14752–7. doi: 10.1073/pnas.232361199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li RF, Stapon A, Blanchfield JT, Townsend CA. Three unusual reactions mediate carbapenem and carbapenam biosynthesis. Journal of the American Chemical Society. 2000;122:9296–9297. [Google Scholar]
- 16.Aguilar L, Martin M, Balcabao IP, Gomez-Lus ML, Dal-Re R, Prieto J. In vitro assessment of the effect of clavulanic acid at concentrations achieved in human serum on the bactericidal activity of amoxicillin at physiological concentrations against Staphylococcus aureus: implications for dosage regimens. Antimicrob Agents Chemother. 1997;41:1403–5. doi: 10.1128/aac.41.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Callaghan CH, Morris A, Kirby SM, Shingler AH. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–8. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sykes RB, Wells JS. Screening for beta-lactam antibiotics in nature. J Antibiot (Tokyo) 1985;38:119–21. doi: 10.7164/antibiotics.38.119. [DOI] [PubMed] [Google Scholar]
- 19.Van Pelt JE, Northrop DB. Purification and properties of gentamicin nucleotidyltransferase from Escherichia coli: nucleotide specificity, pH optimum, and the separation of two electrophoretic variants. Arch Biochem Biophys. 1984;230:250–63. doi: 10.1016/0003-9861(84)90106-1. [DOI] [PubMed] [Google Scholar]
- 20.Ellis KJ, Morrison JF. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–26. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 21.Schowen KB, Schowen RL. Solvent isotope effects of enzyme systems. Methods Enzymol. 1982;87:551–606. [PubMed] [Google Scholar]
- 22.Jencks WP. Catalysis in Chemistry and Enzymology. Dover Publications, Inc; New York: 1987. [Google Scholar]
- 23.Bell RP, Evans PG. Proc. R. Soc. London A. 1966;291:297–323. [Google Scholar]
- 24.Cleland WW. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–38. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 25.Cleland WW. The use of pH studies to determine chemical mechanisms of enzyme-catalyzed reactions. Methods Enzymol. 1982;87:390–405. doi: 10.1016/s0076-6879(82)87024-9. [DOI] [PubMed] [Google Scholar]
- 26.Pocker Y, Janjic N. Enzyme kinetics in solvents of increased viscosity. Dynamic aspects of carbonic anhydrase catalysis. Biochemistry. 1987;26:2597–606. doi: 10.1021/bi00383a028. [DOI] [PubMed] [Google Scholar]
- 27.Blacklow SC, Raines RT, Lim WA, Zamore PD, Knowles JR. Triosephosphate isomerase catalysis is diffusion controlled. Appendix: Analysis of triose phosphate equilibria in aqueous solution by 31P NMR. Biochemistry. 1988;27:1158–67. doi: 10.1021/bi00404a013. [DOI] [PubMed] [Google Scholar]
- 28.Mattei P, Kast P, Hilvert D. Bacillus subtilis chorismate mutase is partially diffusion-controlled. Eur J Biochem. 1999;261:25–32. doi: 10.1046/j.1432-1327.1999.00169.x. [DOI] [PubMed] [Google Scholar]
- 29.Isotope Effects on Enzyme-Catalyzed Reactions. University Park Press; Baltimore, Maryland: 1977. [Google Scholar]
- 30.Quinn DM, Sutton LD. In: Enzyme Mechanism from Isotope Effects. Cook PF, editor. CRC Press; Boston: 1991. pp. 73–126. [Google Scholar]
- 31.Schowen KBJ. Transition States in Biochemical Processes. Plenum: 1978. pp. 225–283. [Google Scholar]
- 32.Toney MD, Kirsch JF. Direct Bronsted analysis of the restoration of activity to a mutant enzyme by exogenous amines. Science. 1989;243:1485–8. doi: 10.1126/science.2538921. [DOI] [PubMed] [Google Scholar]
- 33.Zheng R, Blanchard JS. Identification of active site residues in E. coli ketopantoate reductase by mutagenesis and chemical rescue. Biochemistry. 2000;39:16244–51. doi: 10.1021/bi002134v. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe A, Kurokawa Y, Yoshimura T, Kurihara T, Soda K, Esaki N. Role of lysine 39 of alanine racemase from Bacillus stearothermophilus that binds pyridoxal 5′-phosphate. Chemical rescue studies of Lys39 --> Ala mutant. J Biol Chem. 1999;274:4189–94. doi: 10.1074/jbc.274.7.4189. [DOI] [PubMed] [Google Scholar]
- 35.Adams JA. Insight into tyrosine phosphorylation in v-Fps using proton inventory techniques. Biochemistry. 1996;35:10949–56. doi: 10.1021/bi960613h. [DOI] [PubMed] [Google Scholar]
- 36.Leichus BN, Blanchard JS. Pig heart lipoamide dehydrogenase: solvent equilibrium and kinetic isotope effects. Biochemistry. 1992;31:3065–72. doi: 10.1021/bi00127a006. [DOI] [PubMed] [Google Scholar]
- 37.Xiang B, Markham GD. Probing the mechanism of inosine monophosphate dehydrogenase with kinetic isotope effects and NMR determination of the hydride transfer stereospecificity. Arch Biochem Biophys. 1997;348:378–82. doi: 10.1006/abbi.1997.0439. [DOI] [PubMed] [Google Scholar]
- 38.Cook PF, Yoon MY, Hara S, McClure GD., Jr. Product dependence of deuterium isotope effects in enzyme-catalyzed reactions. Biochemistry. 1993;32:1795–802. doi: 10.1021/bi00058a013. [DOI] [PubMed] [Google Scholar]
- 39.Brouwer AC, Kirsch JF. Investigation of diffusion-limited rates of chymotrypsin reactions by viscosity variation. Biochemistry. 1982;21:1302–7. doi: 10.1021/bi00535a030. [DOI] [PubMed] [Google Scholar]
- 40.Hardy LW, Kirsch JF. Diffusion-limited component of reactions catalyzed by Bacillus cereus beta-lactamase I. Biochemistry. 1984;23:1275–82. doi: 10.1021/bi00301a040. [DOI] [PubMed] [Google Scholar]
- 41.Adams JA, Taylor SS. Energetic limits of phosphotransfer in the catalytic subunit of cAMP-dependent protein kinase as measured by viscosity experiments. Biochemistry. 1992;31:8516–22. doi: 10.1021/bi00151a019. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Adams JA. Is there a catalytic base in the active site of cAMP-dependent protein kinase? Biochemistry. 1997;36:2977–84. doi: 10.1021/bi9619132. [DOI] [PubMed] [Google Scholar]
- 43.Grace MR, Walsh CT, Cole PA. Divalent ion effects and insights into the catalytic mechanism of protein tyrosine kinase Csk. Biochemistry. 1997;36:1874–81. doi: 10.1021/bi962138t. [DOI] [PubMed] [Google Scholar]
- 44.Kramers HA. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica. 1940;7:284–304. [Google Scholar]
- 45.Sierks MR, Sico C, Zaw M. Solvent and viscosity effects on the rate-limiting product release step of glucoamylase during maltose hydrolysis. Biotechnol Prog. 1997;13:601–8. doi: 10.1021/bp9700824. [DOI] [PubMed] [Google Scholar]
- 46.Demchenko AP, Ruskyn OI, Saburova EA. Kinetics of the lactate dehydrogenase reaction in high-viscosity media. Biochim Biophys Acta. 1989;998:196–203. doi: 10.1016/0167-4838(89)90273-2. [DOI] [PubMed] [Google Scholar]
- 47.Somogyi B, Welch GR, Damjanovich S. The dynamic basis of energy transduction in enzymes. Biochim Biophys Acta. 1984;768:81–112. doi: 10.1016/0304-4173(84)90001-6. [DOI] [PubMed] [Google Scholar]
- 48.Larsen TM, Boehlein SK, Schuster SM, Richards NG, Thoden JB, Holden HM, Rayment I. Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product. Biochemistry. 1999;38:16146–57. doi: 10.1021/bi9915768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.