SUMMARY
Annotation of prostate cancer genomes provides a foundation for discoveries that can impact disease understanding and treatment. Concordant assessment of DNA copy number, mRNA expression, and focused exon resequencing in 218 prostate cancer tumors identified the nuclear receptor coactivator NCOA2 as an oncogene in ~11 percent of tumors. Additionally, the androgen-driven TMPRSS2-ERG fusion was associated with a previously unrecognized, prostate-specific deletion at chromosome 3p14 that implicates FOXP1, RYBP and SHQ1 as potential cooperative tumor suppressors. DNA copy-number data from primary tumors revealed that copy-number alterations robustly define clusters of low- and high-risk disease beyond that achieved by Gleason score. The genomic and clinical outcome data from these patients is now made available as a public resource.
INTRODUCTION
Prostate cancer is the most common malignancy in males with ~190,000 new cases diagnosed per year in the United States and ~27,000 deaths. Prostate tumors show tremendous biological heterogeneity, with some patients dying of metastatic disease within 2–3 years of diagnosis whereas others can live for 10–20 years with organ-confined disease, likely a reflection of underlying genomic diversity. Large-scale cancer genome characterization projects studying glioblastoma, lung, colon, pancreas and breast cancers have provided critical new insights into the molecular classification of cancers and have the potential to identify new therapeutic targets (Cancer Genome Atlas Research Network, 2008; Ding et al., 2008; Jones et al., 2008; Parsons et al., 2008; Sjoblom et al., 2006; Weir et al., 2007; Wood et al., 2007). Prostate cancer presents special challenges for such large-scale multicenter genomics projects because of the relatively small tumor size and admixture with stroma that requires careful pathologist-guided dissection.
A number of groups have reported analyses of transcriptomes and copy-number alterations (CNAs) in prostate cancer, but rarely from the same samples and typically from modest numbers of tumors (~50–100 samples) or with lower resolution platforms (Kim et al., 2007; Lapointe et al., 2007; Lapointe et al., 2004; Lieberfarb et al., 2003; Perner et al., 2006; Singh et al., 2002). Consistent and common findings from these reports include the TMPRSS2-ERG fusion in ~50 percent, 8p loss in ~30–50 percent, and 8q gain in ~20–40 percent of cases. The data implicating ERG as a prostate cancer gene is clear (Tomlins et al., 2005), but there has been less progress in defining specific genes targeted by various common amplifications and deletions, in part due to limited availability of complementary transcriptome and exon resequencing data on sufficient patients to narrow the focus to a small list of candidate genes. Numerous transcriptome studies have defined general prostate cancer signatures but, unlike breast cancer (Paik et al., 2004; van de Vijver et al., 2002), these analyses have not identified robust subtypes of prostate cancer with different prognoses (Febbo and Sellers, 2003; Lapointe et al., 2004; Singh et al., 2002).
Here we adopted a comprehensive approach to define transcriptomes and CNAs in 218 prostate tumors (181 primaries, 37 metastases) and 12 prostate cancer cell lines and xenografts, as well as complete exon resequencing and/or focused mutation detection for 157 high interest genes in 80 tumors and 11 cell/xenograft lines (Table 1). After generating a map of CNAs across the dataset, we used matching mRNA and microRNA transcriptome and exon resequencing data to define the frequency of alterations in several common signal transduction pathways, explore various candidate genes within a few selected regions of copy-number gain and loss and correlate genomic alterations to clinical outcome. These data serve as a valuable resource for the cancer genomics community, prostate cancer scientists and clinicians and is readily and freely available through a user-friendly web-based portal (http://cbio.mskcc.org/prostate-portal/).
Table 1.
Summary of clinical characteristics for the study cohort
| Characteristic | Primary tumorsa | Metastasesb |
|---|---|---|
| Age | ||
| Median | 58.3 | 60 |
| Mean | 58.3 | 60 |
| Standard deviation | 7 | 8.6 |
| Min-max | 37.3–83 | 41–82 |
| PSA at diagnosis (ng/ml) | ||
| Median | 6 (IQR 4.4, 9) | 17 (IQR 8.6, 46.6) |
| <4 | 31 (17.2%) | 4 (12.5%) |
| 4–10 | 105 (58.3%) | 6 (18.75%) |
| >10 | 44 (24.5%) | 22 (68.75%) |
| Initial biopsy Gleason score | ||
| 5 | 2 (1%) | -- |
| 6 | 101 (56%) | 2 (6%) |
| 7 | 61 (34%) | 16 (46%) |
| 8 | 11 (6%) | 8 (23%) |
| 9 | 6 (3%) | 9 (25%) |
| Initial clinical stage | ||
| cT1c | 95 (52.4%) | 8 (22%) |
| cT2 | 76 (42%) | 12 (33%) |
| cT3 | 9 (5%) | 3 (8%) |
| cT4 | -- | 1 (3%) |
| Not available | -- | 9 (25%) |
| Ethnicity | ||
| Black | 29 (16.1%) | 2 (5.6%) |
| Asian | 4 (2.2%) | 0 (0%) |
| White hispanic | 0 (0%) | 2 (5.6%) |
| White non-hispanic | 142 (78.5%) | 32 (88.9%) |
| Unknown | 6 (3.3%) | 0 (0%) |
Cohort comprised of aprimary tumors (n=181; 1 patient with metastasis and primary analyzed),
RESULTS
Global copy number and transcriptome profiles define core pathway alterations
We applied rigorous criteria for selecting tumors for genomic analysis that have become the standard in large genomic studies (Cancer Genome Atlas Research Network, 2008) but adapted to address unique challenges posed by prostate cancers. All 218 samples had at least 70% tumor content (Table 1, Figure S1, Table S1). Transcriptome (mRNA and microRNA) and CNA profiling were conducted without amplification, with the exception of exon resequencing, which required whole genome amplification. Because we did not impose a stringent tumor size requirement, the small size of some tumors precluded concurrent analysis across all four platforms (Table 2, Table S2).
Table 2.
Number of primary and metastatic tumors analyzed by each platform
| Data type | Primariesc | Metastases | Cell lines/ xenografts |
All |
|---|---|---|---|---|
| aCGH | 157 | 37 | 13 | 207 |
| mRNA | 131 (29) | 19 | 6 | 156 |
| miRNA | 99 (28) | 14 | 0 | 113 |
| Sequencinga | 75 | 5 | 11 | 98 |
| aCGH, mRNAb | 109 | 19 | 5 | 133 |
| aCGH, mRNA, miRNAb | 79 | 13 | 2 | 94 |
| aCGH, miRNA, sequencinga,b | 72 | 2 | 0 | 74 |
| aCGH, mRNA, miRNA, sequencinga,b | 61 | 1 | 0 | 62 |
Summary of tumors and characterization platforms: asequencing category includes only tumors with matched normals and cell/xenografts lines,
mRNA and miRNA refers to expression profiling data, and
Analysis of known prostate cancer alterations in our dataset indicates successful tumor selection criteria (Figure S1). For example, the frequency of ERG alteration was 52 percent (see Experimental Procedures), consistent with other studies, and chromosome 8p loss and 8q gain were easily detected (Figure 1A). Overt CNAs were observed in 89 percent of tumors, also indicative of high tumor content. Additional histologic and molecular analysis of those tumors without CNA confirmed high tumor content (e.g., detection of TMPRSS2-ERG translocations). To address the possibility that our stringent tumor selection criteria might bias our dataset toward larger, more aggressive prostate cancers, we compared the clinical outcome of the 181 primary tumors (Table S1) in this dataset to 3437 consecutive men with prostate cancer treated by prostatectomy at MSKCC from 2000–2006. The time to biochemical relapse, defined as a rise in serum prostate specific antigen, was somewhat shorter in this study cohort (Table 1, Figure S1A). While this indicates that genomic findings from these samples may be biased toward larger, more aggressive prostate cancers (selected to ensure sufficient nucleic acid yield), this cohort nevertheless includes patients with favorable long-term clinical outcome (24% of patients have greater than 5-year recurrence-free survival).
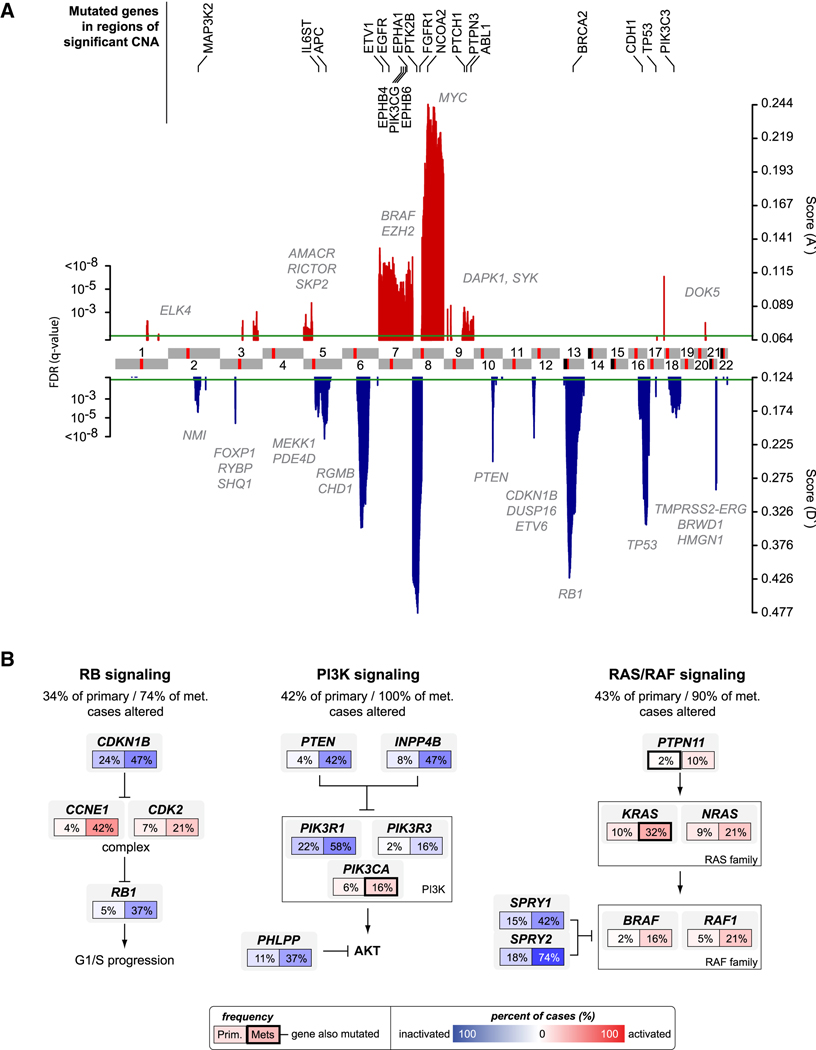
Figure 1. A global view of the prostate cancer genome.
A. Significant genomic aberrations in the prostate oncogenome. Regions of amplification (red) or deletion (blue) with FDR≤10% are plotted, with chromosomes indicated at the center and centromeres in red. Genes in which we detected somatic non-synonymous mutations are listed on top (black). Additional genes of interest targeted by copy-number alterations alone are also indicated (gray). B. Three of the most commonly altered pathways in both primary and metastatic prostate cancers: RB, PI3K, and RAS/RAF signaling. Alteration frequencies are shown for individual genes and for the entire pathway in primary and metastatic tumors. Alterations are defined as those having outlier expression (significant up- or down-regulation) compared to the distribution of expression in normal prostate samples (outlier analysis described in further detail in Experimental Procedures), or by somatic mutations, and are interpreted as activation (red) or inactivation (blue) of protein function. See also Figure S1 and Tables S3 and S6.
Global analysis of CNA data from 194 tumors and 12 cell lines/xenografts revealed broad diversity in alteration levels. Metastatic tumors and cell/xenograft lines harbored the greatest number of whole chromosome, chromosome arm, and focal amplifications and deletions, but primary tumors also displayed a wide range of alteration levels, from tumors appearing metastatic-like in profile to those with fundamentally diploid genomes (Figure S1B). Regions of recurrent CNA were identified using the statistical method RAE (Taylor et al., 2008), revealing 30 focal amplifications and 36 focal deletions as well as recurrent gains and losses of seven chromosome arms (Figure 1A, Tables S3 and S4). The most frequent alteration in the prostate oncogenome was loss of chromosome 8p, a common abnormality in many epithelial tumors that harbors NKX3.1 (He et al., 1997). Interestingly, NKX3.1 mRNA expression did not correlate with copy-number loss, suggesting the possibility of alternative tumor suppressors in this region. Consistent with prior studies, we also found peaks of deletion targeting PTEN on 10q23.31, RB1 on 13q14.2, TP53 on 17p31.1 and the interstitial 21q22.2-3 deletion spanning ERG and TMPRSS2. Other broader deletions included 12p13.31-p12.3, which spans ETV6 and DUSP16 in addition to CDKN1B, the previously reported target of this genomic deletion (Lapointe et al., 2007). The most common amplified loci included MYC on 8q24.21 and a previously unreported NCOA2 amplification on 8q13.3 (discussed further below). Focal amplifications of AR (Xq12) were also common but restricted to metastatic tumors. Other gains span discontinuous regions of 7q, including genes such as BRAF and EZH2, for which we were unable to localize individual target genes. We observed less frequent gains of 5p13.3-p13.1 spanning AMACR, RICTOR, SKP2 as well as 47 other genes and 2 microRNAs.
Eighty tumors were examined for somatic mutations in 138 genes by exon sequencing (Figure 1A, Tables S5 and S6). These and an additional 76 tumors were also profiled for well-known oncogenic mutations in 22 genes by mass spectrometry using the iPLEX Sequenom assay (Table S5). In total, 84 confirmed somatic mutations were detected in 57 different genes (Table S6). Thirty-seven percent of the missense mutations we detected are predicted to affect protein function (Table S6) based on an algorithm that uses a combination of evolutionary information from protein-family sequence alignments and residue placement in known or homology-deduced three-dimensional protein and complex structures (B. Reva, et al., http://www.mutationassessor.org/). Among all mutated genes, including those bearing previously known mutations, the most commonly mutated gene was the androgen receptor (AR), with 4 samples, all metastases. Mutations in 21 other genes were detected in 2 or more samples, but no single gene other than AR had mutations in more than 3 samples. We also confirmed prior data suggesting that common, broadly mutated oncogenes such as PIK3CA, KRAS and BRAF are not commonly mutated in prostate cancer (2 tumors had H1047R and E545K PIK3CA mutations, 2 had G12V and Q61H/L KRAS mutations and one tumor had a BRAF V600E mutation). Mutations in other more recently identified oncogenes such as IDH1 and IDH2 were similarly rare, with only one tumor bearing an IDH2 R172K mutation. Curiously, one tumor with a mutation in the mismatch repair gene MSH6 (V250A) had 11 confirmed somatic mutations versus an average of 2 somatic mutations per tumor, suggestive of a mutator phenotype. Mutations in two other DNA repair genes, BLM and XPC, were each found in a single tumor, but not in association with an increased number of other mutations. Only two tumors had missense mutations in TP53 and none had mutations in PTEN, but both tumor suppressors were commonly altered through hetero- or homozygous copy-number loss (~24 percent and ~21 percent respectively). Comparison of synonymous and non-synonymous changes detected in these samples suggests a low mutation rate in prostate cancer (~0.31 mutations/Mb). Consistent with this notion, the frequency of mutations recovered in our analysis did not exceed the expected background rate (Ding et al., 2008), although the modest number of genes and samples sequenced limits this analysis.
We next integrated the CNA, transcriptome, and mutation data to conduct a core pathways analysis, based on the success of this approach in revealing common pathway alterations in glioblastoma (Cancer Genome Atlas Research Network, 2008). Three well-known cancer pathways were commonly altered – PI3K, RAS/RAF and RB – with frequencies ranging from 34 to 43 percent in primary tumors versus 74 to 100 percent in metastases (Figure 1B). In this analysis, a tumor was considered altered if one or more genes in the pathway was mutated or significantly deregulated at the expression level (outlier expression compared to the distribution of expression in normal prostate samples, see Experimental Procedures). As in glioblastoma, the extremely high frequency of alteration in these pathways became evident only through examination of multiple genes in each pathway since individual genes are affected less commonly. Of particular interest is the PI3K pathway, which was altered in nearly half of primaries and all metastases examined. Loss of PTEN function, through deletion, mutation or reduced expression, has been well documented in prostate cancer with an estimated frequency of ~40 percent (Pourmand et al., 2007), consistent with our findings here. The frequency of PI3K pathway alteration rises substantially when PTEN alterations are considered together with alterations in the INPP4B and PHLPP phosphatases recently implicated in PI3K regulation, the PIK3CA gene itself, and the PIK3CA regulatory subunits PIK3R1 and PIK3R3 (Cancer Genome Atlas Research Network, 2008; Gao et al., 2005; Gewinner et al., 2009; Jaiswal et al., 2009; Ueki et al., 2003). These data provide strong rationale for exploring the clinical activity of PI3K pathway inhibitors, many of which are now in early clinical development, in prostate cancer.
Common genomic alterations in androgen receptor signaling pathway members
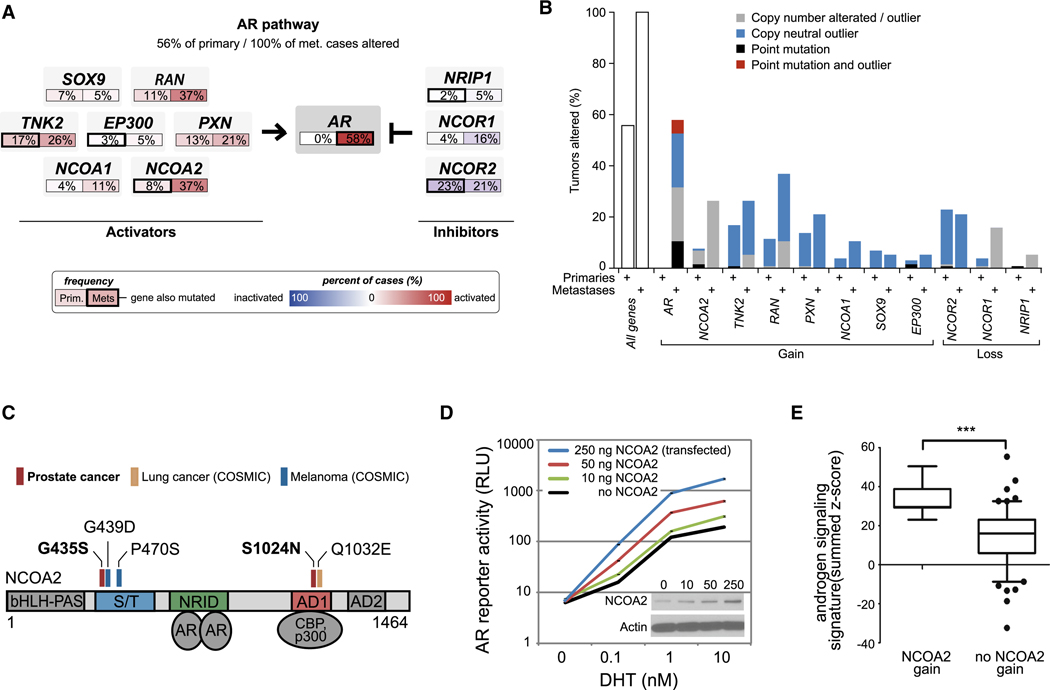
We also conducted a core pathway analysis of AR, which is essential for growth and differentiation of the normal prostate and is responsible for treatment failure in castration-resistant, metastatic disease (Chen et al., 2004; Tran et al., 2009). As expected, alteration of AR through mutation, gene amplification, and/or overexpression was common but occurred exclusively in metastatic samples (58%, Figure 2A). However, AR pathway analysis (including several known AR coactivators and corepressors) revealed alteration in 56 percent of primaries and 100 percent in metastases (Figure 2A, Figure S2). Among AR pathway genes, the most striking finding was a peak of copy-number gain on 8q13.3 (~57 megabases away from the peak at 8q24 commonly attributed to MYC, and of even greater significance) that spans the nuclear receptor coactivator gene NCOA2 (also known as SRC2/TIF2/GRIP1). Seventeen percent of tumors had broad gains of the region spanning NCOA2 on 8q while 6.2% of tumors (1.9% and 24.3% of primary and metastases respectively) harbored focal or high-level amplifications of the locus and these were significantly correlated with elevated NCOA2 transcript levels (p-value < 10−16, Figure S2A). In addition to copy number and expression changes, AR pathway alterations included mutations in NCOA2 (2 confirmed somatic) as well as in NCOR2 (3 tumors), NRIP1, TNK2, and EP300 (1 tumor each). Overall, eight percent of primary tumors and 37 percent of metastases had NCOA2 gain of expression (determined to be outlier expression as described in the Experimental Procedures) or mutation (Figure 2A–B). Including broader gains of 8q, the frequency of NCOA2 alteration may be as high as 20 and 63 percent in primary and metastatic tumors respectively. Of note, NCOA2 mutations have also been reported in melanoma and lung cancer and, in conjunction with the prostate mutations detected here, cluster in two highly conserved regions. These include a serine/threonine-rich stretch (S/T) known to be phosphorylated and an activation domain (AD1) that mediates binding with histone acetyltransferases such as CBP and p300 (Huang and Cheng, 2004) (Figure 2C). A third patient had an A407S NCOA2 substitution that was potentially germline (detected at low frequency in adjacent normal prostate). Interestingly, non-castrate patients with primary tumors harboring NCOA2 mutation, overexpression or high-level amplification had significantly higher rates of recurrence (Figure S2B).
Figure 2. Diversity of androgen signaling pathway alterations in primary and metastatic prostate cancers.
A. Alterations in androgen signaling components where frequencies are shown for AR and selected activators in primary and metastatic tumors. Alterations are defined as those having outlier expression (as described in Figure 1B and Experimental Procedures), or by somatic mutations, and interpreted as activation (red) or inactivation (blue). B. Overall genomic alteration rates in androgen signaling genes. The eleven genes in the AR pathway (panel A) had somatic mutations (black, red) and/or outlier expression (over- or under-expression, see Experimental Procedures), a subset of which was the result of copy-number alterations (gray, broad and focal gain or loss of one or more copies, see Supplementary Information). Primary tumors show moderate levels of alteration in at least one of these eleven AR pathway genes (left), preceding the generally higher alteration rates in metastatic tumors (right). C. The steroid receptor co-activator NCOA2 had two novel somatic point mutations in primary tumors. These clustered near sites of known NCOA2 point mutations in melanoma [G435S in the serine/threonine-rich (S/T) regulatory domain] and lung cancer [S1024N in the transcriptional activation domain 1 (AD1)]. D. Increasing levels of NCOA2 induce increasing androgen-dependent AR transcriptional activity. Increasing amounts of NCOA2 plasmid were transfected into LNCaP cells, resulting in NCOA2 protein expression (inset western blot) and ARE-luciferase reporter activity. Error bars representing the S.E.M (standard error of the mean) are displayed, but represent a very small portion of the signal, and are therefore not visible. E. Non-castrate primary tumors with NCOA2 gain (defined as copy-number amplification greater than single-copy gain, outlier over-expression, or mutation) have higher androgen signaling (student’s t-test, P<0.0001), as assessed by an independent signature of 29 androgen-responsive genes (Hieronymus et al., 2006). See also Figure S2.
Taken together, the high frequency of NCOA2 gain in primary tumors and its known role as an AR coactivator (Agoulnik et al., 2005) suggest that these two genes might collaborate in early prostate cancer progression by enhancing AR transcriptional output. We addressed this possibility by expressing increasing levels of NCOA2 in prostate cancer cells with a fixed endogenous level of non-amplified AR. The range in NCOA2 protein levels is shown by western blot (Figure 2D) and is similar to the ~2 to 4-fold increase in NCOA2 mRNA levels over the mean level seen in the overall prostate cancer cohort. As expected from other work, AR transcriptional output (measured using an androgen-responsive reporter construct) was increased when cells were treated with dihydrotestosterone (DHT) in a dose-dependent fashion, but reached a plateau between 1–10 µM (Figure 2D). Increasing levels of NCOA2 shifted the DHT dose-response curve leftward and upward, indicating that NCOA2 can not only prime AR to respond to lower androgen concentrations but can also boost the total magnitude of AR transcriptional output. One prediction from these in vitro data is that the AR transcriptional output in prostate cancers with NCOA2 gene amplification should be greater than those without. Based on a 29-gene signature of AR transcriptional output previously used to conduct small molecule screens for novel antiandrogens (Hieronymus et al., 2006), NCOA2-amplified primary tumors displayed an increase in AR signaling (Figure 2E). Collectively, the genomic and functional data suggest that NCOA2 functions as a driver oncogene in primary tumors by increasing AR signaling, which is known to play a critical role in early and late stage prostate cancer. In contrast, AR amplification, which is largely restricted to castration-resistant metastatic disease, is more likely a mechanism of drug resistance rather than a natural step in tumor progression. We also propose that NCOA2 and MYC both function as driver oncogenes on the 8q13 and 8q24 amplicons respectively.
Genetic alterations highly associated with TMPRSS2-ERG
The TMPRSS2-ERG fusion is the single most prevalent molecular lesion in prostate cancer (Tomlins et al., 2005). Functional studies of TMPRSS2-ERG, including transgenic expression in the mouse prostate, have shown modest evidence of oncogenic activity (Carver et al., 2009; King et al., 2009; Klezovitch et al., 2008; Tomlins et al., 2008), which raises the possibility that cooperating events are required.
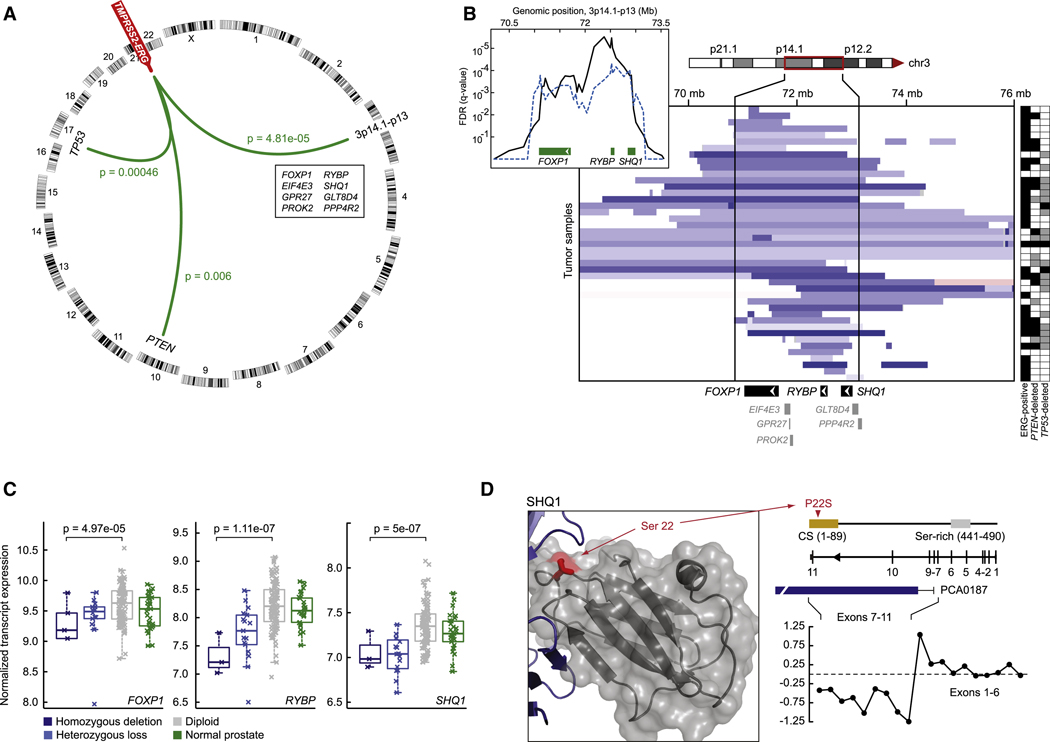
Analysis of 194 tumors for CNAs associated with TMPRSS2-ERG fusion revealed three significant regions of copy-number loss: two spanning the tumor suppressors PTEN and TP53 and a third spanning the multigenic region at 3p14 (Figure 3A). PTEN loss was recently shown to cooperate with TMPRSS2-ERG in transgenic mice and in a prostate tissue reconstitution model (Carver et al., 2009; King et al., 2009; Zong et al., 2009). The 3p14 deletion, whose association with TMPRSS2-ERG was even more significant, has not been previously reported and spans only 8 genes. Further interrogation of 2550 tumors and cell lines spanning 14 tumor types (acute lymphoid leukemia, breast, colorectal, esophageal, glioma, hepatocellular, non-small cell lung, squamous lung, medulloblastoma, melanoma, myeloproliferative, ovarian, renal and prostate) for CNAs in this region suggests this deletion is only found in prostate cancer (Beroukhim et al., 2010). Indeed, the only other focal signal found in this region is an amplicon in melanoma that includes microphthalmia-associated transcription factor (MITF), a previously reported finding (Garraway et al., 2005).
Figure 3. TMPRSS2-ERG-associated deletion of 3p14.1-p13.
A. TMPRSS2-ERG gene fusion co-occurs with genomic aberrations in the prostate cancer genome including deletions of loci encoding TP53 (17p13.1) and PTEN (10q23.31), as well as focal deletion of 3p14.1-p13 (genes listed in genomic order; green lines represent statistically significant associations, FDR ≤ 1%). Chromosomes are shown around the ring. B. Diverse genomic deletions (heterozygous and homozygous deletions, light and dark blue respectively) target a ~2.2mb region of 3p14.1-p13 encoding 8 genes (indicated at bottom). Tumors are rows and those harboring TMPRSS2-ERG fusion or PTEN/TP53 deletion (heterozygous and homozygous deletions are black and grey respectively) are identified at right. Tumors are sorted according to their locus of deletion with focal losses preferentially affecting FOXP1 (top), RYBP and the adjacent gene SHQ1 (bottom), or both loci simultaneously (middle). Inset indicates the pattern of significance of total genomic deletion juxtaposed to the significance of TMPRSS2-ERG-associated deletion (black and dotted blue respectively). C. Transcript expression according to copy-number status for the three genes targeted by the 3p14.1-p13 deletion: FOXP1, RYBP, and SHQ1, all three of which are correlated (p-values as indicated, Anova). EIF4E3 and PPP4R2 expression and copy-number loss were also correlated, but neither of these two genes was focally targeted by 3p14.1-p13 deletion. D. Along with whole-gene deletions of SHQ1 (panel b), we detected a single tumor with a P22S somatic mutation of the CS domain, indicated in red in the three-dimensional structure of the SHQ1 yeast homolog (3eud) and in linear representation of the protein (top right). Also, intragenic deletions (shown here in metastatic sample PCA0187) confer exon-specific loss of expression indicating a truncation event (bottom right).
Closer inspection of the 3p14 deletion in our prostate cohort revealed two distinct peaks of association within the region that, together with expression data and the focal deletion patterns, implicates only three genes: FOXP1, RYBP and SHQ1 (Figure 3B, 3C). Deletions in some tumors spanned FOXP1 only, whereas others included RYBP and SHQ1, but spared FOXP1. FOXP1 encodes a forkhead box transcription factor and functions in motor neuron specification in the spinal cord, as well as early thymocyte development, in collaboration with various HOX genes (Arber, 2008; Pfaff, 2008). A role for FOXP1 in cancer has been proposed based on reduced expression in breast and other cancers, increased expression in some lymphoid malignancies and, remarkably, by translocation-mediated fusion to the ERG homolog ETV1 in at least one prostate cancer (Goatly et al., 2008; Hermans et al., 2008; Koon et al., 2007). Furthermore, recent evidence implicates the FoxP family member FOXP3 as a potential tumor suppressor in prostate cancer (Wang et al., 2009). RYBP (Ring and YY1 Binding Protein) encodes a polycomb group transcriptional repressor implicated in homeotic development and, potentially, as a tumor suppressor through inhibition of MDM2 and subsequent p53 stabilization (Chen et al., 2009). SHQ1 encodes an accessory factor for the assembly of H/ACA ribonucleoproteins (RNP) through direct binding to NAP57, a core RNP subunit. Missense mutations in NAP57 that disrupt interaction with SHQ1 are associated with the bone marrow failure syndrome dyskeratosis congenita, raising a potential link to precancer syndromes (Grozdanov et al., 2009).
To gather further evidence for a potential tumor suppressor role of either of these genes, we searched for point mutations through exon resequencing. We found no mutations in FOXP1 or RYBP, but detected a confirmed somatic mutation in SHQ1 (P22S) in a highly conserved region of the CS domain that is required for SHQ1 function (Singh et al., 2009). A second tumor had a deletion targeting the middle of the SHQ1 gene that, consequently, resulted in production of an aberrant mRNA species truncated at exon 6 (Figure 3D). Although these data further implicate SHQ1 as a tumor suppressor in this locus, the fact that some tumors with 3p14 loss spare SHQ1 (Figure 3B) raises the possibility of multiple tumor suppressors in this region.
Unsupervised clustering of CNAs reveals distinct subgroups with differing risk of relapse after prostatectomy
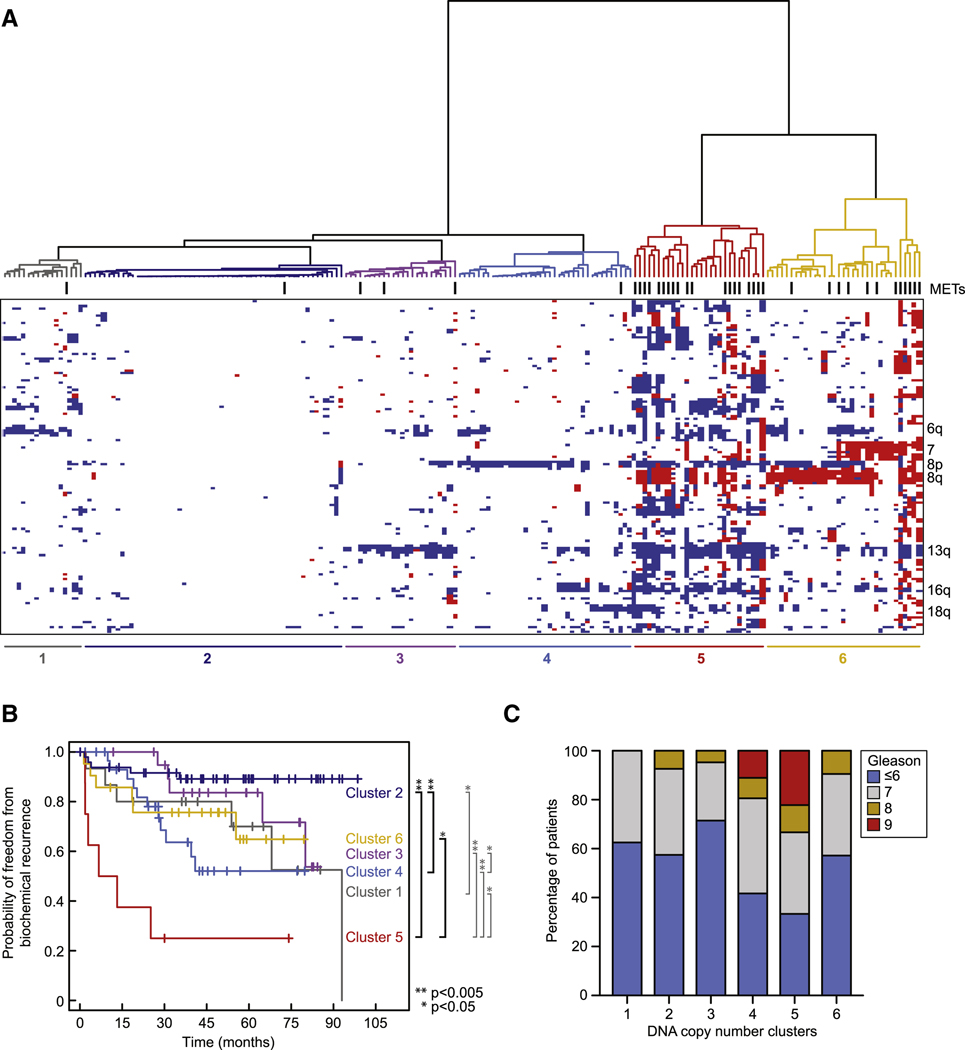
Given the pressing need for biomarkers that distinguish indolent from aggressive prostate cancer, we also examined the genomic data for prognostic significance. It is estimated that 30–50 percent of men diagnosed with prostate cancer could avoid surgery or radiation (and instead be followed by watchful waiting) because they have good-prognosis tumors that are unlikely to progress (Cooperberg et al., 2005). Whereas transcriptome analysis defines breast cancer subgroups with distinct prognoses and treatment outcomes that have changed clinical practice (Paik et al., 2004; van de Vijver et al., 2002), similar studies in prostate cancer have been less clinically useful (Mucci et al., 2008a; Mucci et al., 2008b). The 5-year median clinical followup linked to this tumor set provided an opportunity to address the prognosis question using various forms of oncogenomic data. While unsupervised hierarchical clustering of mRNA and microRNA data failed to identify robust clusters of patients with significant differences in prognosis, the CNA data revealed distinct subgroups with substantial differences in time to biochemical (PSA) relapse (Figure 4A,B, Figure S3A–C). Further attempts to identify individual genes whose expression has prognostic impact through outlier analysis (~1766 genes with over- or under-expressing outliers relative to normal prostate) were only modestly successful and these associations were weak relative to those observed using the CNA data.
Figure 4. Genomic aberrations identify clinically distinct subtypes of prostate cancer.
A. Unsupervised hierarchical clustering of copy-number alterations (heatmap; red is amplification, white is copy-neutral, blue is deletion) indicates six groups of prostate cancers exist. Samples are ordered based on their group membership (dendrogram, groups are colored, metastatic samples are indicated by hashes). Selected genomic regions indicative of group membership are labeled (right). B. There exist significant differences in the risk of biochemical recurrence among the groups of tumors determined by patterns of genomic aberrations (left; p-values as indicated, log-rank test). C. Differences in the risk of biochemical relapse (panel B) were independent of Gleason grade (≤6) in the most diploid clusters (1–4) (no statistically significant association of Gleason grade in clusters 1–4). See also Figure S3 and Table S4.
CNA analysis revealed two notable subgroups of primary tumors – those with minimal CNA (clusters 1–4) and those with substantial CNA (clusters 5–6) that include most of the metastatic samples (Figure 4A). Clusters 5 and 6 are distinguished by the fact that cluster 5 tumors have genome-wide alterations, whereas those in cluster 6 primarily have 8q (NCOA2, MYC) or chromosome 7 gains. Among the tumors with minimal CNA, cluster 2 is characterized by largely unaltered genomes. Using the endpoint of time to biochemical relapse, primary tumors with generally diploid tumors in the minimally altered cluster 2 had an extremely favorable prognosis versus an extremely unfavorable prognosis for the highly altered cluster 5 tumors (Figure 4B).
We next examined whether the prognostic impact of CNA is simply a reflection of genomic instability or the impact of specific genomic alterations. The fact that the two clusters with the highest prevalence of CNAs (5 and 6) have statistically different outcomes supports the latter hypothesis. To explore this question further, we systematically examined the impact of gain or loss of whole chromosome arms or more focal regions of gain or loss across the genome. Combined loss of 13q and 18q, focal amplification of two distinct 5p regions (5p13 or 5p15), and focal deletion of 5q21.1 were each significantly associated with a negative outcome (Figure S3B,C), further supporting the notion that distinct genomic alterations impact prognosis and raising the possibility that genes in these regions play functional roles in prostate cancer.
These findings raise the possibility that CNA assessment at diagnosis may have clinical utility in distinguishing low- from high-risk disease, but only if this data adds to the prognostic impact currently provided by the histology-based Gleason score. Poor prognosis Gleason score (>7) tumors were distributed across clusters 2–6 (albeit with greatest frequency in cluster 5), indicating that histology and CNA are not overlapping (Figure 4C). Furthermore, low-risk Gleason scores (≤6) were not enriched among clusters 1–4. Therefore, Gleason grade cannot fully explain the association with biochemical relapse. These results raise the possibility of a CNA-based test that might guide treatment choice in men with newly diagnosed prostate cancer, though this would require validation in a larger independent data set and confirmation that such information can be obtained from biopsies rather than prostatectomy samples. Such a test might be a genome-wide assessment using array-based CGH or multiple inversion probe (MIP) technology (Wang et al., 2005) or be centered on specific regions of gain or loss identified through further confirmatory studies.
DISCUSSION
The clinical heterogeneity of prostate cancer, coupled with its high prevalence, raises challenges in the management of newly diagnosed patients as well as those with metastatic disease. Genomic-based classification offers the hope of more informed clinical decision-making and may yield novel therapeutic targets. Integrated large scale cancer genomics projects in several tumor types have established the utility of this approach for generating the datasets required to derive such classification schema (Cancer Genome Atlas Research Network, 2008; Chitale et al., 2009; Ding et al., 2008; Jones et al., 2008; Parsons et al., 2008; Sjoblom et al., 2006; Weir et al., 2007; Wood et al., 2007). These reports provide definitive overviews of the genomes of those tumor types and in cases such as TCGA provide easy, web-based access to genomic data that serves as a public resource. The prostate cancer dataset generated here is comparable in size (218 carefully selected, well-annotated tumors) and scope (transcriptome, CNA, exon resequencing) and is linked to clinical outcome. All raw and processed data is freely accessible at http://cbio.mskcc.org/prostate-portal/.
One observation from the exon resequencing data is that somatic point mutations in prostate cancer may be rare relative to other tumor types such as glioblastoma, lung cancer and melanoma (Greenman et al., 2007; Pleasance et al., 2010a; Pleasance et al., 2010b). With the caveat that only 138 genes were examined (selected primarily based on known roles in other cancers), no single gene emerged as commonly mutated. TP53 and PTEN, which are often cited as prostate cancer tumor suppressors (Dong, 2006; Pourmand et al., 2007), were commonly altered, but primarily through copy-number loss rather than point mutation. Ongoing comprehensive sequencing studies (whole-genome or whole-exome capture) will provide more insight into the overall mutation rate in prostate cancer.
Several findings have emerged from our analysis, largely based on the opportunity provided by integrated analysis of multidimensional data. The nuclear receptor coactivator NCOA2 was identified as a highly significant target gene on the 8q13 amplicon and is also subject to mutation in some tumors lacking gene amplification. Functional studies presented here support the hypothesis that increased NCOA2 dosage amplifies AR pathway transcriptional output in primary tumors, providing a mechanism for its potential role as a prostate cancer oncogene. Whereas AR gene amplification or mutation is generally restricted to metastatic, castration-resistant disease (acquired in association with treatment resistance), CNAs or mutations in NCOA2 and other regulators of nuclear receptor function such as NCOR2 are present in primary tumors, thereby extending the potential importance of AR pathway perturbation to disease initiation.
A second finding is a narrow deletion on 3p14 highly associated with TMPRSS2-ERG fusion-positive tumors that appears to be present only in prostate cancers. Integrative analysis of copy number, transcriptome and exon resequencing data implicates three genes within this region (FOXP1, RYBP and SHQ1) as potential context-specific tumor suppressors, either alone or in combination. Our methodology also confirmed prior reports of an association of TMPRSS2-ERG with PTEN loss (Han et al., 2009; Reid et al., 2010), an interaction that has now been validated by in vivo studies in mice (Carver et al., 2009; King et al., 2009; Zong et al., 2009). We found evidence of a possible association with 16q23 deletion previously reported by others (Demichelis et al., 2009), but this did not reach statistical significance in our larger dataset. As has been done with PTEN, these new associations warrant future functional studies and could define unique ERG-specific tumor suppressor interactions.
These findings together with our analysis showing the high impact of CNA data on risk of relapse relative to transcriptome profiling demonstrate the broad utility of this integrated prostate oncogenome dataset. The high prevalence of this important disease and the relative paucity of large comprehensive genomic datasets in prostate cancer make this a unique public resource for the cancer research community.
EXPERIMENTAL PROCEDURES
Specimen collection and annotation
A total of 218 tumor samples and 149 matched normal samples were obtained from patients treated by radical prostatectomy at Memorial Sloan-Kettering Cancer Center. All patients provided informed consent and samples were procured and the study was conducted under Memorial Sloan-Kettering Cancer Center Institutional Review Board approval. Clinical and pathologic data were entered and maintained in our prospective prostate cancer database. Following radical prostatectomy, patients were followed with history, physical exam, and serum PSA testing every 3 months for the first year, 6 months for the second year, and annually thereafter. For all analyses described here, biochemical recurrence (BCR) was defined as PSA ≥ 0.2 ng/ml on two occasions. At the time of data analysis, patient follow-up was completed through December 2008.
Analyte extraction and microarray hybridization
DNA and RNA were extracted from dissected tissue containing greater than 70% tumor cell content as well as from seven cell lines and seven xenografts (see Supplementary Information). Resulting DNA and RNA were hybridized to Agilent 244K array comparative genomic hybridization (aCGH) microarrays, Affymetrix Human Exon 1.0 ST arrays, and/or Agilent microRNA V2 arrays respectively (Table 2). The normalization and statistical analysis of both DNA copy number and expression array data are available in Supplementary Information.
DNA sequencing
In total, 251 million bases in coding exons and adjacent intronic sequences of 138 cancer-related genes in 91 samples were PCR-amplified and sequenced by Sanger capillary sequencing (Table S5). Ninety-five sites of known mutation in 22 genes were also genotyped using the iPLEX Sequenom platform. The details of whole-genome amplification, sequencing, mutation detection pipelines, mutation validation, background mutation rate analysis, and Sequenom genotyping are described in Supplementary Information.
Outlier expression analysis
Outlier profiles for all transcripts and outlier assignments in all tumors were determined from normalized expression data as previously described (Ghosh and Chinnaiyan, 2009). Briefly, in this nonparametric approach an empirical distribution function generated from transcript expression in the 29 normal prostate tissues was used to transform expression in the tumor samples, from which outliers were determined with the criteria described in the Benjamini and Hochberg algorithm (Benjamini and Hochberg, 1995) at an error rate (α) = 0.01.
TMPRSS2-ERG fusion classification
For the purposes of the association analysis among copy-number alterations (described in detail in Supplementary Information), we classified fusion-positive tumors exclusively from aCGH data to maximize our power to detect novel associations in CNA data alone. Tumors were considered fusion-positive if they harbored canonical 21q22.2-3 genomic deletion (D0 or D1 > 0.9 from RAE analysis) with 5’ and 3’ breakpoints in the coding loci of ERG and TMPRSS2 respectively accompanied by interstitial deletion, or those samples with micro-deletions at the expected breakpoint sites in either or both genes in conjunction with intergenic diploidy. This approach under-estimates the true frequency of TMPRSS2-ERG fusion by excluding tumors with balanced rearrangement. Therefore, all other analyses described in this study classified TMPRSS2-ERG status using the subset of cases with both aCGH and expression data. Here, fusion-positive tumors were those having either the genomic deletion described above or whole-transcript outlier expression inferred from exon expression arrays as described above. We note that re-classification of TMPRSS2-ERG status using individual exon expression adjacent to the expected breakpoints in each coding sequence produced similar results.
Pathway analysis
The details of pathway curation and gene selection for the pathway diagrams are described in the Supplementary Information. To determine pathway alteration frequencies, gene alterations were defined by up- or down-regulation compared to normal prostate (outlier expression), or by somatic non-synonymous mutations. A given tumor was considered altered if at least one gene in the pathway was altered. Mutations in genes known to be frequently deleted or down-regulated were considered as inactivating mutations (shades of blue in the figures), while mutations in genes known to be frequently amplified or up-regulated were considered as activating mutations (shades of red). Additionally, the association of NCOA2 gain-of-function alteration (outlier over-expression or copy-number amplification) with androgen signaling was assessed using a 29-gene signature of androgen stimulation (Hieronymus et al., 2006). The significance of this association was tested in non-castrate primary tumors by Student’s t-test.
Hierarchical clustering
Unsupervised hierarchical clustering of discretized copy-number alterations (gain and loss, A0 and D0 ≥ 0.75; otherwise copy-neutral from RAE analysis) assigned to regions of the unified breakpoint profile excluding known CNVs was performed with the Manhattan distance measure and Ward’s linkage.
NCOA2 and AR reporter assay
For NCOA2 assays, pCDNA3-NCOA2 and PSA-Luc reporter were transfected into LNCaP cells that were androgen-starved (20h) and then assayed for growth after another 20h (One-Glo). Additional experimental details available in Supplementary Information.
Data availability and portal
Study data is deposited in NCBI GEO under accession GSE21032. The analyzed data can also be accessed and explored through the MSKCC Prostate Cancer Genomics Data Portal: http://cbio.mskcc.org/prostate-portal/
Full methods are described in the Supplemental Information.
Highlights.
Integrated genomic profiling of 218 prostate tumors provides a unique public resource
Androgen receptor coactivator NCOA2 is amplified in primary and metastatic disease
TMPRSS2-ERG positive tumors associate with 3p14 loss and candidates FOXP1, RYBP, SHQ1
Degree and pattern of CNAs in primary tumors is associated with risk of relapse
SIGNIFICANCE.
Current knowledge of prostate cancer genomes is largely based on small patient cohorts using single modality platforms. We present an integrated oncogenomic analysis of 218 primary and metastatic prostate cancers as well as 12 cell lines and xenografts. Mutations in known, commonly mutated oncogenes and tumor suppressor genes such as PIK3CA, KRAS, BRAF and TP53 are present but rare. However, integrative analysis of mutations, copy-number alterations and expression changes revealed changes in the PI3K, RAS/RAF and androgen receptor (AR) pathways in nearly all metastatic samples and a high frequency of primary samples. These data clarify the role of several known cancer pathways in prostate cancer, implicate several new ones, and provide a blueprint for clinical development of pathway inhibitors.
Supplementary Material
ACKNOWLEDGEMENTS
This work is dedicated to the memory of our colleague and friend William Gerald who initiated this project. We are grateful for the technical assistance and support of A. Viale (MSKCC Genomics Core), K. Huberman, S. Thomas, O. Aminova (Beene Translational Oncology Core), A. Olshen, J. Satagopan (Epidemiology and Biostatistics), L. Vargas, L. Chen (Pathology) and A. Gabow (Bioinformatics Core). This work was supported in part by the MSKCC Prostate SPORE CA092629 and by the David H. Koch Foundation. C.L.S. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- Agoulnik IU, Vaid A, Bingman WE, 3rd, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- Arber S. FoxP1: conducting the Hox symphony in spinal motor neurons. Nat Neurosci. 2008;11:1122–1124. doi: 10.1038/nn1008-1122. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57 [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhang J, Li M, Rayburn ER, Wang H, Zhang R. RYBP stabilizes p53 by modulating MDM2. EMBO Rep. 2009;10:166–172. doi: 10.1038/embor.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale D, Gong Y, Taylor BS, Broderick S, Brennan C, Somwar R, Golas B, Wang L, Motoi N, Szoke J, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28:2773–2783. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperberg MR, Moul JW, Carroll PR. The changing face of prostate cancer. J Clin Oncol. 2005;23:8146–8151. doi: 10.1200/JCO.2005.02.9751. [DOI] [PubMed] [Google Scholar]
- Demichelis F, Setlur SR, Beroukhim R, Perner S, Korbel JO, Lafargue CJ, Pflueger D, Pina C, Hofer MD, Sboner A, et al. Distinct genomic aberrations associated with ERG rearranged prostate cancer. Genes Chromosomes Cancer. 2009;48:366–380. doi: 10.1002/gcc.20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong JT. Prevalent mutations in prostate cancer. J Cell Biochem. 2006;97:433–447. doi: 10.1002/jcb.20696. [DOI] [PubMed] [Google Scholar]
- Febbo PG, Sellers WR. Use of expression analysis to predict outcome after radical prostatectomy. J Urol. 2003;170:S11–S19. doi: 10.1097/01.ju.0000095567.03807.a1. discussion S19–20. [DOI] [PubMed] [Google Scholar]
- Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, Barretina J, Lin WM, Rameh L, Salmena L, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Chinnaiyan AM. Genomic outlier profile analysis: mixture models, null hypotheses, and nonparametric estimation. Biostatistics. 2009;10:60–69. doi: 10.1093/biostatistics/kxn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goatly A, Bacon CM, Nakamura S, Ye H, Kim I, Brown PJ, Ruskone-Fourmestraux A, Cervera P, Streubel B, Banham AH, Du MQ. FOXP1 abnormalities in lymphoma: translocation breakpoint mapping reveals insights into deregulated transcriptional control. Mod Pathol. 2008;21:902–911. doi: 10.1038/modpathol.2008.74. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov PN, Roy S, Kittur N, Meier UT. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA. 2009;15:1188–1197. doi: 10.1261/rna.1532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22:1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He WW, Sciavolino PJ, Wing J, Augustus M, Hudson P, Meissner PS, Curtis RT, Shell BK, Bostwick DG, Tindall DJ, et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- Hermans KG, van der Korput HA, van Marion R, van de Wijngaart DJ, Ziel-van der Made A, Dits NF, Boormans JL, van der Kwast TH, van Dekken H, Bangma CH, et al. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res. 2008;68:7541–7549. doi: 10.1158/0008-5472.CAN-07-5930. [DOI] [PubMed] [Google Scholar]
- Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Huang SM, Cheng YS. Analysis of two CBP (cAMP-response-element-binding protein-binding protein) interacting sites in GRIP1 (glucocorticoid-receptor-interacting protein), and their importance for the function of GRIP1. Biochem J. 2004;382:111–119. doi: 10.1042/BJ20040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, Kan Z, Dbouk HA, Peters BA, Waring P, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, Kumar-Sinha C, Cao X, Dash A, Wang L, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, Nelson PS, Vasioukhin V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koon HB, Ippolito GC, Banham AH, Tucker PW. FOXP1: a potential therapeutic target in cancer. Expert Opin Ther Targets. 2007;11:955–965. doi: 10.1517/14728222.11.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberfarb ME, Lin M, Lechpammer M, Li C, Tanenbaum DM, Febbo PG, Wright RL, Shim J, Kantoff PW, Loda M, et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res. 2003;63:4781–4785. [PubMed] [Google Scholar]
- Mucci LA, Pawitan Y, Demichelis F, Fall K, Stark JR, Adami HO, Andersson SO, Andren O, Eisenstein A, Holmberg L, et al. Testing a multigene signature of prostate cancer death in the Swedish Watchful Waiting Cohort. Cancer Epidemiol Biomarkers Prev. 2008a;17:1682–1688. doi: 10.1158/1055-9965.EPI-08-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucci LA, Pawitan Y, Demichelis F, Fall K, Stark JR, Adami HO, Andersson SO, Andren O, Eisenstein AS, Holmberg L, et al. Nine-gene molecular signature is not associated with prostate cancer death in a watchful waiting cohort. Cancer Epidemiol Biomarkers Prev. 2008b;17:249–251. doi: 10.1158/1055-9965.EPI-07-0722. [DOI] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- Pfaff SL. Developmental neuroscience: Hox and Fox. Nature. 2008;455:295–297. doi: 10.1038/455295a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010a;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010b;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourmand G, Ziaee AA, Abedi AR, Mehrsai A, Alavi HA, Ahmadi A, Saadati HR. Role of PTEN gene in progression of prostate cancer. Urol J. 2007;4:95–100. [PubMed] [Google Scholar]
- Reid AH, Attard G, Ambroisine L, Fisher G, Kovacs G, Brewer D, Clark J, Flohr P, Edwards S, Berney DM, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678–684. doi: 10.1038/sj.bjc.6605554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- Singh M, Gonzales FA, Cascio D, Heckmann N, Chanfreau G, Feigon J. Structure and functional studies of the CS domain of the essential H/ACA ribonucleoparticle assembly protein SHQ1. J Biol Chem. 2009;284:1906–1916. doi: 10.1074/jbc.M807337200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Barretina J, Socci ND, Decarolis P, Ladanyi M, Meyerson M, Singer S, Sander C. Functional copy-number alterations in cancer. PLoS One. 2008;3:e3179. doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki K, Fruman DA, Yballe CM, Fasshauer M, Klein J, Asano T, Cantley LC, Kahn CR. Positive and negative roles of p85 alpha and p85 beta regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J Biol Chem. 2003;278:48453–48466. doi: 10.1074/jbc.M305602200. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY, McNally B, Lin L, Zhou P, Zuo T, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009;16:336–346. doi: 10.1016/j.ccr.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Moorhead M, Karlin-Neumann G, Falkowski M, Chen C, Siddiqui F, Davis RW, Willis TD, Faham M. Allele quantification using molecular inversion probes (MIP) Nucleic Acids Res. 2005;33:e183. doi: 10.1093/nar/gni177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci U S A. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Study data is deposited in NCBI GEO under accession GSE21032. The analyzed data can also be accessed and explored through the MSKCC Prostate Cancer Genomics Data Portal: http://cbio.mskcc.org/prostate-portal/
Full methods are described in the Supplemental Information.
Highlights.
Integrated genomic profiling of 218 prostate tumors provides a unique public resource
Androgen receptor coactivator NCOA2 is amplified in primary and metastatic disease
TMPRSS2-ERG positive tumors associate with 3p14 loss and candidates FOXP1, RYBP, SHQ1
Degree and pattern of CNAs in primary tumors is associated with risk of relapse
SIGNIFICANCE.
Current knowledge of prostate cancer genomes is largely based on small patient cohorts using single modality platforms. We present an integrated oncogenomic analysis of 218 primary and metastatic prostate cancers as well as 12 cell lines and xenografts. Mutations in known, commonly mutated oncogenes and tumor suppressor genes such as PIK3CA, KRAS, BRAF and TP53 are present but rare. However, integrative analysis of mutations, copy-number alterations and expression changes revealed changes in the PI3K, RAS/RAF and androgen receptor (AR) pathways in nearly all metastatic samples and a high frequency of primary samples. These data clarify the role of several known cancer pathways in prostate cancer, implicate several new ones, and provide a blueprint for clinical development of pathway inhibitors.