Abstract
In somatic cells, three major pathways are involved in the repair of DNA double-strand breaks (DBS): Non-Homologous End Joining (NHEJ), Single-Strand Annealing (SSA) and Homologous Recombination (HR). In somatic and meiotic HR, DNA DSB are 5′ to 3′ resected, producing long 3′ single-stranded DNA extensions. Brca2 is essential to load the Rad51 recombinase onto these 3′ overhangs. The resulting nucleofilament can thus invade a homologous DNA sequence to copy and restore the original genetic information. In Arabidopsis, the inactivation of Brca2 specifically during meiosis by an RNAi approach results in aberrant chromosome aggregates, chromosomal fragmentation and missegregation leading to a sterility phenotype. We had previously suggested that such chromosomal behaviour could be due to NHEJ. In this study, we show that knock-out plants affected in both BRCA2 genes show the same meiotic phenotype as the RNAi-inactivated plants. Moreover, it is demonstrated that during meiosis, neither NHEJ nor SSA compensate for HR deficiency in BRCA2-inactivated plants. The role of the plant-specific DNA Ligase6 is also excluded. The possible mechanism(s) involved in the formation of these aberrant chromosomal bridges in the absence of HR during meiosis are discussed.
Introduction
One of the most cytotoxic DNA damage is chromosomal breakage, where a DNA double-strand break (DSB) occurs in the duplex DNA. Failure to repair correctly even one DNA DSB can result in the loss of genetic information, chromosome rearrangement, mutations and lead eventually to cell death. In plants, as in other organisms, cells have developed powerful and rapid cellular responses, leading to cell cycle arrest and DNA DSB repair. In eukaryotes, DNA broken ends can be processed by three major DSB repair pathways that are tightly regulated, depending on cell type and cell cycle phase: Non-Homologous End Joining (NHEJ), Single-strand annealing (SSA), and Homologous Recombination (HR).
In the NHEJ pathway, DNA broken ends are simply joined with little or no further processing. In mammalian cells, the Ku70-Ku80 heterodimer forms a ternary complex with the DNA-PKcs, and binds to the DSB. The binding of this complex prevents excessive degradation and promotes the recruitment of other factors involved in the processing of DNA ends to make them suitable for the ultimate step of ligation by the LigaseIV-Xrcc4 complex [1], [2]. While no ortholog of DNA-PKcs has been found in Arabidopsis, AtKu70, AtKu80, AtLigIV and AtXrcc4 homologs have been identified [3], [4], [5], [6]. Mammalian null mutants affected in the NHEJ pathway present various orders of phenotype severity. For instance, ku mutants are immunodeficient and exhibit an accelerated senescence (in correlation with the deregulation of telomere length), while LigaseIV deficiency leads to embryonic lethality in mice. In Arabidopsis, all characterized nhej mutants are viable but hypersensitive to various DNA damaging agents, except UV [5]. The ku mutants are hypersensitive to menadione (which causes oxidative damage), ionising radiations (X- and gamma-rays) and bleomycin (a radiomimetic), methylmethanesulfonate (MMS, an alkylating agent causing abasic sites and single-strand nicks) [4], [5], [7], [8], [9], [10]. Hypersensitivity to MMS and gamma-irradiation has also been described for ligIV mutants [5], [11]. Direct evidence for their involvement in NHEJ comes from plasmid rejoining assays. In protoplasts derived from ku80 and ku70 mutant plants, the religation efficiency of plasmids linearized by enzymes generating blunt or 5′overhang ends was significantly reduced [9], [10].
The SSA and the HR pathways are homology-dependent processes for repairing DNA DSB. Both are initiated by the 5′ to 3′ resection of the broken DNA ends in order to uncover extensive single-stranded DNA (ssDNA) 3′ overhangs, a critical intermediate in both SSA and HR. These 3′ ssDNA tails are coated by the single-stranded DNA binding protein, RPA.
After DNA resection, the central step of SSA consists of the annealing between complementary single-stranded DNA sequences on either side of the DSB in a RAD52- and RAD59-dependent, but RAD51-independent, manner. Unpaired non-homologous 3′ tails are then cleaved by the Rad1-Rad10 complex (XPF-ERCC1 in mammals), which is also involved in DNA excision repair, in order to complete the DSB repair with DNA synthesis from the newly cleaved ends and their final ligation. In Arabidopsis, mutants affected in AtRAD1 (or UVH1) or AtRAD10 (also called AtERCC1) activities have been identified as gamma- and UV-hypersensitive [12], [13], [14]. In contrast to XPF- or ERCC1-deficient mice, the corresponding single mutant plants are viable in the absence of exogenous DNA damaging agents, grow normally and are fertile. Using a plasmid recombination assay, it was shown that each gene was required for the removal of 3′-ended non-homologous DNA single-stranded tails from SSA intermediates, generated by annealing between direct repeats [15], [16], [17], [18], [19].
In contrast to NHEJ and SSA that are inherently error prone, HR is conservative, as it proceeds via the copy of the missing sequence from a homologous template. Moreover, HR is required during meiosis for correct chromosome segregation and the generation of genetic diversity. Meiotic recombination is initiated by the introduction of programmed DNA DSB catalyzed by the topoisomerase-like transesterase activity of dimeric Spo11. This leads to a covalent link between the catalytic tyrosine of a Spo11 monomer and the 5′ DNA end on both sides of the DSB. In budding and fission yeast, removal of each Spo11 occurs by endonucleolytic cleavage several nucleotides downstream from the 5′ end, catalyzed by the Mre11-Rad50-Xrs2 complex and Sae2. This releases a Spo11 monomer bound to an oligonucleotide, sometimes called a “spolligo” [20], [21], [22], [23].
The repair process of HR in somatic and meiotic cells is initiated by extensive processing of DNA ends, uncovering 3′ ssDNA stretches that become coated by RPA. This resection is essential for the establishment of a recombinase-DNA nucleofilament on the 3′ single strand, which performs the homology search for a target DNA sequence to use as a template to copy, either the sister chromatid in somatic cells or a homologous chromosome in meiotic cells. Two recombinases can be loaded onto the ssDNA extension to mediate the strand displacement and homology search: the ubiquitous Rad51, the eukaryotic RecA homolog, and its homolog Dmc1 that has a specific role during meiosis. Once a homology is found, DSB repair is completed by DNA synthesis using the homologous sequence as a template and religation follows [24].
The displacement of RPA and its replacement by the recombinases rely on mediator proteins, such as the Rad51 paralogs, Rad52 and/or Brca2, which exist in most eukaryotes. In humans, BRCA2 gene mutations are associated with hereditary breast cancer [25], [26] and genome instability [27], [28]. In mice, the knockout of BRCA2 leads to early embryonic lethality associated with chromosomal rearrangements [29]. Structural and biochemical studies have shown the interaction between Rad51 and Brca2 [30], [31], [32]. Together with their co-localization in nuclear foci, after DNA damaging treatment of the cells, this definitively links Brca2 to homologous recombination [33].
Recently, the human Brca2 protein was purified [34], [35], [36]. It appears that one Brca2 molecule binds approximately six Rad51 monomers and that Brca2 stimulates the binding of Rad51 onto ssDNA even when it is covered by RPA. This interaction is mediated through the specific BRC domains which are present in all Brca2 proteins, but in varying numbers depending on species. For example, eight BRC domains are found in human Brca2 [31], whereas only one is present in Brh2 and Ce-BRC2, the Brca2 homologs of Ustilago maydis and Caenorhabditis elegans (C. elegans), respectively [37], [38]. In Arabidopsis, two AtBRCA2 genes have been identified: on chromosomes IV (AtBRCA2(IV), also named AtBRCA2a) and V (AtBRCA2(V) or AtBRCA2b). They encode two proteins of 1511 (AtBRCA2a) and 1155 amino acids (AtBRCA2b), which share 94.5% identity and contain four BRC motifs each. The two Arabidopsis genes are expressed in floral buds and the proteins they encode have been shown to interact with both Rad51 and Dmc1, the meiotic-specific recombinase [39], [40]. Recently, the Brca2-Dmc1 interaction has been confirmed in humans [41]. These data thus linked Brca2 to meiotic recombination for the first time.
The understanding of Brca2 function has been considerably hampered by the early embryonic lethality associated to knocking out BRCA2 in mouse. Clear evidence for the meiotic role of Brca2 came from A. thaliana and C. elegans since the absence of the Brca2 function is viable and only leads to sterility due to meiotic defects in both models [38], [39]. Indeed, RNAi-inactivation of both Arabidopsis BRCA2 genes, specifically during meiosis, caused sterile plants resulting from an improper meiosis with chromosomal aberrations: absence of bivalent formation, chromosomal entangling, bridges and fragmentation. This phenotype was dependent on the formation of meiotic DNA double-strand breaks as it was alleviated in a spo11 mutant [39]. We hypothesized that in A. thaliana the chromosomal abnormalities observed upon depletion of Brca2 at meiosis could be the result of an alternative repair of the meiotic DSB, in the absence of HR [39]. In C. elegans, Martin et al. (2005) showed that the RNAi depletion of LIGIV significantly reduced meiotic chromosome aggregation in Cebrc-2 single mutants and could give rise to chromosomal fragmentation. These observations suggested that NHEJ could be partially responsible for the aberrant chromosome fusions in the absence of CeBRC-2.
In this study, the meiotic defects previously observed in Brca2-inactivated plants were confirmed in brca2 double mutant plants containing a T-DNA insertion in each AtBRCA2 gene. The potential role of alternative DNA repair pathways in the meiotic phenotype was tested by inactivating Brca2 in nhej and/or ssa mutant backgrounds. We demonstrate that neither NHEJ nor SSA were responsible for the observed cytological defects. Moreover, based on the hypothesis that covalent repair is responsible for the observed meiotic chromosomal defects in the absence of Brca2, we tested the role of a recently characterized plant-specific DNA ligase, AtLigase6. Since the abnormal meiotic figures were maintained in lig6 plants inactivated for Brca2, the role of this DNA ligase during meiosis in the absence of HR was excluded.
Results
A brca2 double mutant exhibits the same meiotic phenotype as Brca2-inactivated plants
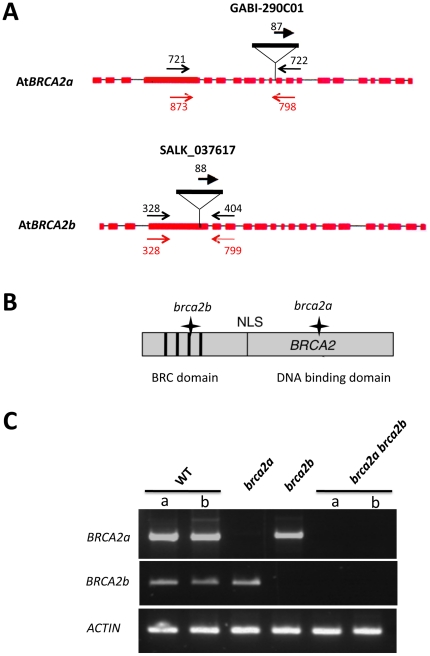
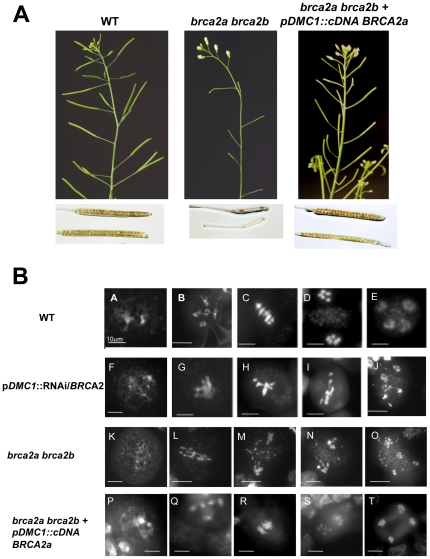
In a previous study, AtBRCA2a and AtBRCA2b expression was inactivated during meiosis by RNAi using an inverted 510 pb-fragment of the BRCA2 cDNA under the control of the meiotic-specific promoter of DMC1 (pDMC1) [39]. In this work, single and double T-DNA insertion mutants for AtBRCA2 were isolated and their phenotype compared to the RNAi-inactivated plants (named pDMC1::RNAi/BRCA2 ). First, brca2 plants mutated in the AtBRCA2 genes via either a T-DNA insertion located in the 10th intron of AtBRCA2a (in the Cter DNA binding domain) or an insertion in the 4th exon of AtBRCA2b (in the Nter domain of the protein, containing the BRC motifs) were isolated (Figure 1A and Figure 1B). AtBRCA2 transcripts were analysed by RT-PCR, using primers flanking the insertion sites in wild-type and in brca2 single mutant plants. Transcripts of the disrupted genes were not detected in the corresponding mutant lines, whereas transcripts of each AtBRCA2 gene were amplified in wild-type plants. This strongly suggested that the two single brca2 lines were null mutants (Figure 1C). Each single mutant showed normal development and fertility. By crossing the single mutants, the double brca2a brca2b mutant was obtained. These latter plants showed no growth defect and behaved as the wild-type under normal greenhouse conditions. However, they were partially sterile producing very short and mostly empty siliques (Figure 2A). Moreover, the presence of meiotic defects was observed after DAPI staining of the chromosomes in the meiocytes. Indeed, all meiotic figures showed chromosomal entangling without bivalent formation, bridges and fragmentation, leading to chromosomal missegregation (Figure 2B) as previously described for pDMC1::RNAi/BRCA2 plants. A transgene containing a full length AtBRCA2a cDNA under the control of the promoter of the meiotic recombinase Dmc1 (pDMC1::cDNA AtBRCA2a) was introduced in 13 brac2a brca2b double mutant plants. 11 transformant plants presented a restored phenotype: 9 were completely fertile as demonstrated by the observation of wild-type siliques content and normal meiosis (Figure 2) and 2 were partially fertile (as they presented some siliques that developed as sterile). Only 1 transformant was sterile with developmental defects. As a control, 11 brca2a brca2b double mutant plants were transformed with a transgene containing the pDMC1::RNAi/0 construct, corresponding to the “empty vector” [39]: all of them were sterile (data not shown). These results reinforce the evidence for the role of AtBRCA2 at meiosis, previously uncovered by our RNAi strategy.
Figure 1. The brca2 single and double mutants.
(A) Position of the T-DNA insertions in AtBRCA2a and AtBRCA2b. The structure of the AtBRCA2a and AtBRCA2b genes is represented by shaded boxes (exons) and thin lines (introns). The T-DNA insertion position is indicated. Each primer pair used to identify the mutants by PCR are compiled on the diagram in black and primer pairs used for RT-PCR analyses are given in red; their localization is correct but not to scale. (B) Schematically represented Brca2 protein with the position of the BRC repeats and the NLS relative to the T-DNA insertions, as indicated by a star. For convenience, and because they share 94.5% of identity, a single Brca2 protein is represented. (C) RT-PCR analysis of AtBRCA2 transcripts in the single and double brca2 mutants. RNA was extracted from young floral buds of wild-type plants (2 different plants, a and b) as well as of brca2a, brca2b and brca2a brca2b (2 different plants, a and b) mutant plants and was then reverse-transcribed. Double-stranded cDNAs were then PCR-amplified using the primer pairs represented in red in Figure 1A. The constitutive ACTIN gene transcript was used as a control.
Figure 2. Meiotic defects in brca2a brca2b mutant plants and in wild-type Brca2-inactivated plants.
(A) Wild-type and brca2 double mutant plants exhibt no growth defect except for sterility. Chloralhydrate discolored siliques are full of seeds in wild-type plants in comparison with the discolored siliques of the brca2 double mutant plants. (B) Observation of meiocytes by DAPI staining in Brca2-deficient plants, transformed or not with the full length cDNA of AtBRCA2a, and in brca2a brca2b homozygous double mutant plants. (A–E) Different stages of meiosis in the wild-type plants. Meiosis is normal. (A) Prophase I stage, (B) diakinesis, the five bivalents are attached by a chiasma, (C) metaphase I with five aligned bivalents, (D) anaphase I, bivalents segregate into two sets of five univalents, (E) anaphase II, with four groups that contain five chromosomes each after sister chromatid separation. (F–J) Different stages of meiosis in wild-type plants transformed with the pDMC1::RNAi/BRCA2 construct. (F) Prophase I, (G) no normal diakinesis phase (H) metaphase I with condensed and entangled chromosomes, (I) anaphase I, with entangled and stretched chromosomes. (J) Anaphase II, with bridges extending between chromosomes. (K–O) Different stages of meiosis in brca2 double mutant plants. (K) Prophase I, (L) anaphase I, entangled and stretched chromosomes. (M) Metaphase II with entangled chromosomes. (N) anaphase II, fragmentated chromosomes. (O) telophase II with chromosome missegregation. (P–T) Different stages of meiosis in brca2 double mutant plants, transformed with the pDMC1::cDNA AtBRCA2a. Meiosis is restored to normal. (P) Prophase I stage, (Q) diakinesis, (R) metaphase I, (S) anaphase I, (T) anaphase II. Bar 10 µm.
Characterization of nhej and ssa mutant plants by RT-PCR and under various genotoxic stress
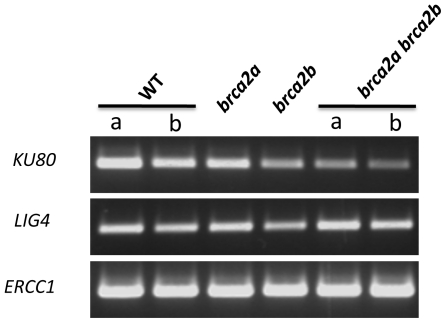
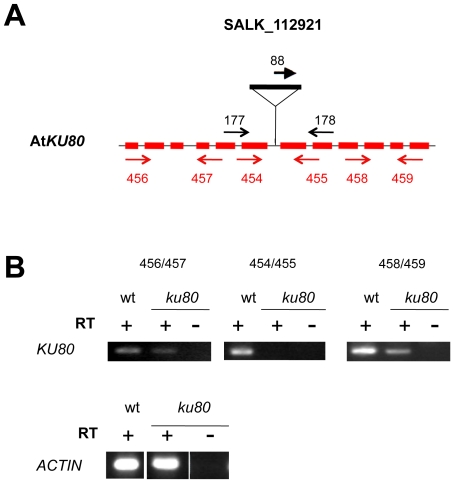
In order to identify the molecular pathways involved in the aberrant cytological phenotype observed in the Brca2-deficient plants during meiosis, mutant plants deficient in either the NHEJ (ku80-/- and ligIV-/-) or the SSA (ercc1-/-) pathways were characterized. Examining amplification of these transcripts specifically in meiocytes was not possible, as meiocytes would have to be specifically dissected which is technically difficult. However, as shown in Figure 3, all these three genes, and thus the pathways they are involved in, were found expressed in young flower buds, where meiosis takes place, in single as well as in double brca2 mutant plants. Two mutant lines have been previously described: SALK_044027, where the T-DNA insertion is in exon 6 of the AtLIGIV gene [42], [43] and SALK_033397 which contains a T-DNA insertion in exon 3 of AtERCC1 [16]. The absence of transcripts corresponding to the affected gene was confirmed for each mutant line by RT-PCR using primers flanking each T-DNA insertion (data not shown). The ku mutant line used in this study (SALK_112921) had not been characterized to date. It contains a T-DNA in the 6th intron of the AtKU80 gene (Figure 4A). RT-PCR analysis of the 5′ and 3′ regions flanking the T-DNA insertion revealed the presence of AtKU80 transcripts in both wild-type and ku80 mutant plants (Figure 4B). However, no transcripts could be detected in ku80 mutant plants when primers flanking the T-DNA insertion were used, suggesting that splicing of the 6th intron did not occur in the ku80 mutant. As the insertion site is positioned in the region encoding the domain involved in hetero-dimerization with Ku70, it is most likely that a putative protein, lacking this domain, would be non-functional. Thus, these ku80 plants were considered as functional null mutants. The mutant plants, whatever the affected DNA repair pathway, exhibited no obvious developmental defects under normal growth conditions and were fertile, as previously described for ercc1, ku80 and ligIV Arabidopsis mutants [9], [13], [16].
Figure 3. RT-PCR analysis of genes involved in NHEJ and SSA in the single and double brca2 mutants.
RNA was extracted from young floral buds and reverse-transcribed, as described in Figure 1C. Double-stranded cDNAs were PCR-amplified using primer pair 454/455 for AtKU80 (see primer positions in Figure 4 and sequences in Table1), 336/445 for AtLIGIV and 452/453 for AtERCC1 (see Table 1 for sequences). The constitutive ACTIN gene transcript used as a control is presented in Figure 1C.
Figure 4. T-DNA insertion and expression in ku80 mutant.
(A) Position of the T-DNA insertion in AtKU80. The structure of the AtKU80 gene is represented by shaded boxes (exons) and thin lines (introns). The T-DNA insertion position is indicated. Each primer pair used to characterize the mutant by PCR are indicated in black and primer pairs used for RT-PCR analyses are given in red; their localization is correct but not to scale. (B) RT-PCR analysis of AtKU80 transcripts in ku80-/- mutant plants. RNA, extracted from floral buds of wild-type or ku mutant plants was reverse-transcribed. Double-stranded cDNAs were amplified by RT-PCR, performed with three different primer pairs: 5′ or 3′ to the T-DNA and flanking the T-DNA insertion. For primer positions, see above (Figure 4A). The constitutive ACTIN gene was used as a control.
We believed that in the absence of HR during meiosis, the different DNA DSB repair pathways could compensate for each other. Thus, nhej mutant plants, ku80 and ligIV, were crossed with ssa mutant plants, ercc1, and double ku80 ercc1 and ligIV ercc1 mutants affected in both pathways were isolated and genotyped. Both double mutants were viable, presented no obvious developmental defects under normal growth conditions and were fertile.
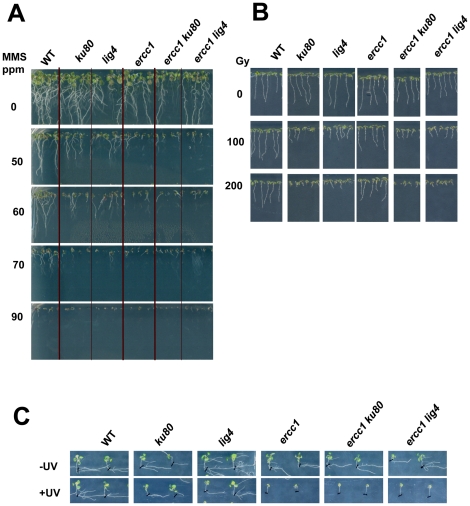
Sensitivity to various DNA damaging agents is a classical assay to characterize DNA repair mutant plants as most of them show no obvious somatic phenotype. To control that our mutants were indeed affected in DNA repair, their sensitivity to MMS, gamma-ray and UV irradiation was assayed. In comparison to wild-type plants, root growth was affected in the nhej plants as well as in the ssa plants in the presence of MMS or after gamma exposure. Indeed, the MMS hypersensitivity was visible at 50 ppm and gamma–ray hypersensitivity was observed at 100 grays for each single mutant line. However, MMS-induced retarded growth was more pronounced in ercc1 than in ku80 and ligIV plants (Figure 5A). MMS is a methylating agent, and due to the occurrence and clustering of modified bases, it can generate both SSB and DSB, which is reflected in the fact that ercc1 mutants (deficient for both SSA and BER) appeared to be more sensitive to this genotoxic treatment. Reciprocally, ercc1 plants were less sensitive to gamma irradiation when compared to ku80 and ligIV (Figure 5B). Ionising radiations mainly give rise to clustered DNA damages (modified bases and abasic sites) that lead to DNA DSB. Such DNA strand breaks are mostly repaired by NHEJ as suggested by the higher hypersensitivity of ku80 and ligIV mutants to gamma-rays. Finally, as expected, only the ercc1 plants were hypersensitive to UV exposure (Figure 5C). All of these results confirmed that the different mutant Arabidopsis lines were affected in DNA DSB repair.
Figure 5. Hypersentivity to MMS, gamma-rays and UV irradiation of nhej, ssa and nhej ssa plants.
Before sowing, all seeds were surface-sterilized. (A) MMS hypersensitivity, 11 days post-germination. Seeds were sown on MS 0.5 agar 1% sucrose supplemented with MMS at various doses. (B) Gamma-irradiation hypersentivity, 7 days post-irradiation. After 48 h at 4°C in darkness, seeds were exposed to various doses of gamma-rays : 0, 100 and 200 grays before being sown on MS 0.5 agar. (C) UV hypersensitivity, 10 days post-irradiation. Seeds were sown in MS 0.5 agar. After 4 days of growth, the plantlets were exposed to UV-C, left in the dark for 3 days to avoid photoreactivation, and then exposed to light.
MMS and gamma-ray sensitivity of the double nhej ssa mutants were assessed in comparison to the single mutant plants (Figure 5). For each stress, we noted that the sensitivity of the double mutant was similar to that observed for the most affected single mutant: ku80 ercc1 and ligIV ercc1 appeared to be hypersensitive to MMS and UV as was the ercc1 single mutant, whereas they showed a similar hypersensitivity to gamma-rays as the nhej single mutant. Therefore, no cumulative effect was observed.
The brca2 meiotic phenotype is maintained during meiosis in nhej and ssa backgrounds
The Brca2 function was inactivated in the nhej and ssa mutant plants by transforming the mutant plants with the previously used pDMC1::RNAi/BRCA2 construct. As a control, mutant plants were also transformed with a pDMC1::RNAi/0 construct containing no insert [39]. No somatic phenotype was observed in any of the transformed plants containing the “empty” construct or the pDMC1::RNAi/BRCA2 construct. When flowers emerged, all plants containing the control construct were fertile, whereas most of the mutant plants transformed with pDMC1::RNAi/BRCA2 were partially sterile in the single nhej or ssa mutants (between 67 to 80%) as well as in the double nhej ssa mutants (between 60 to 78%), as previously observed for wild-type pDMC1::RNAi/BRCA2 transformed plants.
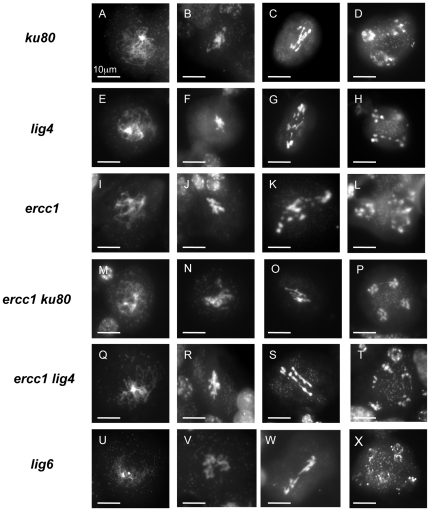
The meiotic behaviour was examined after DAPI staining of the chromosomes in the meiocytes of several independent transformed plants that were inactivated for the Brca2 function: 175 meiotic figures from two ku80, 170 meioses from two ligIV and 34 meioses from two ercc1 lines independently transformed with the pDMC1::RNAi/BRCA2 construct were observed. As a control, they were compared to the meioses of one ku80, one ligIV and one ercc1 plant containing the RNAi/0 construct. All of the observed control plant meiotic figures were normal in the single mutants affected for either NHEJ (ku80, ligIV) or SSA (ercc1), as well as in the nhej ssa double mutant (Supplementary Figure S1). On the other hand, meiosis was profoundly disturbed in meiocytes of these same mutant lines transformed with the pDMC1::RNAi/BRCA2 construct: chromosomal entangling without bivalent formation, fragmentation, and missegregation of chromosomes (Figure 6). Such observations have been previously reported in wild-type pDMC1::RNAi/BRCA2 plants [39]. These observations suggested that, contrary to our hypothesis, in the absence of Brca2 during meiosis, neither NHEJ nor SSA were responsible for an alternative meiotic DSB repair that would have been revealed because of the absence of HR [39]. The impact of the inactivation of both pathways in the absence of Brca2 during meiosis was also examined. Meiotic figures from one pDMC1::RNAi/BRCA2 transformant for ercc1 ku80 (257 meiotic events, among them 80 were post-prophase) and two pDMC1::RNAi/BRCA2 transformants for ercc1 lig4 (115 meiosis, including 63 post-prophase stages) were observed. In all double mutant plants transformed with pDMC1::RNAi/BRCA2, the brca2 meiotic phenotype remained unaltered (Figure 6).
Figure 6. Observation of meiocytes by DAPI staining in nhej, ssa, nhej ssa and lig6 mutant plants transformed with the pDMC1::RNAi/BRCA2 construct.
Different stages of meiosis were observed in plants transformed with pDMC1::RNAi/BRCA2 in nhej mutant plants, ku80 (A–D) or lig4 (E–H), and in ssa mutant plants, ercc1 (I–L), in nhej ssa double mutant plants, ercc1 ku80 (M–P) or ercc1 lig4 (Q–T) and in lig6 mutant plants (U–X). (A, E, I, M, Q, U) prophase I. (B, F, J, N, R, V) metaphase I. (C, G, K, O, S, W) anaphase I. (D, H, L, P, T, X) anaphase II. Bar 10 µm.
All of these results suggest that 1) the aberrant chromosomal figures observed in the absence of Brca2 during meiosis are not due to NHEJ or SSA and 2) the other major DNA DSB repair pathways, in the absence of HR, do not compensate for each other during meiosis.
DNA Ligases in Arabidopsis
Our initial hypothesis was that the chromosomal bridges detected in the “failed” anaphases in the absence of Brca2 were due to covalent DNA links, probably between non-homologous chromosomes. Since our data exclude the role of NHEJ and SSA, all DNA ligases apart from LigaseIV (the NHEJ specific enzyme already studied in this work) could be potentially incriminated. The Arabidopsis genome contains three other sequences encoding DNA ligases: AtLigase1 which is involved in replication and Base Excision Repair (BER), AtLigase1a which shares 71% identity with AtLigase1 but for which no transcripts could be detected (our personal data and transcriptome analyses: http://csbdb.mpimp-golm.mpg.de/csbdb/dbxp/ath/ath_xpmgq.html), suggesting that it may be a pseudogene, and AtLigase6, a plant specific ligase that appears to be involved in seed longevity [44]. AtLigase6 has a highly conserved DNA ligase catalytic domain and a beta-lactamase domain containing a beta-CASP motif found in Artemis and other proteins known to play a role in nucleic acid processing [45], [46]}. Since lig1 mutant plants are embryonic lethal [47], [48], we thus examined whether the plant specific AtLigase6 could be involved in the meiotic phenotype of the Brca2-deficient plants.
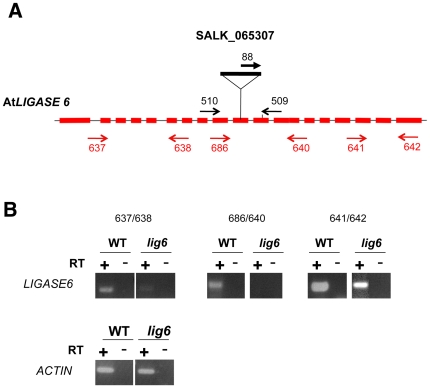
Homozygous lig6 plants containing a T-DNA insertion in exon 11 of the gene were obtained from the SALK collection (SALK_065307) (Figure 7A). All plants grew normally, they were fertile and undertook normal meioses (data not shown). These observations are in agreement with what was previously observed in a different lig6 insertional line (Waterworth et al, 2010) [44]. RT-PCR analyses detected transcripts on both sides of the T-DNA insertion but no transcripts could be found when primers flanking the T-DNA insertion were used (Figure 7B). As the T-DNA insertion is positioned in an exon, 42 bp from the codon of the catalytic lysine just upstream from the conserved motif II [49] lying in the core domain, and more specifically around the nucleotide binding pocket responsible for the nucleotidyl transfer, the catalytic activity of a putatively expressed protein in this mutant is probably non-functional.
Figure 7. T-DNA insertion and expression in lig6 mutant.
(A) Position of the T-DNA insertion in AtLIG6. The structure of the AtLIG6 gene is represented by shaded boxes (exons) and thin lines (introns). The T-DNA insertion position is indicated. Each primer pair used to identify the mutants by PCR are indicated in black while primer pairs used for RT-PCR analyses are given in red; their localization is correct but not to of scale. (B) RT-PCR analysis of AtLIG6 transcripts in lig6-/- mutant plants. RNA, extracted from floral buds of wild-type or lig6 mutant plants was reverse-transcribed. Double-stranded cDNAs were amplified by RT-PCR, performed with three different primer pairs: 5′ or 3′ to the T-DNAand flanking the T-DNA. The position of each primer is given above (Figure 7A). The constitutive ACTIN gene was used as a control.
DNA Ligase6 is not responsible for the brca2 meiotic phenotype
Waterworth et al. (2010) observed a slight but significant growth hypersensitivity of lig6 plants after a 100 gy X-ray irradiation, leading them to suggest that AtLigase6 could play a minor role in the repair of X-ray induced DNA damage. Transformation of our lig6 mutant plants with pDMC1::RNAi/BRCA2 was performed to inactivate the Brca2 function in lig6 plants. 87% of the transformants (26/30) were partially sterile while lig6 plants or pDMC1::RNAi/0 transformed plants (six transformed plants, 57 meioses observed from two independent transformants) were normally fertile (Supplementary Figure S1). After DAPI staining of the chromosomes in the meiocytes of seven lig6 plants, independently transformed with the pDMC1::RNAi/BRCA2 construct (426 meiotic figures, including 219 post-prophase events), the brca2 meiotic phenotype was consistently observed (Figure 6), thus excluding a role of AtLigase6 in this phenotype.
Discussion
In Arabidopsis, RNAi-inactivation of Brca2 during meiosis gave rise to a sterility phenotype due to an aberrant meiosis characterized by an absence of bivalent formation, chromosomal entangling, fragmentation and missegregation. Such defects were Spo11-dependant, therefore an alternative DNA repair process was proposed to be responsible for an aberrant repair of meiotic DSB in the absence of HR. In this study, we show that brca2 double mutant plants exhibit a similar meiotic phenotype when compared to the pDMC1::RNAi/BRCA2 transformed plants. Moreover, our data clearly exclude the role of NHEJ and SSA in the aberrant meiotic chromosomal figures of Brca2-deficient plants.
Phenotypic characteristics of Brca2-deficient plants
In this study, double brca2a brca2b mutant plants were shown to have no obvious phenotype in terms of vegetative growth, contrary to the occasional fasciation described by Abe et al. (2009). This may be explained by the use of different ecotypes. However, the double brca2 mutant displayed the same meiotic phenotype as previously described for pDMC1::RNAi/BRCA2 transformed plants. Each single mutant was fertile, indicating the functional redundancy of the two AtBRCA2 genes at meiosis [50], [51]. This could not have been concluded from the pDMC1::RNAi/BRCA2 transformed plants, as both AtBRCA2 genes were silenced by the RNAi construct. Previously it was found that pDMC1::RNAi/BRCA2 transformed plants produced a few seeds that could have arisen from a partial silencing of the AtBRCA2 genes. However, this does not appear to be the reason since in the present study, a few seeds were also produced by the double mutant plants (Figure 2A). Preliminary experiments showed that the seeds germinated, producing brca2 double mutant plants that developed normally, although not fertile. This could mean that in the absence of Brca2, HR could be partially functional and give rise to some rare normal meiosis events that were not detected in our observations. Alternatively, the abnormal meiosis we observed may not be always detrimental to the chromosomes. It will be of interest to follow the brca2 cumulative phenotypes from generation to generation, to check if their meiotic (and somatic) phenotypes become exacerbated.
NHEJ and SSA do not compensate for HR deficiency during meiosis
Our analyses of the chromosomes in the meiocytes by DAPI staining in the double brca2a brca2b mutants revealed that the depletion of Brca2 during meiosis led to the absence of bivalent formation and to chromosome aggregates, thus confirming our previous study of plants transformed with the pDMC1::RNAi/BRCA2 construct. In anaphase I, aberrant bridges between chromosomes were systematically observed. We hypothesized that these defects were due to covalent repair of meiotic DBS. NHEJ was the main candidate pathway we believed responsible for these aberrant chromosomal figures. FISH experiments would not have proven that the chromosomes involved in these anaphase bridges were covalently linked, just that they were occasionally aberrantly “associated”. Thus, the Brca2 function was inactivated in NHEJ- but also in SSA-deficient plants. In contrast to C. elegans, a role of NHEJ in these meiotic defects can now be excluded, since these aberrant chromosome aggregates were still present in the meiocytes of plants defective in NHEJ [38]. A similar conclusion can be drawn in the case of the SSA-deficient plants. Furthermore, the additive disruption of both the NHEJ and SSA pathways did not modify the brca2 meiotic phenotype. This demonstrates that, in contrast to somatic cells where deletions and translocations can occur in mutants defective in HR due to the error-prone repair of accidental DNA DSB via NHEJ or SSA, neither of these two major DNA DSB repair pathways can compensate for the absence of HR during meiosis in Arabidopsis. During meiosis, as the introduction of DNA DSB is programmed, inhibition of the NHEJ and SSA pathways must be very strong to prevent them to compete for or to replace HR. More generally, it is conceivable that the DNA repair processes that are initiated by programmed DNA DSB must be very carefully controlled. If neither NHEJ nor SSA are responsible for the meiotic defects of Brca2 deficient plants, we cannot exclude the role of alternative DNA DSB repair pathways, such as the backup end-joining pathway involving Xrcc1[52]. Hence, further studies should be addressed to analyse meiosis in triple mutants, deficient for all three pathways, in Brca2-inactivated plants. However, recent data suggest that DNA DSB are still repaired in somatic cells of irradiated plants, defective for HR, SSA, NHEJ and backup end-joining, suggesting that other DNA DSB repair process probably remain to be discovered [53].
Covalent repair or not ?
To better understand the molecular mechanisms responsible for the chromosomal bridges observed during meiotic anaphases in the absence of Brca2, we investigated the putative role of DNA ligases. Four potential DNA ligases have been identified in Arabidopsis: the essential AtLIGASE1 involved in DNA replication and BER, AtLIGASE1a, the NHEJ-specific AtLIGASEIV, and the plant-specific AtLIGASE6. As AtLIGASE1a seems to be a pseudogene, it was excluded from our study. Our results show that the NHEJ specific DNA LigaseIV and the plant-specific Ligase6 were not involved in the meiotic chromosomal defects resulting from the absence of Brca2. Thus, a putative role of a plant DNA ligase activity remains an open question. It is difficult to study the role of DNA Ligase1 as it is essential to DNA replication driving homozygous lig1 mutant plants to be embryo-lethal [47], [48]. In order to by-pass the lethality of the lig1 mutant, Waterworth et al (2009) reduced the expression of AtLIGASE1 using an RNAi construct that was set under the control of the ubiquitous CaMV35S promoter [47]. The partially inactivated plants exhibited precocious flowering but as their growth and development were strongly affected, it was difficult to describe them as clearly fertile. Hence, it would be interesting to undertake a meiosis-specific inhibition of AtLIGASE1 expression using the pDMC1 meiotic promoter, as previously carried out for Brca2. Otherwise, another possibility to consider is that the chromosomes are not covalently linked and that other proteins involved in chromatid cohesion and synapsis could help maintain the aberrant chromosome associations observed in the absence of Brca2.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana Colombia ecotype were used in this study. Mutant lines were identified in the T-DNA express database of the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu). The insertional mutant affected in AtERCC1 (line SALK_033397) and in AtLIGIV (line SALK_044027) have been described previously [16], [42], [43] as well as the AtBRCA2b insertion line (SALK_ 037617) [50]. The newly characterized mutant lines were GABI_290C01 for AtBRCA2a, SALK_112921 for AtKU80 and SALK_065307 for AtLIGASE6. Wild-type and mutant Arabidopsis plants were cultivated in a greenhouse at 23°C under long day conditions (16 h light, 8 h dark, humidity 75%).
Isolation of genomic DNA and genotyping of the plants
Plants were genotyped by PCR performed on genomic DNA extracted from leaves of 2–3 week-old plants in Edwards' buffer [54]. 1/50 of the extracted DNA was used as a template for PCR with two gene-specific primers and one primer specific for the left border of the T-DNA (Salk or Gabi-Kat, depending on the mutant lines) in separate reactions (see Table 1 and Figures 1, 4 and 7). The wild-type allele was amplified with oligonucleotides 721/722 for the AtBRCA2a locus, 328/404 for AtBRCA2b, 177/178 for AtKU80, and 510/509 for AtLIGASE6. The mutant allele was detected using primer 88 (LBa1) for SALK T-DNA lines or 87 (08409) for Gabi-Kat T-DNA lines and primer 772 for the AtBRCA2a locus, 404 for AtBRCA2b, 178 for AtKU80, or 509 for AtLIGASE6. PCR reactions were performed in a 20 µl final volume, with 0.2 mM of dNTPs, 5 mM MgCl2, 1 µM of each primer, 1U of Taq DNA polymerase (Invitrogen™). They were incubated in a 2720 Thermocycler (Applied Biosystem) at 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 50°C for 30 s and 72°C for 1 min, except for the PCR on the AtBRCA2 genes where the annealing step was performed at 52°C for 30 s. The PCR samples were then visualized after migration on 0.7% agarose gels in the presence of ethidium bromide.
Table 1. Sequence and use of primers in this study.
| Name | Gene | DNA sequence (5′-3′) | Use |
| 87 | Gabi T-DNA o8409 | ATATTGACCATCATACTCATTGC | genotyping |
| 88 | Salk T-DNA LBa1 | TGGTTCACGTAGTGGGCCATCG | genotyping |
| 721 | AtBRCA2a (At4g00020/10) | GATTGTGCTCTGAATGCTAC | genotyping |
| 722 | AtBRCA2a (At4g00020/10) | CAATTTCTTTACCTTGAGGA | genotyping |
| 873 | AtBRCA2a (At4g00020/10) | ATGAGACCGATTGTGCTCTGAATGC | RT-PCR |
| 798 | AtBRCA2a (At4g00020/10) | CCAATTTCTTTACAGAAGCCTAGTCG | RT-PCR |
| 328 | AtBRCA2b (At5g01630) | GCTCTGAATATCAGTAAACCTGCT | genotyping and RT-PCR |
| 404 | AtBRCA2b (At5g01630) | TGTATCACACGATACAACAGACA | genotyping |
| 799 | AtBRCA2b (At5g01630) | TACAACAGACAAACCACTTGAAGCTTGCT | RT-PCR |
| 177 | AtKU80 ((At1g48050) | TGTCTTTTGCTTGTTGTGCAG | genotyping |
| 178 | AtKU80 ((At1g48050) | GCAGAAGGTGCAAGGTCAAG | genotyping |
| 456 | AtKU80 ((At1g48050) | ATGGCACGAAATCGGGAGGGTTTG | RT-PCR |
| 457 | AtKU80 ((At1g48050) | ACGATCAAGAAAGTCTCCAGCTAC | RT-PCR |
| 454 | AtKU80 ((At1g48050) | GAAGATTAAGGTGTGGGTTTATAAG | RT-PCR |
| 455 | AtKU80 ((At1g48050) | GTAAAACGAATCAGGAGTATCATCTC | RT-PCR |
| 458 | AtKU80 ((At1g48050) | CAAGGAGAATCCAAAGTTGAAGAAGG | RT-PCR |
| 459 | AtKU80 ((At1g48050) | CGTCTACTATATCACTGTCCGCTG | RT-PCR |
| 510 | AtLIGASE6 (At1g66730) | TCATTGCAGAATTGCTAAGGG | genotyping |
| 509 | AtLIGASE6 (At1g66730) | GAAGACGCAGACTTCAACCTG | genotyping |
| 637 | AtLIGASE6 (At1g66730) | AGAGCACGCTTGTTGGAGGG | RT-PCR |
| 638 | AtLIGASE6 (At1g66730) | TAAATTACGGGCCAATGTTCTAACAAG | RT-PCR |
| 686 | AtLIGASE6 (At1g66730) | GAGGGTGTTTCTGCTGCAGTAGTTGAGGCTTACAA | RT-PCR |
| 640 | AtLIGASE6 (At1g66730) | AAGAGCCAACAGCTGTTCTCCA | RT-PCR |
| 641 | AtLIGASE6 (At1g66730) | TTCATGGCTCAAGGTTAAGCGAGAT | RT-PCR |
| 642 | AtLIGASE6 (At1g66730) | GTTTGAGCATGAAACATCTCTGCGA | RT-PCR |
| Act-464 | AtACT2 (At3g18780) | TGAGACCTTTAACTCTCCCG | RT-PCR |
| Act-465 | AtACT2 (At3g18780) | GATGGCATGAGGAAGAGAGA | RT-PCR |
| 336 | AtLIGIV (At5g57160) | TTGCTGCTGAGGTATTGCAACGTAGAC | RT-PCR |
| 445 | AtLIGIV (At5g57160) | CCATCAAGGATACACTTGTCCACCAAT | RT-PCR |
| 452 | AtERCC1 (At3g05210) | CCCACAGTTCAAGCCAAACGCATC | RT-PCR |
| 453 | AtERCC1 (At3g05210) | ACATTCTGTCATGCTCCAGGCAC | RT-PCR |
RNA isolation and RT-PCR analyses
Total RNA was extracted from leaves or floral buds of 2–3 week-old individual plants with the NucleoSpin® RNA Plant (Macherey-Nagel) according to the manufacturers' specifications. 2 µg of total RNA was used as a template for reverse-transcription with the RT ImProm II™ (Promega) and oligod(T) as a primer. 1/20 of the RT reactions were used as a template for PCR in a total volume of 50 µl. The quality of the RT reaction was controlled by examining actin expression by PCR using primers act-464 and act-465. For AtBRCA2a and AtBRCA2b cDNAs, specific primers flanking the T-DNA insertion were designed: 873/798 and 328/799 respectively (see Table 1 and Figure 1). For AtKU80, specific primers were designed 5′ to the insertion (456/457), flanking the T-DNA insertion (454/455) and 3′ to the T-DNA (458/459) (see Table 1 and Figure 4). For AtLIGIV and AtERCC1 cDNAs, specific oligonucleotides were designed flanking their respective T-DNA insertion site: the 336/445 pair for AtLIGIV and the 452/453 pair for AtERCC1 (see Table 1 for sequences). For AtLIGASE6, specific primers were designed 5′ to the insertion (637/638), flanking the T-DNA insertion (686/640) and 3′ to the T-DNA (641/642) (see Table 1 and Figure 7). The PCR was as follows: 94°C for 3 min, followed by 35 cycles at 94°C for 30 s, 50°C or 52°C for 30 s and 72°C for 1 min, except for the ACTIN gene (see Table1 for sequences of the ACT2 primers) where the elongation step was performed at 58°C for 30 s. 20 µl of the RT-PCR reaction were then loaded onto a 3% agarose gel (NuSieve) in the presence of ethidium bromide for visualization.
pDMC1:: cDNA AtBRCA2a construct
The full length cDNA of AtBRCA2a was previously cloned in pUC18 as described in [39]. It was subsequently subcloned first into pKannibal [55] and then into the XhoI–SpeI-restricted pPF408 to be set under the pDMC1 promoter control [39].
In vitro assays for sensitivity to MMS, gamma-rays and UV
Seeds were surface-sterilized with a solution containing 50% bleach diluted in EtOH. Sterilized seeds were sown on MS 0.5 agar media (Kalys) containing 1% sucrose and supplemented with 0, 50, 60, 70, 80 or 90 ppm of MMS and then set at 4°C in the dark for 48 h to synchronize germination, before being placed vertically in the growth chamber for 14 days to allow the roots to grow along the agar surface. For irradiation experiments, sterilized seeds stored at 4°C in the dark were exposed to 0, 100 or 200 Grays from a 137Cs source at a dose rate of approximately 50 gy.min−1 (IBL-637 (CIS-BioInternational), Institut Curie, Orsay). After irradiation, they were sown on MS 0.5 agar media and set vertically in a growth chamber. After 11 days, root growth was observed. For UV experiments, sterilized seeds were sown on MS 0.5 agar and after 4 days of vertical growth, the plantlets were exposed to 540 J.m−2 of UV-C (254 nm). The plates were then 90 degrees rotated, set in the dark for 3 days to avoid photoreactivation and then exposed 3 days to light to observe the recovery of main root growth of each seedling during two weeks.
Plant crosses
Since all single mutants used in this study were fertile, double mutants were obtained by crossing two homozygous mutants affected in the gene of interest. Double mutants were identified by PCR of the F2 population obtained by self-fertilisation of F1 plants heterozygous for both genes.
Transformation of plants with RNAi constructs
The RNAi constructs aimed at silencing both BRCA2 genes and the control without any insert were previously described in [39]. Plant transformations were carried out by floral dip as described previously [56]. T1 transformants were selected on sand supplemented with Basta® and transferred to soil pots. Approximately one to two weeks after, the selected transformed plants were sprayed with Basta® (4% ammonium glufosinate) for a second control of their resistance.
DAPI staining and cytology
The flower buds or the siliques were fixed in a solution of absolute ethanol and acetic acid (3/1 v/v) at room temperature. Chromosome spreads were prepared as in [57]. Photographs were captured using a Photometrics CoolSNAP EZ camera driven by Metavue 7.0 r4 software.
Fixed siliques were placed in ethanol 70% during 2 h, and then in a chloralhydrate solution (8 g/3 ml glycerol 66%) during a night in the dark. Images were captured on a Zeiss stereo-microscope Stemi SV1 with a SONY camera driven by Zeiss Axiovision Software.
All images were further processed with Adobe Photoshop CS2.
Supporting Information
Observation of meiocytes by DAPI staining in nhej , ssa , nhej ssa and lig6 mutant plants transformed with the RNAi/0 control construct. Normal meiotic progression in plants transformed with pDMC1::RNAi/0 in nhej mutant plants, ku80 (A–B) and lig4 (C–D), in the ssa mutant ercc1 (E–F), in double nhej ssa mutants ku80 ercc1 (G–H) and lig4 ercc1 (I–J), and in lig6 mutant plants (K–L). Bivalents were correctly associated during the first meoitic phase (diakinesis (A–E–G–K) and metaphaseI (C, I). Segregation of homologous chromosomes and during the second division, sister chromatid separation occurred normally without chromosomal bridges or fragmentation (metaphase II or early anaphase II (L), anaphase II (B–D–H–J) plants, and telophase II (F)). Bar 10 µm.
(TIF)
Acknowledgments
We are grateful to Thierry Lagrange and Sylvie Lamy for kindly providing the T-DNA insertion line disrupted in AtBRCA2a (GK_290C01) and PCR genotyping conditions, Sophie Blanchet for the ACTIN primers (ACT2) and PCR conditions for RT-PCR. The service of Dr V. Favaudon at Institut Curie (Orsay) is thanked for access to the 137Cs irradiator. We also thank Michael Hodges and Mireille Bétermier for critical reading of the manuscript, and Guillaume Blayo and Patricia Racine for their technical contribution.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Centre National de la Recherche Scientifique and the Université Paris Sud-11. M.D. is the recipient of a French Ministère de L'Enseignement Supérieur et de la Recherche doctoral fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 2.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H. Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA double-strand breaks. Plant J. 2002;29:771–781. doi: 10.1046/j.1365-313x.2002.01258.x. [DOI] [PubMed] [Google Scholar]
- 4.Riha K, Watson JM, Parkey J, Shippen DE. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. Embo J. 2002;21:2819–2826. doi: 10.1093/emboj/21.11.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friesner J, Britt AB. Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 2003;34:427–440. doi: 10.1046/j.1365-313x.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- 6.West CE, Waterworth WM, Jiang Q, Bray CM. Arabidopsis DNA ligase IV is induced by gamma-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 2000;24:67–78. doi: 10.1046/j.1365-313x.2000.00856.x. [DOI] [PubMed] [Google Scholar]
- 7.West CE, Waterworth WM, Story GW, Sunderland PA, Jiang Q, et al. Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. Plant J. 2002;31:517–528. doi: 10.1046/j.1365-313x.2002.01370.x. [DOI] [PubMed] [Google Scholar]
- 8.Bundock P, van Attikum H, Hooykaas P. Increased telomere length and hypersensitivity to DNA damaging agents in an Arabidopsis KU70 mutant. Nucleic Acids Res. 2002;30:3395–3400. doi: 10.1093/nar/gkf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallego ME, Bleuyard JY, Daoudal-Cotterell S, Jallut N, White CI. Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 2003;35:557–565. doi: 10.1046/j.1365-313x.2003.01827.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang YK, Chang WC, Liu PF, Hsiao MK, Lin CT, et al. Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair. Plant Mol Biol. 2010;74:453–466. doi: 10.1007/s11103-010-9685-5. [DOI] [PubMed] [Google Scholar]
- 11.van Attikum H, Bundock P, Overmeer RM, Lee LY, Gelvin SB, et al. The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 2003;31:4247–4255. doi: 10.1093/nar/gkg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Preuss SB, Jiang CZ, Baik HK, Kado CI, Britt AB. Radiation-sensitive Arabidopsis mutants are proficient for T-DNA transformation. Mol Gen Genet. 1999;261:623–626. doi: 10.1007/s004380050004. [DOI] [PubMed] [Google Scholar]
- 13.Hefner E, Preuss SB, Britt AB. Arabidopsis mutants sensitive to gamma radiation include the homologue of the human repair gene ERCC1. J Exp Bot. 2003;54:669–680. doi: 10.1093/jxb/erg069. [DOI] [PubMed] [Google Scholar]
- 14.Fidantsef AL, Mitchell DL, Britt AB. The Arabidopsis UVH1 gene is a homolog of the yeast repair endonuclease RAD1. Plant Physiol. 2000;124:579–586. doi: 10.1104/pp.124.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubest S, Gallego ME, White CI. Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep. 2002;3:1049–1054. doi: 10.1093/embo-reports/kvf211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubest S, Gallego ME, White CI. Roles of the AtErcc1 protein in recombination. Plant J. 2004;39:334–342. doi: 10.1111/j.1365-313X.2004.02136.x. [DOI] [PubMed] [Google Scholar]
- 17.McWhir J, Selfridge J, Harrison DJ, Squires S, Melton DW. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat Genet. 1993;5:217–224. doi: 10.1038/ng1193-217. [DOI] [PubMed] [Google Scholar]
- 18.Weeda G, Donker I, de Wit J, Morreau H, Janssens R, et al. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol. 1997;7:427–439. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- 19.Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol. 2004;24:1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, et al. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol. 2009;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole F, Keeney S, Jasin M. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 2010;24:1201–1207. doi: 10.1101/gad.1944710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 25.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 26.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 28.Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–8993. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 29.Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, et al. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 30.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 31.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 32.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 33.Tarsounas M, Davies AA, West SC. RAD51 localization and activation following DNA damage. Philos Trans R Soc Lond B Biol Sci. 2004;359:87–93. doi: 10.1098/rstb.2003.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, et al. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- 38.Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siaud N, Dray E, Gy I, Gerard E, Takvorian N, et al. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. Embo J. 2004;23:1392–1401. doi: 10.1038/sj.emboj.7600146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dray E, Siaud N, Dubois E, Doutriaux MP. Interaction between Arabidopsis Brca2 and its partners Rad51, Dmc1, and Dss1. Plant Physiol. 2006;140:1059–1069. doi: 10.1104/pp.105.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorslund T, Esashi F, West SC. Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. Embo J. 2007;26:2915–2922. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heacock ML, Idol RA, Friesner JD, Britt AB, Shippen DE. Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 2007;35:6490–6500. doi: 10.1093/nar/gkm472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka S, Ishii C, Hatakeyama S, Inoue H. High efficient gene targeting on the AGAMOUS gene in an Arabidopsis AtLIG4 mutant. Biochem Biophys Res Commun. 2010;396:289–293. doi: 10.1016/j.bbrc.2010.04.082. [DOI] [PubMed] [Google Scholar]
- 44.Waterworth WM, Masnavi G, Bhardwaj RM, Jiang Q, Bray CM, et al. A plant DNA ligase is an important determinant of seed longevity. Plant J. 2010;63:848–860. doi: 10.1111/j.1365-313X.2010.04285.x. [DOI] [PubMed] [Google Scholar]
- 45.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonatto D, Revers LF, Brendel M, Henriques JA. The eukaryotic Pso2/Snm1/Artemis proteins and their function as genomic and cellular caretakers. Braz J Med Biol Res. 2005;38:321–334. doi: 10.1590/s0100-879x2005000300002. [DOI] [PubMed] [Google Scholar]
- 47.Waterworth WM, Kozak J, Provost CM, Bray CM, Angelis KJ, et al. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009;9:79. doi: 10.1186/1471-2229-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreuzza S, Li J, Guitton AE, Faure JE, Casanova S, et al. DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development. 2010;137:73–81. doi: 10.1242/dev.041020. [DOI] [PubMed] [Google Scholar]
- 49.Timson DJ, Singleton MR, Wigley DB. DNA ligases in the repair and replication of DNA. Mutat Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 50.Wang S, Durrant WE, Song J, Spivey NW, Dong X. Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc Natl Acad Sci U S A. 2010;107:22716–22721. doi: 10.1073/pnas.1005978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abe K, Osakabe K, Ishikawa Y, Tagiri A, Yamanouchi H, et al. Inefficient double-strand DNA break repair is associated with increased fasciation in Arabidopsis BRCA2 mutants. J Exp Bot. 2009;60:2751–2761. doi: 10.1093/jxb/erp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charbonnel C, Gallego ME, White CI. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 2010;64:280–290. doi: 10.1111/j.1365-313X.2010.04331.x. [DOI] [PubMed] [Google Scholar]
- 53.Charbonnel C, Allain E, Gallego ME, White CI. DNA Repair (Amst) in press; 2011. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. [DOI] [PubMed] [Google Scholar]
- 54.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helliwell C, Waterhouse P. Constructs and methods for high-throughput gene silencing in plants. Methods. 2003;30:289–295. doi: 10.1016/s1046-2023(03)00036-7. [DOI] [PubMed] [Google Scholar]
- 56.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 57.Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, et al. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell. 1999;11:1623–1634. doi: 10.1105/tpc.11.9.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Observation of meiocytes by DAPI staining in nhej , ssa , nhej ssa and lig6 mutant plants transformed with the RNAi/0 control construct. Normal meiotic progression in plants transformed with pDMC1::RNAi/0 in nhej mutant plants, ku80 (A–B) and lig4 (C–D), in the ssa mutant ercc1 (E–F), in double nhej ssa mutants ku80 ercc1 (G–H) and lig4 ercc1 (I–J), and in lig6 mutant plants (K–L). Bivalents were correctly associated during the first meoitic phase (diakinesis (A–E–G–K) and metaphaseI (C, I). Segregation of homologous chromosomes and during the second division, sister chromatid separation occurred normally without chromosomal bridges or fragmentation (metaphase II or early anaphase II (L), anaphase II (B–D–H–J) plants, and telophase II (F)). Bar 10 µm.
(TIF)