Abstract
Human induced pluripotent stem cells (hIPSCs) represent a unique opportunity for regenerative medicine since they offer the prospect of generating unlimited quantities of cells for autologous transplantation as a novel treatment for a broad range of disorders1,2,3,4. However, the use of hIPSCs in the context of genetically inherited human disease will require correction of disease-causing mutations in a manner that is fully compatible with clinical applications3,5. The methods currently available, such as homologous recombination, lack the necessary efficiency and also leave residual sequences in the targeted genome6. Therefore, the development of new approaches to edit the mammalian genome is a prerequisite to delivering the clinical promise of hIPSCs. Here, we show that a combination of zinc finger nucleases (ZFNs)7 and piggyBac8,9 technology in hIPSCs can achieve bi-allelic correction of a point mutation (Glu342Lys) in the α1-antitrypsin (A1AT, also called SERPINA1) gene that is responsible for α1-antitrypsin deficiency (A1ATD). Genetic correction of hIPSCs restored the structure and function of A1AT in subsequently derived liver cells in vitro and in vivo. This approach is significantly more efficient than any other gene targeting technology that is currently available and crucially prevents contamination of the host genome with residual non-human sequences. Our results provide the first proof of principle for the potential of combining hIPSCs with genetic correction to generate clinically relevant cells for autologous cell-based therapies.
Currently available methods for gene targeting rely on positive selection to isolate rare clones that have undergone homologous recombination. To remove the unwanted selection cassettes, Cre/loxP or Flp/FRT recombination systems are used, which leave behind single loxP or FRT sites10,11. These small ectopic sequences have the potential to interfere with transcriptional regulatory elements of surrounding genes12, most of which are not fully characterized in the human genome. An alternative method to remove selection cassettes is to convert them into transposons. The most suitable transposon for this purpose is piggyBac, a moth-derived DNA transposon, which can transpose efficiently in mammalian cells including human embryonic stem cells (hESCs)9,13. A remarkable feature of this mobile element is seamless excision, which enables removal of transgenes flanked by piggyBac inverted repeats without leaving any residual sequences9,14.
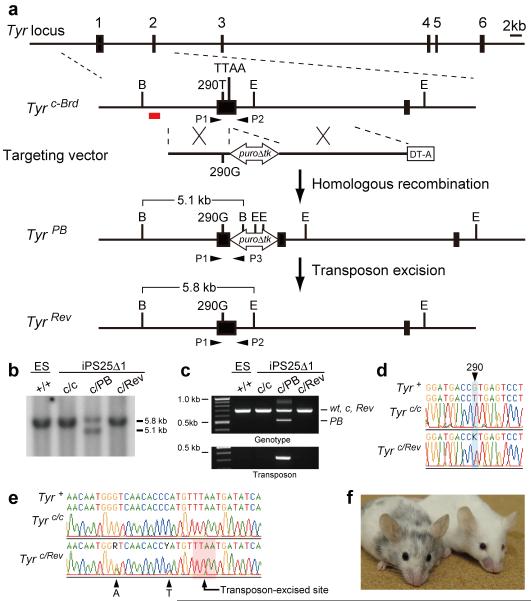
To explore the use of piggyBac for the correction of point mutations, we designed a vector to correct an albino mutation (G290T substitution in the Tyr gene) in mouse induced pluripotent stem cells (mIPSCs) isolated from fibroblasts of the C57Bl6-Tyrc-Brd strain15. The targeting vector was constructed, carrying a wild-type 290G sequence and a PGK-puroΔtk cassette flanked by piggyBac repeats into the TTAA site (Fig. 1a). Following isolation of targeted clones, the selection cassette was excised from the mIPSCs genome by transient expression of the piggyBac transposase and subsequent FIAU selection. Genomic modification was verified by Southern blot and PCR analyses (Fig. 1b, c). The correction of the G290T mutation and seamless piggyBac excision were confirmed by sequence analyses (Fig. 1d, e). Two introduced silent mutations were observed, confirming that the T290G substitution was mediated by gene correction, not by spontaneous reversion (Fig. 1e). The function of the reverted allele was tested by injecting the corrected mIPSCs into albino mouse blastocysts. The resulting chimeric mice displayed a black coat color, indicating phenotypic correction of the albino mutation (Fig. 1f). These results collectively demonstrate that the piggyBac transposon can be used as a versatile tool for highly precise modification (e.g. correction or mutation) of the mammalian genome at a single base-pair level.
Figure 1. Correction of the G290T mutation in the Tyr gene in mIPSCs.
a, The strategy for precise genome modification using the piggyBac transposon. Top line, structure of the Tyr gene; red line, 5′ external probe for Southern blot analysis; open arrow, piggyBac transposon carrying a PGK-puroΔtk cassette; P1, P2 and P3, PCR primers; B, BamHI; E, EcoNI. b, c, Southern blot (b) and PCR analyses (c) showing insertion (c/PB) and excision (c/Rev) of the piggyBac transposon. ES, mouse ESCs as a control. d, e, Sequence analyses revealed correction of the G290T mutation (d) and seamless excision of the piggyBac transposon (e). Note that two silent mutations (A and T indicated by arrowheads) introduced near the TTAA site were also detected. f, A chimeric mouse generated by injecting corrected Tyr c/Rev mIPSCs (left) displays black coat color. Right, a non-injected albino mouse.
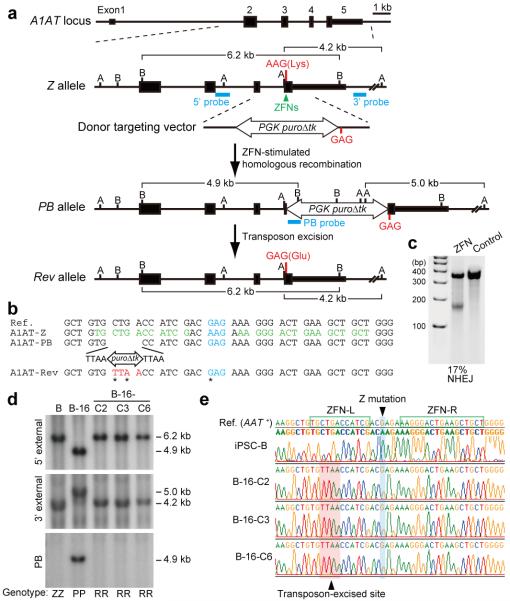
We next explored whether this approach could be used to correct a mutation in hIPSCs derived from individuals with α1-antitrypsin deficiency (A1ATD)16. A1ATD is an autosomal recessive disorder found in 1 out of 2000 individuals of North European descent and represents the most common inherited metabolic disease of the liver17,18. It results from a single point mutation in the A1AT gene (the Z allele; Glu342Lys) that causes the protein to form ordered polymers within the endoplasmic reticulum of hepatocytes17,18. The resulting inclusions cause cirrhosis for which the only current therapy is liver transplantation. The increasing shortage of donors and harmful effects of immunosuppressive treatments impose major limitations on organ transplantation, making the potential of hIPSC-based therapy highly attractive. Since homologous recombination is relatively inefficient in hESCs6, we employed ZFN technology, which stimulates gene targeting in hESCs as well as hIPSCs7,10,19. ZFN pairs were designed to specifically cleave the site of the Z mutation (Fig. 2a-c, Supplementary Table 1 and Supplementary Note). A targeting vector was constructed from isogenic DNA with piggyBac repeats flanking the PGK-puroΔtk cassette (Fig. 2a). To minimize the distance between the mutation and the piggyBac transposon, a CTG leucine codon, 10 bp upstream of the mutation, was altered to a TTA leucine codon, generating the TTAA sequence, which would be left in the genome following piggyBac excision (Fig. 2b).
Figure 2. Correction of the Z mutation in A1ATD-hIPSCs.
a, The strategy for precise genome modification using ZFNs and the piggyBac transposon. Top line, structure of the A1AT gene; blue lines, Southern blot probes; thin and thick boxes, non-coding and coding exons, respectively; open arrow, piggyBac transposon; B, BamHI; A, AflIII. b, Sequences of wild-type (Reference), Z, PB, and Rev alleles. Amino acid position 342 (blue), recognition sites for ZFNs (green), piggyBac excision site (red) are shown. Sequence changes in Rev allele from Z allele were indicated by asterisks. c, Surveyor nuclease assay showing the cleavage of Z mutation in ZFNs-transfected K562 cells. Non-transfected cells were used as a control. d, Southern blot analysis showing bi-allelic piggyBac insertion (B-16) and bi-allelic excision (B-16-C2, -C3 and -C6) during correction of the A1ATD-hIPSCs line B. Genomic DNA was digested by BamHI (5′ and PB probes) or AlfIII (3′ probe). Genotype: ZZ, homozygous for Z allele; PP, homozygous for insertion of piggyBac; RR, homozygous for reverted allele. e, Sequence analysis showing correction of Z mutation in 3 corrected hIPSC lines. Wild-type sequence (top line) and A1ATD-hIPSC (second line).
Puromycin-resistant hIPSC colonies obtained after co-electroporation of ZFN expression vectors and the targeting vector were screened for targeted clones by PCR. A1ATD-hIPSC lines derived from 3 different patients yielded targeted clones (Table 1). Remarkably, 54% of the puromycin-resistant colonies were targeted on one allele, while 4% were the result of simultaneous targeting of both alleles (Supplementary Fig. 1).
Table 1.
Summary of PCR genotyping of ZFN-stimulated gene targeting
| A1ATD-iPSC line | Clones analyzed |
Het.a | Homo. / Hemi. b |
Het. + additonal integrations c |
Homo. / Hemi. + additonal integrations c |
Non-targeted d |
|---|---|---|---|---|---|---|
| A | 84 | 45 | 3 | 23 | 8 | 5 |
| B | 18 | 10 | 2 | 3 | 3 | 0 |
| C e | 216 | 112 | 9 | 52 | 21 | 22 |
|
| ||||||
| Mean frequency [%] | 54 | 6 | 23 | 12 | 5 | |
Het., clones heterozygous for PB allele.
Homo./Hemi., clones homozygous or hemizygous for PB allele. Cells with one targeted allele and deletion of the other allele are undistinguishable from correctly targeted homozygous clones by PCR. Such cells are designated as hemizygotes.
Vector backbone integration was analyzed by PCR.
Clones showing incorrect PCR bands are included.
A sum of 2 independent experiments.
To remove the piggyBac-flanked selection cassette from these modified clones, we transiently transfected two homozygously targeted clones (B-16 and C-G4) with a hyperactive form of the piggyBac transposase8 and subjected them to FIAU selection. The genotype of the resulting FIAU-resistant colonies was analyzed by PCR and confirmed by Southern blot (Fig. 2d and Supplementary Fig. 2a). Bi-allelic excision was observed in 11% of FIAU-resistant colonies (Table 2). Sequence analyses demonstrated that the Z mutation was corrected on both alleles and that transposon excision yielded a TTAA sequence as initially planned (Fig. 2b, e and Supplementary Fig. 2b). The resulting corrected A1ATD-hIPSC (c-hIPSC) lines maintained the expression of pluripotency markers for more than 20 passages and their abilities to differentiate into cells expressing markers of the three germ layers (Supplementary Fig. 3), indicating that genome modification did not alter the pluripotency of c-hIPSCs.
Table 2. Frequencies of bi-allelic piggyBac excision.
| Bi-allelic excision w/o re-integration |
Bi-allelic excision w/ re-integration |
||||
|---|---|---|---|---|---|
| Cell line | analyzed | No. of clones | Frequency [%] | No. of clones | Frequency [%] |
| B-16 | 88 | 15 | 17 | 33 | 38 |
| C-G4 | 94 | 5 | 5 | 19 | 20 |
|
| |||||
| Mean frequency [%] | 11 | 29 | |||
Genomic instability is known to be associated with prolonged culture of hESCs20,21 and those arising during genome modification would be another concern for clinical application of hIPSCs. Therefore, we analyzed the genomic integrity of the hIPSC lines using comparative genomic hybridization (CGH) (Supplementary Table 2a-c). Two out of three A1ATD-hIPSC primary lines differed from their parental fibroblasts, showing amplifications or deletions ranging from 20 kb to 1.3 Mb, including a gain of 20q11.21, a frequently amplified region in hESCs22,23 (see Supplementary Analysis and Supplementary Fig. 4). Line A retained a normal genome content compared to its parental fibroblast. Reassuringly, we found that after ZFN-stimulated targeting, four out of six homozygous clones had unaltered genomes compared to their parental hIPSC lines. Sixteen cell lines with bi-allelic piggyBac excision were compared with their corresponding primary hIPSCs and 12 had unaltered genomes. We also analyzed the hIPSC lines by SNP arrays to check for loss of heterozygosity and found that all lines analyzed retained heterozygosity throughout their genome (Supplementary Fig. 5). This observation demonstrates that bi-allelic gene correction was the result of simultaneous homologous recombination followed by simultaneous excision at both alleles and that mitotic recombination was not involved in this process.
ZFN off-target cleavage and imprecise excision after multiple piggyBac transposition might introduce mutations into the genome. In order to investigate these possibilities at a single basepair resolution, we sequenced exomes of the corrected B-16-C2 line and its parental fibroblast. Comparison of these exomes identified 29 mutations (Supplementary Table 3). The genesis of these mutations was determined by analysis of the primary hIPSC line and the homozygously targeted intermediate. Twenty-four point mutations and one 1-bp deletion were detected in the primary hIPSC line and four mutations arose during genetic correction: one during targeting and three during piggyBac excision. These mutations appeared to arise during culture since their genomic signatures were inconsistent with ZFN off-target sites or piggyBac integration sites (Supplementary Analysis). Taken together, we conclude that the combination of ZFNs with piggyBac provides a new method for rapid and clean correction of a point mutation in hIPSCs without affecting their basic characteristics.
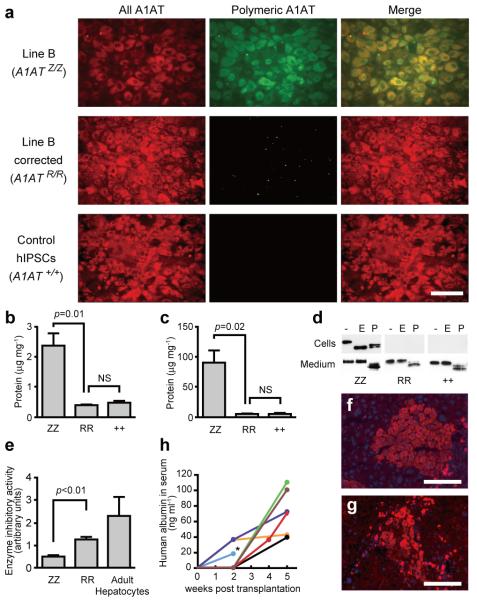
To confirm that the genetic correction of hIPSCs resulted in the expected phenotypic correction, hIPSCs were differentiated in vitro into hepatocyte-like cells, the main cell type affected by the disease A1ATD. Differentiation of the corrected lines occurred as expected, resulting in a near homogenous population of hepatocyte-like cells (Supplementary Fig. 6a-c). Remarkably, CGH analysis of differentiated cells showed that hepatic differentiation neither increases the number of genetic abnormalities nor selects for cells with abnormal karyotype (Supplementary Table 2d). The resulting cells shared key functional attributes of their in vivo counterparts including glycogen storage, LDL-cholesterol uptake, albumin secretion and Cytochrome P450 activity (Supplementary Fig. 6d-g). Importantly, immunofluorescence and ELISA both confirmed the absence of mutant polymeric A1AT in c-hIPSCs-derived hepatocyte-like cells that instead efficiently secreted normal endoglycosidase–H-insensitive monomeric A1AT (Fig. 3a-d). In addition, secreted A1AT displayed an enzymatic inhibitory activity that was comparable to that obtained from normal adult hepatocytes (Fig. 3e), thereby suggesting that physiological restoration of enzyme inhibitory activity could be achieved.
Figure 3. Functional analysis of restored A1AT in c-hIPSCs-derived hepatocyte-like cells.
a, Immunofluorescence showing the absence of polymeric A1AT protein in hepatocyte-like cells generated from c-hIPSCs. All forms of A1AT (left panels) and misfolded polymeric A1AT (middle panels). b, c, ELISA to assess the intracellular (b) and secreted (c) levels of polymeric A1AT protein in hepatocyte-like cells derived from A1ATD-hIPSCs (ZZ), c-hIPSCs (RR) and control hIPSCs (++). d, Endoglycosidase H (E) and peptide:N-glycosidase (P) digestion of A1AT immunoprecipitated from uncorrected (ZZ), corrected (RR) and control (++) hIPSC-derived hepatocyte-like cells (upper panels) and corresponding culture medium (lower panels). e, Chymotrypsin ELISA showing that corrected cells (RR) have A1AT enzymatic inhibitory activity that is superior to uncorrected cells (ZZ) and close to adult hepatocytes. f, g, Immunofluorescence of transplanted liver sections detecting human albumin (f) and A1AT (g). DNA was counterstained with DAPI. h, ELISA read-out of human albumin in the mouse serum longitudinally followed for each mouse. Asterisk, the mouse was subjected to histology analysis. Scale bars, 100 μm. Data in b, c and e are shown as mean ± s.d. (n=3). Student’s t-test was performed. NS, not significant.
Finally, the in vivo function of c-hIPSCs-derived hepatocyte-like cells (B-C16-2 line) was assessed following transplantation into the liver of Alb-uPA+/+;Rag2−/−;Il2rg−/− mice via intra-splenic injection. Livers harvested 14 days after injection were colonized by human cells identified using antibodies specific to human albumin and A1AT (Fig. 3f, g). These human hepatocyte-like cells were distributed throughout the liver lobes and were seen to be integrated into the existing mouse parenchyma (Fig. 3f, g). In addition, human albumin was detected in the serum of transplanted animals for at least 5 weeks (Fig. 3h), while no tumor formation was detected in any mice. Therefore, c-hIPSCs-derived hepatocyte-like cells were able to colonize the liver in vivo and display functional activities characteristic of their human ESC-derived counterparts24. Collectively these analyses demonstrate that genetic correction of the Z mutation resulted in functional restoration of A1AT in patient-derived cells.
All experimental evidence above strongly support the applicability of genetic correction in patient-specific iPSCs for cell-based therapy of A1ATD. We therefore repeated the genetic correction in more clinically relevant cells using patient-specific iPSCs reprogrammed from fibroblasts with Sendaiviral vectors, an integration-free method25 (Supplementary Fig. 7a-f). One primary hIPSC line with an intact genome by CGH analysis (Supplementary Fig. 7e and Supplementary Table 4) was corrected by the method described above. The final product, iPSC-3-G5-A7, had the corrected A1AT, had an intact genome compared to the parental fibroblast, and expressed normal A1AT protein when differentiated to hepatocyte-like cells (Supplementary Fig. 8 and Supplementary Table 4). This is the first demonstration of the generation of mutation-corrected patient-specific iPSCs, which could realize the therapeutic promise of hIPSCs.
In the present study, we demonstrate that ZFNs and piggyBac transposon enable simultaneous bi-allelic correction of diseased hIPSCs. No residual ectopic sequences remain at the site of correction and the genome appears to be undisturbed elsewhere. Although we could readily obtain cell lines without large genomic alterations during genetic modification, the resulting corrected hIPSCs carry 29 mutations in protein coding exons, of which 22 were non-synonymous or splice site mutations. The likely impact of this mutation load needs to be considered in the context of their likely functional impact, taking into account the normal germ-line load, accumulated somatic variation, the presence of compensating normal gene copies and the requirement for the gene product in the derived differentiated cells. From this point of view, only eight mutations might affect gene functions in hepatocyte-like cells (Supplementary Table 3). Nevertheless, the corrected iPSCs could efficiently differentiate to hepatocyte-like cells and engraft into the animal model for liver injury without tumor formation. Therefore, limited genomic abnormalities might have restricted biological consequences. Careful screening of primary and corrected hiPSCs using deep sequencing analyses would contribute to the safe use of hIPSCs in clinical applications.
hIPSCs derived from different patients were effectively corrected, demonstrating that this method could be applied to a large number of A1ATD-hIPSC lines. Since the bi-allelic correction could be carried out in less than 4 months, our approach may be compatible with large-scale production of corrected patient-specific hIPSCs not only for A1ATD but also for other monogenic disorders.
Methods summary
A1ATD-hIPSCs were described previously16. 2 × 106 hIPSCs were co-transfected with ZFN expression vectors and the donor template, and subjected to puromycin selection (1 μg ml−1) initiated 4 days after transfection. For transposon excision, targeted cells were transfected with pCMV-hyPBase8, cultured for 4 days, replated and selected in 250 nM 1-(2-Deoxy-2-fluoro-beta-D-arabinofuranosyl)-5-indouracil (FIAU). To increase clonogenicity, cells were treated with ROCK inhibitor26, Y-27632 (10 μM) 4 hours prior to dissociation and 24 hours post plating. Resulting colonies were picked 2 weeks later, analyzed by PCR and further verified by Southern blot analysis. Primer sequences are listed in Supplementary Table 5.
Supplementary Material
Acknowledgements
We thank Sir Aaron Klug and Dr Michal Minczuk for their advice, M.A. Li for comments on the manuscript, P. Ellis, N. Hammond and C. McGee for CGH analysis, the Sanger Institute sequencing facility for exome sequencing, N. Conte and S. Rice for assistance with bioinformatic analysis, M. Alexander for her help with cell culture reagents. We also thank L. Zhang, S. Hinkley and the production group for ZFN assembly and validation, K. Tong and X. Meng for technical assistance, J.C. Miller and E. Leung for ZFN off-target site analysis and S. Abrahamson and P.D. Gregory for careful reading of the manuscript. This work was supported by Wellcome Trust (WT077187; A.B.), MRC Senior non-clinical fellowship and the Cambridge Hospitals National Institute for Health Research Biomedical Research Center (L.V.), the Medical Research Council and Papworth NHS Trust (D.A.L), Bill and Melinda Gates Foundation, Inserm and Institut Pasteur (H.S), and Japan Science and Technology Agency (N.F). K.Y. is supported by a postdoctoral fellowship of Japan Society for the Promotion of Science. S.T.R and F.R. are Wellcome Trust Clinical Training Fellows. I.V. is supported by a fellowship from the International Human Frontiers Science Program Organization.
Appendix
Methods
Plasmid construction
Gateway-adapted piggyBac transposon vectors: A destination vector pPB-R1R2-NP was constructed as follows. The attR1 and attR2 sites were PCR-generated and digested by NheI/HindIII and XhoI/SpeI, respectively. EM7-neo was PCR-generated and digested by HindIII/XhoI. These 3 fragments were then cloned into the NheI-SpeI site of pPB-LR527, resulting in pPB-R1R2-Neo. An EcoRI-XbaI fragment containing PheS was excised from pR6K-R1R2-ZP28, blunt-ended and cloned into the blunted XhoI site of pPB-R1R2-Neo, resulting in pPB-R1R2-NP. An entry vector pENTR-PGKpuroΔtk was constructed by cloning a KpnI-NotI PGK-puroΔtk fragment into the KpnI-NotI site of pENTR-2B.
A targeting vector for Tyr: The targeting vector was constructed using BAC recombineering. A BAC clone RP24-221M7 was introduced into Escherichia coli strain EL350 29. A mini targeting vector was first constructed to modify the Tyr gene on the BAC. Left and right homology arms were PCR-generated and digested by AscI/BsiWI and NsiI/PacI, respectively. The transposon fragment was excised from pPB-R1R2-NP by NsiI/BsiWI digestion. These 3 fragments were then cloned into AscI/PacI site of pMCS, resulting in pMCS-Tyr-NP. An AscI-PacI fragment was excised from pMSC-Tyr-NP and used for BAC targeting. A retrieving vector was constructed by cloning PCR-generated left and right homology arm into the XhoI/AscI site of pMSC-DTA, following AscI/HindIII and XhoI/HindIII digestion of the left and right arm, respectively. The retrieving vector was linearized by HindIII digestion and used to retrieve 3.0-kb 5′ arm, the transposon and 6.5-kb 3′ arm. Finally, the Neo-PheS cassette was replaced with the PGK-puroΔtk cassette by Gateway cloning, resulting in pDTA-TyrPB. The targeting vector was linearized by AscI prior to electroporation into the albino mIPSCs.
A donor template vector for A1AT: A 2-kb fragment, which contained 1 kb at both side of Z mutation, was first PCR-amplified using genomic DNA from A1ATD-hIPSC line B as a template and cloned into pCR4-blunt-TOPO (Invitrogen), resulting in pCR4-AAT_Z. To construct a donor template with corrected sequence and a piggyBac transposon, the 5′ arm and 3′ arm were PCR-amplified and digested with AscI/NsiI and BsiWI/PacI, respectively. The NsiI-BsiWI fragment containing a piggyBac transposon with the Neo-PheS cassette was excised from pPB-R1R2-NP. The digested fragments were cloned into the AscI-PacI site of pMCS, resulting in pMCS-AAT-PB:NP. The Neo-PheS cassette was subsequently replaced with a PGK-puroΔtk cassette by Gateway cloning, resulting in the final donor vector, pMCS-AAT-PB:PGKpuroΔtk.
The plasmids (pPB-R1R2-NP, pENTR-PGKpuroΔtk, pMCS-AAT-PB:PGKpuroΔtk) have been deposited in the Wellcome Trust Sanger Institute Archives and available upon request (http://www.sanger.ac.uk/technology/clonerequests/).
Cell culture
Appropriate ethical approval and patient consent have been obtained (Ethics reference no. 08/H0311/201; R&D no. A091485). A1ATD-hIPSCs (ref. 16; A, patient 2 line 1; B, patient 1 line1; C, patient 3 line1) were cultured on MEF-feeder layers in hESC medium: DMEM/F12 supplemented with 20 % knockout serum replacement, 1 mM GlutaMax, 0.1 mM 2-mercaptoethanol, 1x non-essential amino acid and 4 ng ml−1 FGF2 (Invitrogen). Subculture was performed every 5-7 days by detaching hIPSCs by incubation in 0.5 mg ml−1 dispase and 0.5 mg ml−1 collagenase type IV for 1 hr at 37 °C, collecting detached hIPSC colonies, breaking down into small clamps and plating them onto new feeder plates. Mouse embryonic fibroblasts (CF1 or B6129F1) were cultured in DMEM containing 10 % FCS, 2 mM Glutamine, 0.1 mM 2-melcaptoethanol and 1x non-essential amino acid. mIPSCs (iPS25Δ1; ref. 15) were cultured on MEF-feeder layers in mESC medium: KO-DMEM supplemented with 15 % FBS, 1 mM GlutaMax, 0.1 mM 2-mercaptoethanol, 1x non-essential amino acid and 1000 unit ml−1 LIF (Millipore).
Gene Targeting and transposon excision in mouse IPSCs
1 × 107 cells were electroporated with 25 μg of a linearized targeting vector in 800 μl of HEPES-buffered saline using a Gene Pulser II electroporator (230 V, 500 μF) and plated onto three 10-cm dishes. The next day, puromycin selection (1 μg ml−1) was initiated. Resulting colonies were picked and screened by PCR. Targeted clones were expanded and further verified by Southern blot analysis. Correctly targeted clones were then subjected to transposon excision. 2 × 106 cells were electroporated with 40 μg of pCMV-hyPBase in 800 μl of HEPES-buffered saline using a Gene Pulser II electroporator (230 V, 500 μF) and plated onto one well of a 6-well plate. After passage once, cells were replated on day 4 at 5 × 105 cells per 10-cm dish. On the following day, FIAU (0.2 μM) selection was initiated. On day 5 of selection, FIAU was withdrawn. Resulting colonies were picked at day 7 and screened by PCR. Primer sequences to detect homologous recombination are shown in Supplementary Table 5.
ZFNs-mediated gene targeting in A1ATD-hIPSCs
On the day of electropolation (day 0), near-confluent cells were pre-treated with a ROCK inhibitor26 (Y-27632, Sigma) at 10 μM for 3-4 hrs prior to electroporation. Cells were then washed with PBS once, detached by Accutase (Millipore; 10 min at 37 °C) and mixed with DMEM/F12 containing 10% FCS. Cells were dissociated into single-cell suspension by vigorous pipetting and counted. 2 × 106 cells were pelleted and mixed with 5 μg of a 5′-ZFN expression vector, 5 μg of a 3′-ZFN expression vector and 2 μg of the donor template in 100 μl of hESC solution 1 (Lonza). The cell suspension was transferred to a cuvette and electroporated using the Amaxa Nucleofector device (Lonza) with program A23. The electroporated cells were plated onto one or two 10-cm feeder dishes in MEF-conditioned hESC medium containing 10 μM Y-27632. hESC medium without any drug was used for daily medium change between day 1-3. On day 4, puromycin selection (1 μg ml−1) was started. On day 6, medium was changed to MEF-conditioned hESC medium containing 0.5 μg ml−1 puromycin, which was used for medium change at every other day until picking colonies. Resulting colonies were picked on day 13-17. Colonies was cut into 2 pieces. One half was transferred onto a well of 24-well feeder plate and the other half was lysed and used for PCR-genotyping. PCR-positive clones were further expanded and homologous recombination was verified by Southern blot analysis.
Transposon excision in homozygously targeted hIPSCs
Homozygously targeted clones (B-16, C-G4, SeV-1-C3 and SeV-3-G5) were used for transposon removal. Line A-derived clones were omitted because this line displayed a lower capability of differentiating into endodermal lineages. Cells were prepared as described above. 2 × 106 cells were mixed with 10 μg of the hyperactive piggyBac transposase expression vector (pCMV-hyPBase8) in 100 μl of hESC solution 1 and electroporated using the Nucleofector device with the program A23. Electroporated cells were plated onto 6-well plate in 1:2, 1:3 and 1:6 dilutions in MEF-conditioned hESC medium containing 10 μM Y-27632. Note that ROCK inhibitor was added to the culture medium until day 6 in this experiment. On day 2, cells with ~80 % confluency were passaged using Accutase at a split ratio 1:2, 1:3 and 1:6 into 6-well plates. On day 4, cells with ~80 % confluency were washed with PBS, detached with Accutase, suspended in hESC medium and pelleted. Cells were resuspended in hESC medium into single-cell level and counted. 1 × 104 cells were then plated onto one 10-cm dish in hESC medium containing 10 μM Y-27632. 16-18 hrs after plating (day 5), medium was changed to hESC medium containing 0.25 μM FIAU and 10 μM Y-27632. On day 6, medium was changed to hESC medium containing 0.25 μM FIAU and then medium was changed every other day. Genotype and deletion of the piggyBac transposon were analyzed by PCR and further verified by Southern blot analysis.
CGH analysis
Genomic DNA was extracted using a DNeasy kit (Qiagen). Agilent 244K human genome arrays were used following the manufacturer’s protocol. The arrays were scanned with an Agilent microarray scanner and data were generated by Agilent Feature Extraction software. CGH calls were made with Agilent’s DNA analytics software using the ADM2 algorithm (6.0 threshold) with a minimum of 5 probes in the region as a filter.
SNP analysis
An Illumina HumanCytoSNP-12 SNP array was used following the manufacturer’s protocol. Genotype calls were performed by Illumina’s GenomeStudio. B allele frequency and log R ratio were analyzed by KaryoStudio. CNVpatition v2.4.4 bundled in KaryoStudio was used for copy number analysis.
ZFN design
ZFNs were designed against a region containing Z mutation in the A1AT gene (see Fig. 2a, b) and assembled as previously described30. The amino acid residues at positions ‘−1’ to ‘+6’ of the recognition alpha helix31,32 of each of the zinc finger DNA-binding domain for each DNA triplet target are shown in Supplementary Table 2. The ZFNs were linked to wild type FokI catalytic domain. The activity of the ZFN at the endogenous target site was determined using the Surveyor Nuclease assay as previously described33.
hIPSCs-derived hepatocyte-like cell transplantation in immunodeficient uPA transgenic mice
All mice were housed in pathogen-free conditions and animal studies were approved by the committee on animal experimentation of the Institut Pasteur and by the French Ministry of Agriculture. Differentiated cells (5 × 105 cells per animal in 50 μl DMEM) were injected into the spleens of 3- to 4-week-old Alb-uPA+/+;Rag2−/−;Il2rg−/− mice (n=7). The recipient mouse was sacrificed 2 weeks after transplantation for histological analysis. Blood samples were collected and human albumin in plasma was quantified by ELISA (Bethy Laboratories). Frozen liver sections were analyzed by immunofluorecence with human albumin (Dako) or human A1AT (Dako) specific antibodies. Non-transplanted mice were used as controls.
Exome sequencing
The corrected iPSC line, B-16-C2, and its parental fibroblasts were analyzed. Exome sequencing and analysis were performed as described previously34 with minor modification. Exome pulldown was performed using an Agilent SureSelect Human All Exon 50Mb Kit according to the manufacturer’s instructions. Enriched DNA was sequenced on an Illumina HiSeq2000 (75-bp paired-end sequencing). 90.32% (Fibroblast-B) and 90.72% (B-16-C2) of total targeted regions were covered with more than 10x sequencing depth, covering 93.01% and 93.35% of CCDS exons, respectively. Substitutions in the coding sequence were called as positions with at least 20% of reads reporting a different base with respect the reference human sequence (GRCh37). Additionally, somatic mutations were identified by comparing the sequence with the control fibroblasts, and removing the common polymorphisms described in dbSNP and in the 1000 Genomes Project35. Small insertions and deletions were identified using samtools, as the ones not present in the control cell line and that had at least 20x of coverage and 20% of the reads reporting the mutation. Validation of mutations was carried out by Sanger capillary sequencing on parental Fibroblast-B, A1ATD-hIPSC line B, the homozygously targeted B-16 cells and the piggyBac-excised B-16-C2 cells.
Sendaiviral regrogramming, RT-PCR, quantitative RT-PCR, bisulfite sequencing, immunostaining, flow cytometric analysis, ELISA and EndoH analysis
These experiments were performed as described previously16,24,25,36
- 27.Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 31.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 32.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 33.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 34.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium TGP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seki T, et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Exome sequence data have been deposited at the European Genome-Phenome Archive (http://www.ebi.ac.uk/ega/) hosted by the European Bioinformatics Institute under accession EGAS00001000055. CGH and SNP array data have been deposited with EBI ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MEXP-3316 and with Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE31035, respectively. The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature. Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of this article at www.nature.com/nature. Requests for the ZFNs used in this study should be sent to pgregory@sangamo.com.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 5.Fairchild PJ. The challenge of immunogenicity in the quest for induced pluripotency. Nat Rev Immunol. 2010;10:868–875. doi: 10.1038/nri2878. [DOI] [PubMed] [Google Scholar]
- 6.Tenzen T, Zembowicz F, Cowan CA. Genome modification in human embryonic stem cells. J Cell Physiol. 2010;222:278–281. doi: 10.1002/jcp.21948. [DOI] [PubMed] [Google Scholar]
- 7.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 8.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci U S A. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Weyden L, Adams DJ, Bradley A. Tools for targeted manipulation of the mouse genome. Physiol Genomics. 2002;11:133–164. doi: 10.1152/physiolgenomics.00074.2002. [DOI] [PubMed] [Google Scholar]
- 12.Meier ID, et al. Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J. 2010;24:1714–1724. doi: 10.1096/fj.09-140749. [DOI] [PubMed] [Google Scholar]
- 13.Lacoste A, Berenshteyn F, Brivanlou AH. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 15.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rashid ST, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perlmutter DH. Autophagic disposal of the aggregation-prone protein that causes liver inflammation and carcinogenesis in alpha-1-antitrypsin deficiency. Cell Death Differ. 2009;16:39–45. doi: 10.1038/cdd.2008.103. [DOI] [PubMed] [Google Scholar]
- 18.Gooptu B, Lomas DA. Conformational pathology of the serpins: themes, variations, and therapeutic strategies. Annu Rev Biochem. 2009;78:147–176. doi: 10.1146/annurev.biochem.78.082107.133320. [DOI] [PubMed] [Google Scholar]
- 19.Zou J, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitalipova MM, et al. Preserving the genetic integrity of human embryonic stem cells. Nat Biotechnol. 2005;23:19–20. doi: 10.1038/nbt0105-19. [DOI] [PubMed] [Google Scholar]
- 21.Baker DE, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 22.Lefort N, et al. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat Biotechnol. 2008;26:1364–1366. doi: 10.1038/nbt.1509. [DOI] [PubMed] [Google Scholar]
- 23.Spits C, et al. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat Biotechnol. 2008;26:1361–1363. doi: 10.1038/nbt.1510. [DOI] [PubMed] [Google Scholar]
- 24.Touboul T, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 25.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.