Abstract
Aims
Since many biomarkers of both the tumor and its microenvironment are expected to involve differential expression of divalent proteins capable of protein or peptide ligand interaction, we are developing multivalent nanodevices for the identification of biomarkers in prostate cancer.
Patients & Methods
We compared a multivalent thioredoxin-targeted nanodevice with monovalent thioredoxin in binding to human prostate cell line(s) and freshly frozen tissue specimens obtained after resection from patients with biopsy-proven prostate cancer.
Conclusion
The nanodevice binds specifically with enhanced avidity to tumor microenvironment-associated stromal cells in prostate cancer tissue specimens. Cells that bind the nanodevice also reacted with antibodies to dimeric thioredoxin reductases 1 and 2, suggesting the utility of the nanodevice as a potentially specific and functional marker of tumor stromal cells.
Keywords: nanoassembly, nanotechnology, nano-tool, prostate cancer, reactive stroma, thioredoxin, thioredoxin reductase
Most cell surface receptors bind ligands as homo- or hetero-dimers [1–3] with possible oligomerization exhibited by G-protein-coupled receptors [4]. Thus, nanoscale designs for multiple ligand presentation are preferred for cell targeting and visualization since they can produce high-affinity cooperative interaction with their cellular targets. Multivalency can be achieved with a variety of systems. Dendrimers have the advantage of being nanoscale (i.e., less than 100 nm in diameter) and capable of multivalent ligand presentation. For example, fluorescently labeled polyamidoamide dendrimers have been successfully targeted to folate receptors on tumor cells by chemically linking multiple folate ligands at the G5 (8 nm) round of extension [5]. Moreover, growth factors chemically conjugated to fluorescein-labeled G5 dendrimers permit visualization of tumor cells in vitro [6]. A significant advantage of dendrimers in this application is their capacity to conform to an irregular cell surface by flattening upon binding [7]. However, the requirement for chemical modification can diminish the effectiveness of peptide and protein binding.
Quantum dots also permit multivalent substitution; for example, biotinylated EGF linked through streptavidin-functionalized quantum dots permits levels of surface substitution approaching 30:1 [8]. However, the surface arrangement and degree of flexibility are unknown and the effects of biotinylation are expected to diminish the native binding capacity of any protein or peptide ligand.
Protein–protein interaction provides a third route to multivalency. For example, the barnase– barstar interaction permits the assembly of a trivalent antibody system with the expected cooperative enhancement of binding avidity [9]. Here again, radiolabeling required chemical modification of the assembly, with associated drawbacks. The Cowpea mosaic virus can also serve as a multivalent nanodevice. It offers a selective site for chemical modification in each of the capsid subunits. The single reactive amino acid on the native capsid subunit [10], or a reactive cysteine sulfhydral inserted by in vitro mutagenesis [11] permit site-specific chemical modification of capsid subunits. However, ligand presentation will be constrained by the rigid icosahedral symmetry of the capsid and by ligand structure and will be influenced by the necessity for chemical linkage.
We have developed a modular addressing system based on selective targeting of DNA– methyltransferase fusion proteins to DNA scaffolds [12–14] that facilitates the construction of multivalent nanoscale assemblies. With this system, the nucleic acid scaffolds and protein components are all created separately and then assembled under physiological conditions. Flexibility in the design allows us to tailor the nanodevice to the application. This system permits the display of multiple ligands at preselected positions on a DNA scaffold so as to take advantage of the dimeric or multimeric nature of most receptors and enzymes. Moreover, the fusion ligands can be either identical or nonidentical depending on the intended design. The fusion ligands are expressed and purified under native conditions to maintain the native conformation and retain native activity. The DNA itself can be labeled with a variety of chromophores or functional groups during synthesis, adding increased versatility to the approach. A schematic of the process used for the construction of the trivalent nanodevice (ND-Trx3) displaying bacterial thioredoxin (Trx) [15] as a targeting ligand and a molecular model of ND-Trx3 is depicted in Figure 1. Previous work, in which prostate cancer cell lines were targeted with this system [15], suggested it might selectively bind to human prostate cancer tissues specimens. Since human and bacterial thioredoxin are structural isomers [15], each of the four well-characterized thioredoxin interacting proteins in human tissue: thioredoxin reductase 1 (TxnRD1), thioredoxin reductase 2 (TxnRD2), thioredoxin-glutathione reductase (TxnRD3/TGR) and thioredoxin binding protein 2 (Txnip/VDUP-1), is a candidate for ND-Trx3 target. In this report, we compare the binding of ND-Trx3 with a fluorescently labeled thioredoxin bioconjugate (Trx-F) for their ability to bind specific cell surface receptor targets on cell lines and tumor specimens.
Figure 1. Construction of a self-assembling thioredoxin-targeted nanodevice.
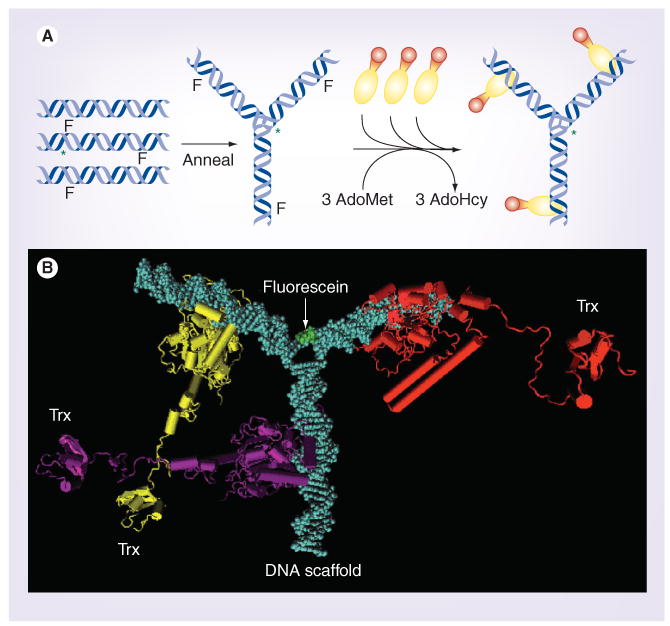
(A) The self-assembly sequence for the ND-Trx3. Oligodeoxynucleotides are synthesized separately and annealed to form a DNA Y-junction that carries a 5FdC moiety at methylation sites recognized by M•EcoRII on each arm of the Y-junction. Fluorescein-dT is added near the center of one of the three oligodeoxynucleotides during synthesis. Purified M•EcoRII-Trx fusion proteins are mixed with the Y-junction and AdoMet. Incubation at 37°C permits the methylation reaction to trap the fusion proteins at the methyltransferase recognition sites. (B) Molecular model of the nanodevice (ND-Trx3). The DNA scaffold is colored blue, the three M•EcoRII-Trx fusion proteins are colored yellow, purple and red, and the fluorescein, which is attached to the DNA scaffold, is green.
AdoHcy: S-adenosylhomocysteine; AdoMet: S-adenosylmethionine; ND-Trx3: Nandodevice; Trx: Thioredoxin.
Patients & methods
Freshly frozen prostate tissue specimens were obtained after resection from biopsy-confirmed cases of prostate cancer with Gleason scores of: <7 (n = 12); 7 (n = 19); >7 (n = 6) and benign prostate hyperplasia (BPH; n = 4) from City of Hope Pathology Core Facility, Duarte, CA, USA. The histologic classification of the prostate cancer was determined according to the Gleason criteria [16]. The Institutional Review Board (IRB) of the City of Hope Hospital approved the use of tissue specimens for this study, in accordance with the ethical policy and procedure. The initial diagnosis was reconfirmed by morphologic examination of newly sectioned and hematoxylin and eosin (H&E)-stained tissue specimens.
Molecular modeling
Molecular modeling methods have been described previously [15]. The ND-Trx3 model was refined in Insight II (Acellerys Inc, CA, USA) to demonstrate that the peptide linkers that join the methyltransferases to the thioredoxins have the flexibility needed to allow two of the three ligands to bind a putative homodimeric target protein. The models of ND-Trx3 were rendered in Chimera (Computer Graphics laboratory, University of California San Francisco, CA, USA). The trivalent ND-Trx3 was modeled in association with a homodimeric interacting protein (Escherichia coli Thioredoxin Reductase, Protein Data Base ID: 1F6M) using Swiss PBD Viewer and the image of the complex was rendered using Chimera (Figure 2).
Figure 2. Model of the binding interaction between the nanodevice (ND-Trx3) and a dimeric target.
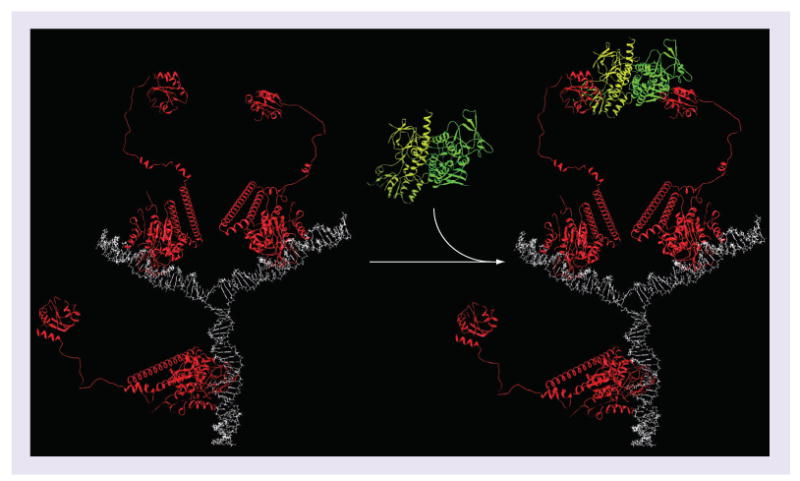
The trivalent (ND-Trx3) was modeled in association with a homodimeric interacting protein (Escherichia coli Trx reductase) in order to illustrate its potential for cooperative binding. The nanodevice displays three ligands on long flexible peptide linkers. The flexibility of these linkers allows each ligand to bind the homodimeric binding protein as a pair. The maximum distance of the two tethered ligands is approximately 20 nm, which can easily accommodate human Trx reductase 1 in which the Trx binding sites are approximately 12 nm apart. Each ligand can partner with one of the other two ligands to form the complex. If one ligand binds the homodimeric interacting protein, this brings the second ligand-binding site within range of the second ligand and increases the likelihood of another ligand hitting the binding site. This increases the apparent binding affinity (i.e., the avidity).
ND-Trx3: Nanodevice; Trx: Thioredoxin.
ND-Trx3 assembly
The methods used to assemble ND-Trx3 have been described previously [15]. Briefly, the technology takes advantage of the mechanism of action of DNA methyltransferases [14]. During catalysis, the C6 of cytosine or 5-fluorocytosine undergoes nucleophilic attack from all known cytosine methyltransferases. This breaks the 5–6 double bond in the ring and activates C5 for methyl transfer. After the methyl group is transferred from S-adenosylmethionine (AdoMet) to C5, the normal progress of the reaction is to remove the proton at C5 and the enzyme nucleophile from C6 by β-elimination. However, when fluorine is present at C5 this cannot occur due to the strength of the fluorine–carbon bond. Thus, the progress of the reaction is stalled and a covalent complex is formed between the enzyme and the cytosine ring targeted by the enzyme. In the case of M•EcoRII this is the internal cytosine in the CC(A/T)GG recognition sequence in DNA.
To construct the DNA scaffold, three strands of DNA were synthesized using the convertible F-dC phosphoramidite (TMP-F-dU-CE Phosphoramidite, Glen Research, VA, USA) at one of the M•EcoRII targets on each arm. The complementary strand carries a 5-methylcytosine residue at the M•EcoRII target. A fluorescenated phosphoramidite (5′-fluorescein phosphoramidite, Glen Research) is added to one strand at the predicted junction point in the Y-junction. Trx-M•EcoRII fusion proteins were cloned and purified as previously described [15]. Spontaneous Y-junction assembly was obtained by mixing equimolar amounts of each of the three oligodeoxynucloetides. Once the DNA scaffold was assembled, Trx-M•EcoRII fusion proteins were added along with S-adenosylmethionine and incubation was continued for 2 h at 37°C. Since the 5-fluorocytosine substituted recognition sequences can form covalent links with the methytransferases, the sites activated by fluorocytosine substitution retain the covalently bound fusion proteins in a preselected order set down during DNA synthesis. Assembly of the DNA strand and the fusion proteins was monitored with microfluidics [12].
Reaction of Trx with fluorescein-N-hydroxysuccinimide ester to produce the fluoresceinated Trx bioconjugate (Trx-F)
Trx-F was prepared from E. coli Trx (Promega Corp., WI, USA), with the EZ-Label™ Fluorescein Protein Labeling Kit 53000 (Thermo Fischer, IL, USA). Purification of the bioconjugate was carried out as described by the manufacturer. Protein concentration was determined by the method described in [17]. Activity was determined by reacting the Trx and Trx-F with insulin [18] and spectroscopically measuring absorbance at 650 nm for 120+ min at 5 min intervals. Fluoresceination did not alter measured activity, and activity was retained in Trx-F stored in 10% glycerol at -20°C. Since bacterial Trx has a molecular weight of only 11.7 kD, intact stoichiometry of the bioconjugate was determined from molecular ions observed by mass spectroscopy.
ND-Trx3 & Trx-F binding to cell lines
DU-145 cells were grown on cover slips as previously described [15] and incubated for 1 min in medium containing phosphate buffered saline (PBS), 20 nM and 1 μM Trx-F or ND-Trx3. The slides were processed and analyzed by fluorescent microscopy as previously described [15].
ND-Trx3 & Trx-F binding to frozen tissue sections
Frozen tissues were cut into 5 μm sections, placed on microscope slides and stored at -80°C until used. The slides were brought to room temperature and washed with PBS. After washing, the specimens were incubated for 1 min with 200 nM Trx-F, 20 nM ND-Trx3 in 200 μl of ice cold PBS or with 200 μl of PBS alone. Slides were exposed to ND-Trx3, washed and analyzed by fluorescent microscopy as previously described [15]. A tiled image of the entire tissue slice was obtained to identify regions of ND-Trx3 binding. The tissue slice was photographed at 100× magnification by taking a series of photographs of the tissue slice using the automated microscope stage and tiling the pictures together to create an overall composite image of the specimen.
Imunohistochemistry
Adjacent sections (5-μm thick) of tissue biopsy specimens of prostate cancer or BPH were stained with H&E, and immunohistochemically with monoclonal antibodies to α-smooth muscle actin (α-SMA; BMD 1004–04, Accurate Chemical & Scientific Corp., Wesbury, NY, USA), TxnRD1 (HPA 001395, Sigma-Aldrich, St Louis, MO, USA) or TxnRD2 (HPA 003323, Sigma-Aldrich) as described recently [19]. For each experiment, a control that consisted of an irrelevant monoclonal antibody of the same immunoglobulin subclass and concentration was included to determine specificity of each antibody. Moreover, tissue biopsy sections of prostate cancer with known reactivity with these antibodies were used as positive controls. A pathologist (S.A.I.) independently reviewed the immunostained sections in a fully blinded manner. The data for immunostaining analysis were stratified in two categories, namely strong or weak, based upon the intensity of staining with the antibodies. Slides were photographed using an A×70 automated microscope.
Results
In principle, Trx-F could be used in place of ND-Trx3 in this application if the targets were monomeric. However, three of the four candidate molecules (TxnRD1, TxnRD2, TxnRD3) are known to be homodimers, suggesting that cooperative interaction with the trivalent ND-Trx3 should increase its avidity and generate enhanced performance in binding to freshly frozen tissue specimens. Given our previous demonstration [15] that human and bacterial Trx are structurally homologous and the demonstration that human thioredoxin reductases recognize bacterial Trx in vitro [20], ND-Trx3 is expected to bind most effectively to human thioredoxin reductases. Molecular modeling of ND-Trx3 bound to bacterial thioredoxin reductase (Figure 2) showed that there were no steric constraints that would prevent cooperative binding by any two of the ligands displayed by ND-Trx3. The ligands are tethered to ND-Trx3 via a flexible peptide linker that is long enough to accommodate the binding of the ligands to receptors where the binding sites are a maximum of 20 nm apart. In previous work, ND-Trx3 was shown to bind to MCF-7 breast cancer cells and LnCaP prostate cancer cells, but not PC-3 or primary cultures of prostate epithelial cells when they were incubated with a 1 μM solution for 1 min [15]. In more recent work, a third prostate cancer cell line (DU-145) was also found to bind ND-Trx3. Binding and uptake of ND-Trx3 was observed to be different in each of the cell lines (data not shown). Since the three cancer cell lines are representative of the heterogeneity of the disease, the result suggested that ND-Trx3 binding in frozen prostate tumor specimens might distinguish between aggressive and indolent tumors. In principle, the fluoresceinated ligand could also be used for this purpose; however, as can be seen in Figure 3, the reaction with NHS-fluorescein has the potential to attack ten surface lysine amino groups and thereby significantly alter the surface properties of the molecule. Nevertheless, only two of the ten possible fluorescein attachment sites are close enough to the residues required for binding to interfere with its binding activity, if we assume that modification does not significantly disrupt the proper folding of the molecule. Thus, if the loss of activity of the ligand required the modification of a particular set (q) of sites, and average substitution stoichiometry (s) out of (n) possible substitution sites, combinatorics [21] can be used to obtain the probability that activity would be lost. The probability is given by:
Figure 3. Reaction of thioredoxin with fluorescein-N-hydroxysuccinimide ester to produce a fluorescently labeled thioredoxin bioconjugate.
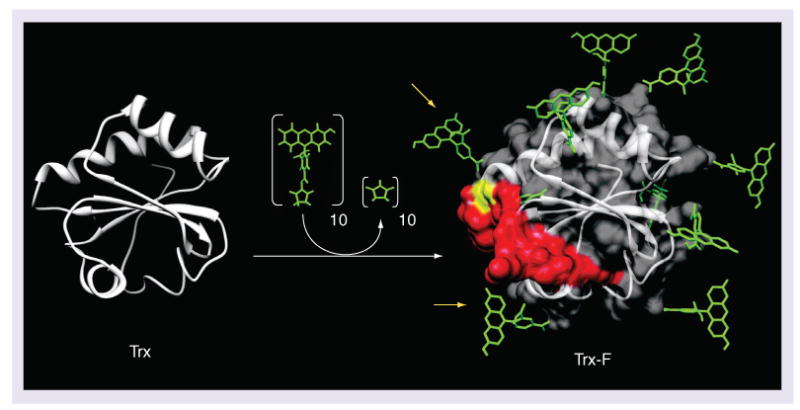
Trx-F was created by reacting fluorescein-N-hydroxysuccinimide with Trx. Escherichia coli Trx contains ten surface lysines that are shown with coupled fluoresceins in the model of the fully substituted molecule on the right. Residues in Trx that are critical for binding to Trx reductase are colored red in the Trx-F model. The amino acid cysteine 32 in Trx (yellow) forms a disulfide bond with Trx reductase. At least two fluoresceins (yellow arrows) could inhibit binding of the Trx to the Trx reductase.
Trx: Thioredoxin; Trx-F: Fluorescently labeled thioredoxin bioconjugate.
Mass spectroscopic measurements (data not shown) yielded an average stoichiometry of one fluorescein per molecule of Trx-F with 16.7% of the molecules labeled. If we take q = 1, s = 1 and n = 10, only approximately 10% of the fluoresceinated molecules are expected to be inactive, and the remaining activity due to fluoresceination would be essentially unchanged at 98.3% of its original level. Consistent with this prediction, we noted that that there was no significant loss in Trx activity due to fluoresceination, as measured by the Trx catalyzed reduction of insulin (data not shown) [18].
We performed a series of dilutions of ND-Trx3 and the Trx-F on tumor specimens and cell lines. When incubated with DU-145 cells, ND-Trx3 was observed to have much higher avidity compared with the fluoresceinated ligand at the same concentrations (Figure 4, lower panels). Significant binding was observed at 500 nM with very strong binding observed at 1 μM. Conversely, weak binding of the Trx-F, was first observed at 1 μM with significant binding at 4 μM (Figure 4, upper panels). Increasing the concentration of the Trx-F to 8 μM did not increase the observed fluorescence (data not shown). The difference in avidity between the Trx-F and ND-Trx3 was even more dramatic when tumor specimens were examined microscopically (Figure 5). In this and subsequent experiments where tissue sections were visualized, a series of 204 fluorescent images were taken at 200× magnification using an automated microscope stage. In order to visualize the fluorescent image of the entire tumor section, the images were tiled into one large file.
Figure 4. ND-Trx3 and fluorescently labeled thioredoxin bioconjugate binding to cell lines.
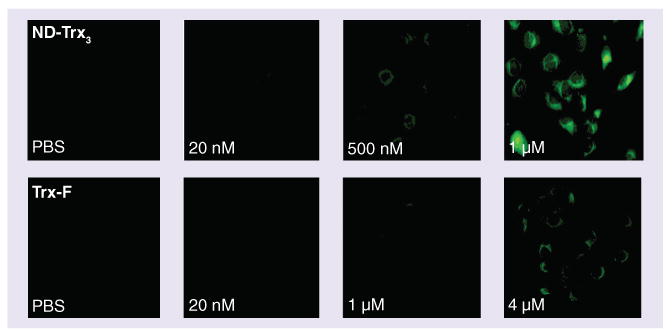
DU-145 cells were incubated for 1 min in growth medium containing PBS with ND-Trx3 or Trx-F over the indicated concentration ranges. Increasing the concentration of Trx-F to 8 μM did not enhance fluorescence beyond that observed with 4 μM.
ND-Trx3: Nanodevice; PBS: Phosphate-buffered saline; Trx-F: Fluorescently labeled thioredoxin bioconjugate.
Figure 5. ND-Trx3 and Trx-F binding to frozen tissue sections.
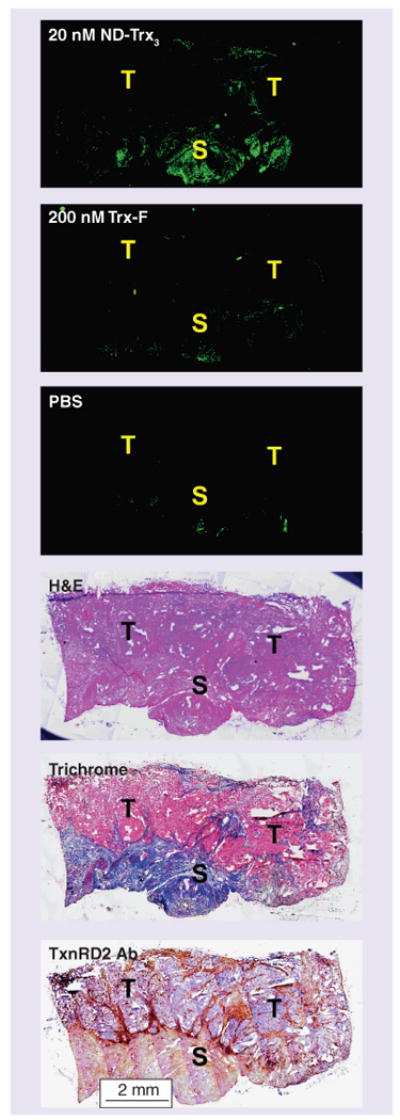
Frozen prostate specimens from regions containing high-grade tumor lesions were incubated or stained as indicated. Tiled images photographed at 100× magnification allow visualization of the entire tumor specimen after size reduction. Tumor (T) and stromal (S) regions are indicated in each panel.
H&E: Hematoxylin and eosin;
ND-Trx3: Nanodevice; PBS: Phosphate-buffered saline; S: Stromal region; T: Tumor region; Trx-F: Fluorescently labeled thioredoxin bioconjugate; TxnRD2 Ab: Thioredoxin reductase 2 antibody.
Based on these results and our previous results with cell lines [15], we began staining frozen tissue sections with 500 nM ND-Trx3. At 500 nM ND-Trx3, strong binding was observed over the entire specimen (data not shown). Dilution titration of ND-Trx3 demonstrated that 20 nM yielded selective binding of ND-Trx3 to frozen prostate tissue sections. This end point was determined as the concentration at which selective fluorescence above background binding was observed in the specimen (Figure 5, ND-Trx3). When adjacent sections of the specimen were incubated with the Trx-F at 10× the concentration of ND-Trx3, it failed to yield the same fluorescence intensity as ND-Trx3 (Figure 5, Trx-F), although it was significantly stronger than the autofluorescence from the PBS control (Figure 5, PBS). This suggests that the avidity of Trx-F towards the Trx targets is low due to the low level of fluoresceination in the chemically modified Trx (16.7%) and the potential 1.7% loss of binding activity during chemical fluoresceination, significantly diminish its fluorescent signal. If we assume a multiple equilibrium [22] for bound ligand where the targeted dimers do not exhibit cooperativity, then at equivalent input concentrations (C) the bound fraction would be proportional to:
for a trivalent ND-Trx3 compared with C(.167) (.9) for Trx-F, where (n) is the number of ligands presented and (s) is the number of binding sites on the interacting protein. When C is much lower than the intrinsic association constant for the binding reaction, the expected ratio is approximately 40:1 in favor of fluorescence from ND-Trx3.
Although fluorescent binding was achieved in certain regions of the specimen with ND-Trx3 (Figure 5, ND-Trx3), it was difficult to determine its source based on fluorescence alone. To determine the origin of the fluorescence, adjacent serial sections of the tumor specimens were stained with both ND-Trx3 and H&E. When tiled, high magnification images were reduced in size so as to display the entire specimen, both the lesion and stromal regions could be identified (Figure 5).
Using this method it was possible to identify the lesion (Figure 5, H&E) and the region surrounding the tumor as reactive stroma (Figure 5, Masson's trichrome). Moreover, the region identified as reactive stroma correlated with the region of ND-Trx3 fluorescence, and thioredoxin reductase antibody staining (Figure 5, TxnRD2 Ab).
As noted above, our initial experiments with frozen human specimens at 500 nM ND-Trx3 gave strong fluorescence from the region in the specimens identified above as the cancer lesion; however, the fluorescence from the lesion was overshadowed by the vastly more intense binding of ND-Trx3 to the reactive stroma surrounding the lesion (data not shown). This suggested that ND-Trx3 had comparable affinities for the cells in the cancer lesion and the DU145 cell line, but that it had a much stronger affinity for the cells in the reactive stromal regions. This is consistent with the cell line data at 20 nM where fluorescence from DU145 is not observed (Figure 5, ND-Trx3). As expected, fluorescence from the cancer lesion was not observed at this concentration, while fluorescence from the surrounding reactive stroma was still clearly visible (Figure 5, ND-Trx3).
At high magnification, it was possible to visualize detailed cellular components near the lesion and in matched regions of BPH (Figure 6). Using these methods we were able to correlate the regions of high fluorescent intensity (Figure 6A) with the same regions in the H&E stained section (Figure 6C) of the adjacent tumor section. While ND-Trx3 showed negligible binding to regions of BPH (Figure 6B & 6D) it showed strong binding activity with the stroma adjacent to the tumor cells, but not the tumor itself (Figure 6A & 6C).
Figure 6. Binding of thioredoxin-targeted ND-Trx3 mirrors binding of antibodies to thioredoxin reductases 1 and 2.
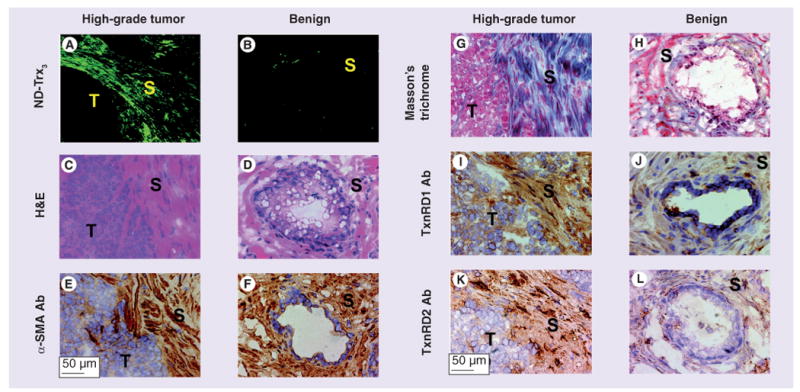
Sections (5-μm thick) of freshly frozen tissue specimens obtained after resection from patients with biopsy-proven prostate cancer (A) or benign prostatic hyperplasia (BPH) (B) were stained with ND-Trx3 or immunostained with antibodies to α-SMA, TxnRD1 or TxnRD2. Serial sections of each case were stained with H&E to confirm the presence of cancer (C), BPH (D) and their cellular characteristics in the particular section. Anti-α-SMA antibody showed strong reactivity with stromal cells in cancer (reddish brown staining [E]) and BPH tissue specimens (reddish brown staining [F]). Masson's trichrome staining was strongly positive for reactive stromal cells near the tumor lesion (G) but not with the stromal cells in BPH specimens (H). Trichrome-stained stromal cells (G) also showed reactivity with anti-TxnRD1 antibody (reddish brown staining [I]) and ND-Trx3 (fluorescence [A]) in cancer, but not BPH tissue specimens (absence of reddish brown staining [J] and fluorescence [B]). A similar specificity and pattern of α-SMA-positive stromal cells with anti-TxnRD2 antibody (reddish brown staining [K]) and ND-Trx3 (fluorescence [A]) was observed in cancer or BPH tissue specimens [L & B]). The sections were counterstained with Harris's hematoxylin (blue nuclear staining). Original magnification: 100×. Tumor (T) and stromal (S) regions are indicated in each panel.
Ab: Antibody; α-SMA: α-smooth muscle actin; H&E: Hematoxylin and eosin; ND-Trx3: Nanodevice; S: Stromal region; T: Tissue region; TxnRD: Thioredoxin reductase.
Frozen tissue sections from 41 patients who underwent robot assisted-prostatectomy were studied. A total of nine of the specimens were too poorly preserved to permit analysis. 19 of the frozen sections, representing 19 excised tumor specimens, were found to lack a cancer lesion. Of the remaining 13 frozen sections representing 13 different patients, nine showed significant fluorescence from the reactive stroma surrounding the tumor lesion when exposed to ND-Trx3 (Figure 6A). In control experiments, we repeated ND-Trx3 binding using benign prostate tissue from sections exhibiting BPH and observed very little binding of the ND-Trx3 (Figure 6B).
In order to determine the identity of the stromal cell type with preferential reactivity with ND-Trx3, tissue sections from randomly selected sets of prostate cancer and BPH were immunostained with anti-α-SMA antibody, a known marker of smooth muscle and myofibroblasts or cancer-associated fibroblasts [23]. The antibody showed strong reactivity with stromal cells in prostate cancer (Figure 6E) and with their counterparts in BPH (Figure 6F) in tissue specimens. The result demonstrated that α-SMA-positive stromal cells that were located directly adjacent to tumor lesions showed binding reactivity with ND-Trx3 (Figure 6A & 6E), whereas α-SMA-positive stromal cells in BPH exhibited no detectable binding reactivity with ND-Trx3 (Figure 6B & 6F).
To determine whether or not ND-Trx3 was binding to the reactive stroma, the sections were stained with Masson's trichrome. This technique has been used to differentiate reactive prostatic stroma from normal prostatic stroma as the smooth muscle cells of prostatic stroma stain red and show orientation of the cells [24]. By contrast, in reactive stroma the cellular fibers become disorganized, are smaller and stain predominantly blue. Images obtained from sections of tumor specimens stained with Masson's trichrome clearly showed that ND-Trx3 was binding to reactive stromal regions (Figure 6A & 6G). The BPH stroma showed a negligible staining with Masson's trichrome (Figure 6H).
In order to attempt to identify the ND-Trx3 target, we evaluated the binding reactivity of antibodies directed at several candidate Trx-interacting proteins. When a TxnRD1 antibody was used to stain tissue sections adjacent to those studied with ND-Trx3 using immunohistochemistry, the stain (Figure 6I) was found to overlap the regions stained by ND-Trx3 (Figure 6A), therefore allowing identification of this dimeric reductase as a target of ND-Trx3 in stromal cells in tumor, but not in their counterparts in BPH tissue specimens (Figure 6B & 6J). An antibody to TxnRD2 (Figure 6K & 6L) produced a pattern of immunoreactivity, which was similar to that obtained with the antibody to TxnRD1, whereas an antibody to Trx-glutathione reductase did not exhibit reactivity with the tissue specimens (data not shown). These data identify dimeric TxnRD1 and TxnRD2 as targets of ND-Trx3 that are overexpressed in the prostate tumor microenvironment-associated stromal cells.
Discussion
We have tested a multivalent nanodevice designed to target homodimeric interacting proteins. Our results suggest that many cancer-specific biomarkers can be identified with native protein or peptide ligands. Moreover, the multivalent nanotechnology described here is superior to ligands carrying bioconjugated chromophores in this application. The technology we describe here is potentially useful for distinguishing prostate tumor microenvironment-associated stromal cells from those present in BPH based on native protein or peptide ligand binding affinities. Furthermore, its capacity for multivalent design makes it particularly useful for identifying dimeric targets. ND-Trx3 showed preferential binding to reactive stroma in prostate tumor specimens and identified dimeric thioredoxin reductases as specific biomarkers that are overexpressed in the tumor microenvironment.
Reactive stroma is a component of the tumor microenvironment that appears to be essential for the growth and maintenance of the tumor [23,25–29]. Most of the stromal cells present in normal prostate tissue exhibit a smooth muscle phenotype that is said to be involved in the normal glandular functions of the prostate. During the development of prostate cancer, there is a stromal response similar to that associated with wound repair in which the normal stromal phenotype changes from a smooth muscle to a myofibroblastic phenotype characterized by blastic cells [30] in what is called reactive stroma. These changes in the stroma are indicative of a change in the microenvironment caused by the presence of the cancer cells [26]. The stromal cells also undergo biochemical changes, including the secretion of growth factors (e.g., TNF-α, TGF-β, VEGF and bFGF) [31] that promote angiogenesis [32]. Moreover, the myofibroblasts express extracellular matrix components and proteases that result in remodeling of the extracellular matrix that may stimulate cancer growth and migration [26], suggesting that these cells may be a key component of reactive stroma. Tumor-associated macrophages are also a component of reactive stroma and may increase the level of oxidative stress in the tumor microenvironment by the secretion of H2O2. Our results suggest that the stromal cells overexpress thioredoxin reductases in response to the locally increased oxidative environment induced by the abortive wound response. In addition to suggesting that thioredoxin reductases may be useful biomarkers of the presence of prostate cancer-associated stromal cells, our findings also raise the possibility of utilizing ND-Trx3 to target therapeutics to the tumor microenvironment. ND-Trx3 could be adapted to deliver nucleic acid therapeutics that induce apoptosis directly to tumor or stromal cells. In addition, reactive stroma is present in most of the solid tumors, thus increasing the range of cancers that may benefit from these findings.
Conclusion
We have developed a nanodevice that binds selectively to the reactive stroma in freshly frozen prostate tissue biopsy specimens obtained from robot-assisted prostatectomies. The modular design of ND-Trx3 indentified TxnRD1 and TxnRD2 as potential biomarkers of reactive stroma in the prostate tumor microenvironment. Moreover, ND-Trx3 has potential to be utilized as prognostic marker of prostate cancer and a vehicle to therapeutically target the prostate cancer microenvironment.
Future perspective
Given the recent progress in our understanding of the tumor microenvironment and its biomarkers, one can anticipate the emergence of noninvasive prognostic tools for prostate cancer diagnosis and prognosis based on this work and the work of others in the field within the next 5–10 years. Moreover, the use of these biomarkers in personalized medicine, surrogate end points and targets in prostate cancer therapy is also near at hand.
Executive summary.
A multivalent nanodevice (ND-Trx3) can be designed to bind homodimeric targets.
The design increased the avidity of the nanodevice for prostate cancer tissue and cell lines compared with the isologous monovalent ligand.
ND-Trx3 bound preferentially to cells in the prostate cancer tumor microenvironment.
Thioredoxin reductases were identified as primary targets of ND-Trx3, thus identifying them as specific biomarkers of tumor microenvironment-associated stromal cells in human prostate cancer.
The results suggest that thioredoxin reductases 1 and 2 may serve as diagnostic biomarkers in prostate cancer, and as targets in the delivery of therapeutics to the tumor microenvironment by ND-Trx3.
Acknowledgments
The authors would like to acknowledge the Synthetic and Biopolymer Chemistry Core, the Pathology Core Facility and the Microscopy Core of the City of Hope Comprehensive Cancer Center (P30 CA33572) for their help and advice.
Molecular graphics images were produced using the UCSF Chimera package from the Computer Graphics Laboratory, University of California San Francisco (supported by NIH P41 RR-01081). This study was supported by grants CA102521 and CA136055 from the US National Cancer Institute of the NIH, and by grant W81XWH-08–1-0517 from the Congressionally Directed Medical Research Program of the US ARMY to S.S.S.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research: The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Dalrymple MB, Pfleger KD, Eidne KA. G protein-coupled receptor dimers: functional consequences, disease states and drug targets. Pharmacol Ther. 2008;118(3):359–371. doi: 10.1016/j.pharmthera.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Berque-Bestel I, Lezoualc'h F, Jockers R. Bivalent ligands as specific pharmacological tools for G protein-coupled receptor dimers. Curr Drug Discov Technol. 2008;5(4):312–318. doi: 10.2174/157016308786733591. [DOI] [PubMed] [Google Scholar]
- 3.Burgess AW. EGFR family: structure physiology signalling and therapeutic targets. Growth Factors. 2008;26(5):263–274. doi: 10.1080/08977190802312844. [DOI] [PubMed] [Google Scholar]
- 4.Prezeau L, Rives ML, Comps-Agrar L, Maurel D, Kniazeff J, Pin JP. Functional crosstalk between GPCRs: with or without oligomerization. Curr Opin Pharmacol. 2010;10(1):6–13. doi: 10.1016/j.coph.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Myc A, Patri AK, Baker JR., Jr Dendrimer-based BH3 conjugate that targets human carcinoma cells. Biomacromolecules. 2007;8(10):2986–2989. doi: 10.1021/bm700727g. [DOI] [PubMed] [Google Scholar]
- 6.Thomas TP, Shukla R, Kotlyar A, Kukowska-Latallo J, Baker JR., Jr Dendrimer-based tumor cell targeting of fibroblast growth factor-1. Bioorg Med Chem Lett. 2010;20(2):700–703. doi: 10.1016/j.bmcl.2009.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7▪.Mecke A, Lee I, Baker JR, Jr, Holl MM, Orr BG. Deformability of poly(amidoamine) dendrimers. Eur Phys J E Soft Matter. 2004;14(1):7–16. doi: 10.1140/epje/i2003-10087-5. Documents flexibility of targeted dendrimers. [DOI] [PubMed] [Google Scholar]
- 8.Lidke DS, Nagy P, Heintzmann R, et al. Quantum dot ligands provide new insights into ERBB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22(2):198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 9▪.Deyev SM, Waibel R, Lebedenko EN, Schubiger AP, Pluckthun A. Design of multivalent complexes using the barnase*barstar module. Nat Biotechnol. 2003;21(12):1486–1492. doi: 10.1038/nbt916. Discusses effects of use of multivalency in engineered antibody biotechnology. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. Natural supramolecular building blocks. Wild-type cowpea mosaic virus. Chem Biol. 2002;9(7):805–811. doi: 10.1016/s1074-5521(02)00165-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Lin T, Johnson JE, Finn MG. Natural supramolecular building blocks. Cysteine-added mutants of cowpea mosaic virus. Chem Biol. 2002;9(7):813–819. doi: 10.1016/s1074-5521(02)00166-7. [DOI] [PubMed] [Google Scholar]
- 12.Clark J, Shevchuk T, Swiderski PM, et al. Mobility-shift analysis with microfluidics chips. Biotechniques. 2003;35(3):548–554. doi: 10.2144/03353rr01. [DOI] [PubMed] [Google Scholar]
- 13.Clark J, Singer EM, Korns DR, Smith SS. Design and analysis of nanoscale bioassemblies. Biotechniques. 2004;36(6):992–996. 998–1001. doi: 10.2144/04366RV01. [DOI] [PubMed] [Google Scholar]
- 14.Smith SS, Niu L, Baker DJ, Wendel JA, Kane SE, Joy DS. Nucleoprotein-based nanoscale assembly. Proc Natl Acad Sci USA. 1997;94(6):2162–2167. doi: 10.1073/pnas.94.6.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15▪▪.Singer EM, Smith SS. Nucleoprotein assemblies for cellular biomarker detection. Nano Lett. 2006;6(6):1184–1189. doi: 10.1021/nl060549h. Discusses interaction of ND-Trx 3 with cell lines. [DOI] [PubMed] [Google Scholar]
- 16.Gleason DF. Histologic grading of prostate cancer: a perspective. Hum Pathol. 1992;23(3):273–279. doi: 10.1016/0046-8177(92)90108-f. [DOI] [PubMed] [Google Scholar]
- 17.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 18.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254(19):9627–9632. [PubMed] [Google Scholar]
- 19.Saha B, Chaiwun B, Imam SS, et al. Overexpression of E-cadherin protein in metastatic breast cancer cells in bone. Anticancer Res. 2007;27(6B):3903–3908. [PubMed] [Google Scholar]
- 20.Gromer S, Arscott LD, Williams CH, Jr, Schirmer RH, Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J Biol Chem. 1998;273(32):20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 21.Cameron PJ. Combinatorics: topics, techniques, algrithms. Cambridge University Press; Cambridge, UK: 1994. [Google Scholar]
- 22.Tanford C. The Physical Chemistry of Macromoleucles. John Wiley & Sons, Inc; NY, USA: 1961. [Google Scholar]
- 23.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 24▪▪.Ayala G, Tuxhorn JA, Wheeler TM, et al. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res. 2003;9(13):4792–4801. Discusses the prognostic utility of reactive stroma. [PubMed] [Google Scholar]
- 25.Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. J Urol. 2001;166(6):2472–2483. [PubMed] [Google Scholar]
- 26.Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res. 2002;8(9):2912–2923. [PubMed] [Google Scholar]
- 27.Chung LW, Baseman A, Assikis V, Zhau HE. Molecular insights into prostate cancer progression: the missing link of tumor microenvironment. J Urol. 2005;173(1):10–20. doi: 10.1097/01.ju.0000141582.15218.10. [DOI] [PubMed] [Google Scholar]
- 28.Galie M, Sorrentino C, Montani M, et al. Mammary carcinoma provides highly tumourigenic and invasive reactive stromal cells. Carcinogenesis. 2005;26(11):1868–1878. doi: 10.1093/carcin/bgi158. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa H, Liyanarachchi S, Davuluri RV, et al. Role of cancer-associated stromal fibroblasts in metastatic colon cancer to the liver and their expression profiles. Oncogene. 2004;23(44):7366–7377. doi: 10.1038/sj.onc.1208013. [DOI] [PubMed] [Google Scholar]
- 30▪▪.Yanagisawa N, Li R, Rowley D, et al. Stromogenic prostatic carcinoma pattern (carcinomas with reactive stromal grade 3) in needle biopsies predicts biochemical recurrence-free survival in patients after radical prostatectomy. Hum Pathol. 2008;39(2):282–291. doi: 10.1016/j.humpath.2007.04.025. Discusses role of reactive stromal grading approach to preoperative prognosis. [DOI] [PubMed] [Google Scholar]
- 31.Sunderkotter C, Steinbrink K, Goebeler M, Bhardwaj R, Sorg C. Macrophages and angiogenesis. J Leukoc Biol. 1994;55(3):410–422. doi: 10.1002/jlb.55.3.410. [DOI] [PubMed] [Google Scholar]
- 32.Yang F, Tuxhorn JA, Ressler SJ, Mcalhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65(19):8887–8895. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]


