Abstract
Radiotherapy and chemotherapy are effective cancer treatments due to their ability to generate DNA damage. The major lethal lesion is the DNA double-strand break (DSB). Human cells predominantly repair DSBs by non-homologous end joining (NHEJ), which requires Ku70, Ku80, DNA-PKcs, DNA ligase IV and accessory proteins. Repair is initiated by the binding of the Ku heterodimer at the ends of the DSB and this recruits DNA-PKcs, which initiates damage signaling and functions in repair. NHEJ also exists in certain types of bacteria that have dormant phases in their life cycle. The Mycobacterium tuberculosis Ku (Mt-Ku) resembles the DNA-binding domain of human Ku but does not have the N- and C-terminal domains of Ku70/80 that have been implicated in binding mammalian NHEJ repair proteins. The aim of this work was to determine whether Mt-Ku could be used as a tool to bind DSBs in mammalian cells and sensitize cells to DNA damage. We generated a fusion protein (KuEnls) of Mt-Ku, EGFP and a nuclear localization signal that is able to perform bacterial NHEJ and hence bind DSBs. Using transient transfection, we demonstrated that KuEnls is able to bind laser damage in the nucleus of Ku80-deficient cells within 10 sec and remains bound for up to 2 h. The Mt-Ku fusion protein was over-expressed in U2OS cells and this increased the sensitivity of the cells to bleomycin sulfate. Hydrogen peroxide and UV radiation do not predominantly produce DSBs and there was little or no change in sensitivity to these agents. Since in vitro studies were unable to detect binding of Mt-Ku to DNA-PKcs or human Ku70/80, this work suggests that KuEnls sensitizes cells by binding DSBs, preventing human NHEJ. This study indicates that blocking or decreasing the binding of human Ku to DSBs could be a method for enhancing existing cancer treatments.
Introduction
Genomic DNA is damaged by endogenous reactive oxygen species on a daily basis and has to be repaired to maintain the integrity of the DNA sequence. Damage includes oxidative base damage, abasic sites, single-strand breaks (SSBs) and double-strand breaks (DSBs). These same types of damages are introduced by radiotherapy (1) and certain chemotherapies, but these cancer treatments are believed to be more effective at killing cells due to the clustering of damage, resulting in DSBs and/or more complex lesions (for review, see ref. 2). Without repair, DSBs can result in the loss of genetic information, and aberrant repair can generate chromosomal translocations, deletions and aberrations (3–5). DSBs, or DSBs with near-by oxidative damage, are potentially the most lethal lesions. Mammalian cells have predominantly two mechanisms to repair DSBs: non-homologous end joining (NHEJ) and homology-directed repair, which includes homologous recombination and single-strand annealing. More recently, a “back-up” mechanism was identified that repairs DSBs when the classical NHEJ is not functioning or is overwhelmed, and this is called alternative NHEJ (for review, see ref. 6).
Homologous recombination (for review, see ref. 7) requires an intact copy of the DNA, and hence this type of repair occurs during late S phase and the G2 phase of the cell cycle when sister chromatids are in close proximity to provide homologous sequences. The first stage of repair is resection of the DNA from the DSB to create a long single-stranded region of DNA. A number of nucleases such as Mre11-Rad50-Nbs1 (MRN), CtIP and hExo1 are involved in this step. The single-stranded DNA is then stabilized by the binding of RPA prior to formation of a Rad51 nucleoprotein filament. The nucleoprotein filament searches for homology and performs strand invasion, and DNA synthesis occurs from the invading strand copying the homologous sequence. Repair is therefore accurate. If strand invasion does not occur, then the resected DNA ends can be repaired by single-strand annealing resulting in deletions. Alternative NHEJ also requires DNA resection to reveal microhomology regions at the termini that are then used by DNA ligase III to complete repair. This alternative pathway also results in deletions.
NHEJ (for review, see ref. 8) in mammalian cells occurs during all phases of the cell cycle and is initiated by the binding of the Ku70/80 heterodimer to the DSB termini. Ku acts like a docking protein on the DNA for repair proteins such as the DNA-PKcs: Artemis complex and DNA polymerase λ or μ. Once the termini have been processed, the ligase complex consisting of DNA ligase IV, XLF and XRCC4 binds to complete repair. Each of the NHEJ repair enzymes is able to function independently in a purified in vitro system, even without Ku. However, when there are <4 bp of homology between the two termini Ku stimulates ligation (9), and in the cell Ku is believed to protect the DNA termini from excessive degradation and stabilize the ligase complex with DNA. Ku is therefore an essential component of NHEJ. In fact, cells deficient in Ku80 are sensitive to damaging agents generating DSBs (10,11). NHEJ in mammalian cells is believed to be important for the repair of DSBs generated by radiotherapy and cancer treatments; where as homologous recombination is important for the repair of DSBs during replication. Of course, DSBs can occur during S phase due to the stalling of replication at damage in the DNA and collapse of the replication forks (12,13).
The development of complementary treatments to manipulate the cancer cell’s DNA repair capacity is a way to improve clinical outcomes of well-established therapies. Small molecule inhibitors have been developed to block ataxia telangiectasia mutated (ATM) signaling, DNA-PKcs, PARP, DNA ligases and the MRN complex (for review, see refs. 14,15). By targeting specific enzymes, specific repair pathways can be compromised and these inhibitors have proven successful at sensitizing cells to cancer treatments. However, the targeted tumor expression of a molecule to bind DSBs and block all the DSB repair pathways by preventing human Ku binding and/or inhibiting DNA end resection could also be an effective method for enhancing cell sensitivity to radiotherapy and chemotherapy. We hypothesized that the bacterial Ku from Mycobacterium tuberculosis could bind DSBs in the mammalian genome and sensitize human cancer cells to DNA damaging agents. NHEJ in M. tuberculosis requires two proteins (16,17): a Ku protein (Mt-Ku) and an ATP-dependent ligase (Ligase D). The Mt-Ku protein is only ∼30 kDa and forms a homodimer that binds DNA termini (18). This binding recruits the Ligase D via the N-terminus of the ligase (19,20). Bacterial Ku therefore functions in a similar way to human Ku; binding DNA termini and recruiting the next protein required for repair. Bacterial Ku is similar in structure to the human Ku core DNA binding domain but lacks the N- and C-termini of human Ku80 and Ku70 (18). Since the C-terminus of Ku80 binds DNA-PKcs (21–23), we hypothesized that this bacterial Ku would be unable to communicate with DNA-PKcs and possibly other human Ku interacting repair proteins. In the work presented here, we examine the ability of Mt-Ku to interact with human NHEJ proteins, to bind DSBs in the mammalian genome and to sensitize human cells to DNA damage.
Materials and methods
Bacterial strains
A wild-type Escherichia coli strain (BW35-Hfr KL16(PO-45) thi-1relA1 spoT1 e14− λ−), previously obtained from Dr Susan S. Wallace (University of Vermont, Burlington, VT, USA), provides the background for the strains used in this work unless stated otherwise. BWKuLig#2 and BWLig express Mt-Ku and Mt-Ligase D and Mt-Ligase D, respectively (17). BW23474 (17,24) was obtained from Coli Genetics Stock Center (Yale University, New Haven, CT, USA) and used to propagate the derivatives of pLA2.
Mammalian cells
Xrs 6 and U2OS cells were grown in α-MEM with 10% fetal bovine serum at 37°C and 5% CO2. U2OS cells expressing EGFPnls or KuEnls were grown in media supplemented with 200 μg/ml Geneticin sulfate (G418).
Oligodeoxyribonucleotides
The oligodeoxyribonucleotides (oligonucleotides) used were purchased from Operon Technologies Inc. (Alameda, CA, USA) or the DNA facility at Iowa State University. The oligonucleotides EGFP-1, EGFP-2, KuNLS3 and KuNLS4 contained 5′ phosphates. The sequences for the oligonucleotides were as follows: EGFP-1, d(GTACAAGCTTGAATTCTGCAGTCGACGGTACACCG); EGFP-2, d(GATCCGGTGTACCGTCGACTGCAGAATTCAAGCTT); KuNLS3, d(AGCTTGGGAAAGTTACCAAGAGAAAACACGATAATGAAGGTTCTGGAAGCAAAAGGCCCAAGGTGTGAG); KuNLS4, d(GATCCTCACACCTTGGGCCTTTTGCTTCCAGAACCTTCATTATCGTGTTTTCTCTTGGTAACTTTCCCA). The following primers were used for PCR of the Mt-Ku coding sequence: 5′, d(GAATTCGCTAGCGGTACCATATGCGAGCCATTTGGACG); 3′, d(GGTAGCGCTAGCGGAGGCGTTGGGACGTT). The following primers were used to sequence the Mt-Ku mammalian expression construct: EGFP3, d(CAACGAGAAGCGCGATCAC) and EGFP4, d(TGACGCAAATGGGCGGT), and CRIM1, d(ATCCATAAGATTAGCGG) was used to sequence the Mt-Ku bacterial expression construct. The primers (24) used for PCR to check for integration of pLAKuEnls and to confirm the presence of the Mt-Ligase D expression construct were: P1λ, d(GGCATCACGGCAATATAC); P4λ, d(TCTGGTCTGGTAGCAATG); P1HK, d(GGAATCAATGCCTGAGTG); P4HK, d(GGCATCAACAGCACATTC); P2, d(ACTTAACGGCTGACATGG); P3, d(ACGAGTATCGAGATGGCA). The following primers were used to generate the double-stranded 53 mer by PCR for the in vitro binding studies: d(GTGCTAGAGCTTGCTACGAC) and d(TGGAGAATCCCGGTGC).
Plasmids
pLA2 and pINT-ts, were obtained from the Coli Genetics Stock Center (Yale University, New Haven, Connecticut). pINT-ts encodes carbenicillin resistance, while pLA2 encodes kanamycin resistance in E. coli. pLA2 and its derivatives were replicated in bacterial strain BW23474. The two plasmids used in the end-joining assay were pBestluc (Promega, Madison, WI, USA) that expresses firefly luciferase and confers carbinicillin resistance (100 μg/ml) and pACYC184 (New England Biolabs, Beverly, MA, USA) that encodes chloramphenicol and tetracycline resistance genes. Bacteria containing pACYC184 were grown on LB containing 34 μg/ml chloramphenicol.
pEGFP-Actin and pDSRED2nuc were obtained from Clontech (Mountain View, CA) and the plasmid expressing a fusion of YFP-Ku80 is a derivative of pCDNA3 (25). These plasmids express fluorescent proteins from a cytomegalovirus immediate early promoter and confer kanamycin resistance to bacteria and neomycin resistance to mammalian cells.
The Mt-Ku coding region was amplified using primers (5′ and 3′) that contained an Nhe I site. The coding region was amplified from a pET16b plasmid carrying the RV0937c coding region, which was obtained from Dr Aidan Doherty (University of Sussex, UK). The fragment was digested with NheI and inserted into the pDSRED2nuc plasmid at Nhe I to generate pKuDSREDnuc. This plasmid was used as a source for the Mt-Ku insert but could not be used for mammalian expression studies due to the formation of aggregates in the nucleus of mammalian cells.
To generate an expression vector to express EGFP in mammalian cells, pEGFP-Actin was digested with BsrG I and BamH I to remove the actin coding sequence. The oligonucleotides EGFP-1 and EGFP-2 were annealed and ligated to the BsrG I-BamH I linear DNA to obtain pEGFP. The addition of this sequence introduced a Hind III restriction site and positioned the EGFP coding sequence in-frame with a translation stop codon. To target the expression of EGFP to the nucleus, the KuNLS3 and KuNLS4 oligonucleotides were annealed and ligated to the Hind III-BamH I sites in pEGFP to create pEGFPnls. This double-stranded oligonucleotide encodes the human Ku70 539-556 peptide, which is a nuclear localization signal. DNA sequencing confirmed the in-frame addition of the nuclear localization signal and a translation stop codon to EGFP.
To generate a mammalian expression vector to express a fusion of Mt-Ku and EGFP in the nucleus, the Mt-Ku coding sequence was isolated on a DNA fragment with Nhe I termini from pKuDSREDnuc and ligated into the Nhe I site 5′ to the ATG of EGFP in pEGFPnls. The DNA encoding the KuEnls protein was sequenced to make sure mutations had not been introduced during the production of the vector.
To generate a construct to test the function of the KuEnls protein in bacteria, the coding sequence for KuEnls was isolated from pKuEnls on a DNA fragment with a 5′ Nde I overhang and a blunt 3′ terminus. This fragment was ligated downstream of the arabinose-inducible promoter in pLA2 at the Nde I and Sma I sites to generate pLAKuEnls.
Integration of pLAKuEnls into the E. coli chromosome
Integrating pLAKuEnls into the E. coli chromosome was performed as described by Haldimann and Wanner (24). Briefly, the BWLig strain (17) was transformed with pInt-ts, which only replicates at 30°C. pInt-ts expresses an integrase that recognizes the lambda bacteriophage attachment site in the E. coli chromosome and pLAKuEnls. BWLig bacteria containing pInt-ts were grown and prepared for electroporation. Bacteria were transformed with 100 ng of pLAKuEnls vector and grown at 37°C for 1 h, followed by 42°C for 30 min. Growth at these temperatures prevents the replication of pInt-ts and activates the integrase to promote integration of pLAKuEnls into the host chromosome. A portion of the culture was grown overnight at 37°C on solid medium containing 10 μg/ml kanamycin. To ensure the antibiotic resistance gene was integrated, the colonies were then grown on solid medium without antibiotic before replating again on solid medium containing 10 μg/ml kanamycin. The new strain (BWKuEnlsLig) was tested for carbenicillin resistance to ensure that pInt-ts had been “cured” from the bacteria.
Primers P1, P2, P3 and P4 were used to PCR the BWKuEnlsLig strain to determine successful single-copy vector integration at the lambda site. The presence of the Mt-Ligase D vector at the HK022 site in the genome was also confirmed by PCR. PCR was performed as previously described (17,24).
Western analysis of bacterial extracts
Bacteria were grown, induced with 0.2% l-Arabinose for 1 h and cell-free extracts prepared as previously described (20). The protein concentration was determined according to the Bradford method using the Bio-Rad Protein Assay reagent (Rio-Rad, Hercules, CA, USA). Protein (50 μg) was electrophoresed through a tris-glycine 4–20% gradient sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred by electroblotting to 0.2-μm nitrocellulose membrane. Two membranes were prepared: one was probed with an anti-His-tag monoclonal antibody (final concentration 0.2 μg/ml; Novagen, Madison, WI, USA) and the other with an anti-EGFP antibody (diluted to 1: 1000; Roche, Madison, WI) according to the manufacturer’s recommendations. A secondary horseradish peroxidase antibody (diluted 1:3000; GE Amersham, Piscataway, NJ, USA) and chemiluminescent substrate (ECL-plus substrate, GE Amersham) was subsequently used to visualize the His-tagged and the EGFP-fusion proteins using autoradiography.
Plasmid end-joining assay
The end-joining assay was performed using Cla I-linearized pBestluc. The digested DNA was purified twice through a 0.8% agarose gel using the Qiaquick Gel Extraction kit (Qiagen Inc., Valencia, CA, USA). To prepare electrocompetent bacteria, the strains were grown as previously described (17,20). Two cultures were prepared: one grown only in LB and one grown for 1 h in LB containing 0.2% l-Arabinose to induce expression of the NHEJ proteins. Linearized pBestluc (50 ng) and 0.1 ng pACYC184 were co-transformed into the bacteria using the Bio-Rad Gene Pulser Xcell Electroporation System (Hercules, CA, USA) at 2.5 KV, 200 Ω and 25 μF, transferred to 1.5 ml LB and grown for 1.5 h at 37°C and 250 r.p.m. A portion of the cultures was plated in triplicate on solid medium containing either 100 μg/ml carbenicillin or 34 μg/ml chloramphenicol and grown overnight at 37°C. Re-circularization of pBestluc in the bacteria results in carbenicillin-resistant colonies. The colonies were counted and a ratio of the carbenicillin-resistant (CarbR) colonies/chloramphenicol-resistant (CmR) colonies was calculated for each transformation. Since pACYC184 encodes chloramphenicol resistance, calculating this ratio enables each sample to be normalized for transformation efficiency. This ratio corresponds to the total repair. The CarbR colonies were then transferred to nylon membranes and sprayed with firefly luciferase substrate to determine the number of colonies that either expressed active (Luc+) or inactive (Luc−) firefly luciferase. Luc+ colonies are due to accurate repair of pBestluc, while Luc− colonies are generated by inaccurate repair. A ratio of CarbR/CmR was determined for each transformation for the Luc+ and Luc− colonies.
The CarbR/CmR ratios from the end-joining assay were compared to the corresponding BWKuLig#2 data using the Instat3 program and the Mann–Whitney test. All P-values <0.05 were considered statistically significant.
Transfection of mammalian cells
The xrs 6 cell line was transiently transfected with 2 μg of pEGFPnls, pKuEnls or YFP-Ku80 using Fugene 6 (Roche). Imaging and laser micro-irradiation were performed after 48 h.
To generate stably transfected cell lines, U2OS cells were transfected with Mlu I-linearized pEGFPnls or pKuEnls using Fugene 6. After 24 h, cells were trypsinized and grown in media containing 400 μg/ml G418. Single colonies were replated after 13 days and grown to generate the U2OS cell lines expressing KuEnls or EGFPnls. Cells were analyzed using flow cytometry to determine the percentage of green fluorescent cells in the population. The KuEnls-expressing cell line was very stable with a green fluorescent population of ≥95%, while with continued passage the vector control cell line expressing EGFPnls contained a decreasing percentage of green fluorescent cells. This may have been due to a high expression level per cell of EGFPnls, as pEGFPnls contained a Kozak consensus translation initiation site upstream of the protein-coding sequence. The Kozak sequence was not present on the pKuEnls vector. Only cell populations with ≥90% of cells fluorescing green were used in experiments. Cell extracts were prepared from stable cell lines and western analysis performed as described above. The anti-EGFP antibody (diluted to 1: 1000; Roche) was used to detect EGFPnls and KuEnls, while anti-Ku86, anti-Ku70 and anti-DNA-PKcs antibodies (all diluted 1:200, Santa Cruz Biotechnology, Inc. Santa Cruz, CA) were used to detect Ku80, Ku70 and DNA-PKcs, respectively. The anti-actin antibody (BD Biosciences Pharmingen, San Diego, CA) was used at a dilution of 1:5000.
Micro-irradiation of cells
Cells were cultured in 35-mm glass-bottom culture dishes and the growth medium replaced with carbon dioxide-independent medium before analysis. During imaging, micro-irradiation and analysis, cells were maintained at 37°C. DSBs were introduced into the genomic DNA of the cultured cells using a pulsed nitrogen laser (365 nm, 10 Hz pulse) as previously described (25). The Micropoint Ablation Laser System (Photonic Instruments, Inc.) was directly coupled to the epifluorescence path of the microscope (Axovert 200M; carl Zeiss MicroImaging Inc.) and focused through a Plan-Apochomat 63×/NA 1.4 oil immersion objective. The output of the laser was set at 75% of the maximum.
Survival analysis
A colony-forming assay was used to determine survival of cells following treatment with the damaging agent. Briefly, subconfluent cells were trypsinized, serially diluted, and attached to 100-mm tissue culture dishes. The appropriate cell number for each dose was added to the dishes to obtain an optimal number of surviving colonies (70–150). Dishes were prepared in triplicate at two cell densities per dose point. Cells were incubated for 4 h to allow the cells to attach. To treat with bleomycin sulfate or hydrogen peroxide, the chemical was added to the medium of the cells. After 24 h, the medium containing bleomycin sulfate was replaced with fresh medium. For treatment with hydrogen peroxide, the medium was not replaced due to the rapid degradation of hydrogen peroxide. To treat with ultraviolet radiation, cells were allowed to attach to the dish. The medium was then replaced with 2 ml PBS prior to irradiation with 254-nm light. The PBS was replaced with cell medium post-irradiation. Cells were allowed to grow for 12 days. Colonies were stained with 1% gentian violet, 70% ethanol and 5% formaldehyde. Cell toxicity was measured by the reduction in colony-forming ability. At least three experiments were performed for each agent with each cell line. Data were fitted to a linear-quadratic model with a non-linear regression in which the ln(Surviving fraction) of all the data versus dose was fitted by polynomial regression. The Student t-test was used to compare the α and β values of the KuEnls- and EGFPnls-expressing cell lines. The survival data for the cell lines were also compared by calculating the area under the curve for each cell line using the Trapezoidal Rule, and the Student t-test was used to compare the values. These statistical analyses were performed using SAS 9.2 (SAS Inc. Gary, NC).
Growth analysis
Cells were plated at 5 × 104/35-mm tissue culture dish and incubated at 37°C and 5% CO2. Increase in cell number was measured as a function of time. Every 24 h, cells were trypsinized from triplicate dishes and counted using a hemocytometer. Linear regression was used to analyze the exponential portion of growth (48–120 h) to obtain the cell doubling time. This experiment was repeated three times for each cell line.
The Instat3 program and the Mann–Whitney test were used to compare the doubling times of the cell lines. P-values <0.05 were considered statistically significant.
Electromobility shift assay
pGEM-3Z/601 reverse was used as the template for PCR to generate a double-stranded 53 mer for the electromobility shift assays. The DNA was labeled on the 3′ terminus using α32P-dGTP and T4 DNA polymerase. Human Ku (Ku70/Ku80) was expressed using recombinant baculovirus in Sf9 insect cells and purified according to Ono et al. (26), and Mt-Ku was expressed from pET16b in E. coli and purified according to Weller et al. (18). Human DNA-PKcs was purified from HeLa cells according to Cary et al. (27). Binding reactions (10 μl) containing human Ku, Mt-Ku, DNA-PKcs or combinations of these proteins with the 32P-labeled double-stranded DNA probe (6–400 nM) were performed in buffer containing 25 mM Tris–HCl, pH 7.9, 25 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM DTT and 5% glycerol. Binding reactions were performed for 30 min at room temperature, placed on ice and loading buffer (0.04% bromophenol blue, 0.04% xylene cyanol, 5% glycerol) added to the reactions. Samples were subjected to electrophoresis through a 4% Tris-glycine native polyacrylamide gel prior to analysis using a phosphorimager.
Co-immunoprecipitation
Pure human Ku (10 μg) and pure his-tagged Mt-Ku (10 μg) were mixed in 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT. Anti-Ku70 monoclonal antibody (generated by the Chen laboratory, UT Southwestern) or Ni-NTA beads were added to the pure proteins, followed by protein G agarose. Samples were incubated at 4°C for 5 h with agitation prior to washing to remove non-specific bound protein. The bound proteins were finally released by boiling and subjected to electrophoresis through a 10% SDS-polyacrylamide gel. The proteins were transferred to a membrane and western analysis performed with an anti-Ku70 monoclonal antibody, anti-his HRP antibody (Pierce, Rockford, IL) or an anti-Ku80 antibody (generated by the Chen laboratory, UT Southwestern).
Results
Generation of a plasmid to express an EGFP-tagged Mt-Ku in the nucleus of mammalian cells
Our previous experiments using the SV40 nuclear localization signal resulted in Mt-Ku protein aggregate formation in the nucleus of mammalian cells. Human Ku70 has a C-terminal peptide sequence (amino acids 539–556 KVTKRKHDNEGSGSKRPK) that is essential for the nuclear localization of Ku70 (28) but is not required for binding Ku80 (29). The sequence encoding the nuclear localization signal (nls) from human Ku70 was therefore used to target EGFP and the EGFP fusion with Mt-Ku to the nucleus.
Wang et al. (30) previously demonstrated that addition of the Bacillus subtilis YkoV (Ku) protein to the N-terminus of EGFP produced a YkoV-EGFP protein that could localize to the nucleoid during spore germination. Our early experiments also determined that a fusion of EGFP at the N-terminus of Mt-Ku resulted in an EGFP-Mt-Ku protein that would not bind DNA damage. The Mt-Ku coding sequence was therefore ligated upstream of EGFP in pEGFPnls. This created a vector to express the Mt-Ku fusion protein (KuEnls) (Figure 1A) in the nucleus of mammalian cells. The nuclear localization of KuEnls and EGFPnls was confirmed following transient transfection of the vectors into mouse fibroblasts (data not shown).
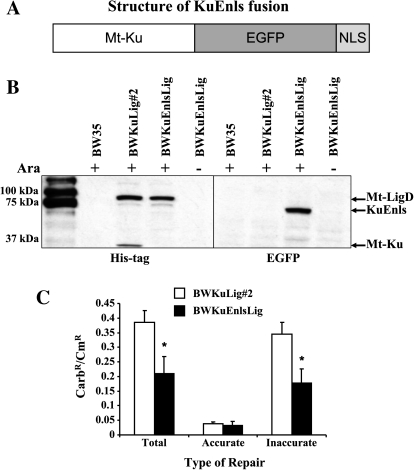
Fig. 1.
The KuEnls protein is able to function in bacterial NHEJ. An open reading frame was generated to express KuEnls, which consists of the Mycobacterium tuberculosis Ku protein fused to the N-terminus of EGFP and the nuclear localization signal from Ku70 fused to the C-terminus of the EGFP protein (A). This open reading frame was transferred into a vector downstream of the bacterial araB promoter and integrated into the Escherichia coli chromosome of BWLig (17) to create the BWKuEnlsLig. BW35 is the parental strain and BWKuLig#2 can be induced to express histidine-tagged M. tuberculosis Ku (Mt-Ku) and Ligase D (Mt-LigD). Bacteria were grown with or without arabinose and extracts examined by western analysis to detect expression of Mt-Ku, Mt-LigD and KuEnls (B). A plasmid re-circularization assay was used to check for DSB repair activity of BWKuLig#2 and BWKuEnlsLig (C). The results are shown for total, accurate and inaccurate repair of Cla I-linearized pBestluc for bacteria grown in the presence of arabinose. * indicates P < 0.05 compared to BWKuLig#2. At least four transformations were performed. The mean and standard deviation are shown.
The KuEnls fusion protein can function with Mt-Ligase D to re-circularize linear plasmid in E. coli
Previously, we generated E. coli strains expressing Mt-Ku (BwKu), Mt-Ligase D (BWLig) or Mt-Ku and Mt-Ligase D (BWKuLig#2; ref. 17) and demonstrated that both Mt-Ku and Mt-Ligase D were required to re-circularize plasmid DNA. To test whether the KuEnls protein could bind DSBs and function like Mt-Ku, we integrated an arabinose-inducible KuEnls expression vector (pLAKuEnls) into the lambda integration site in the genome of strain BWLig. This created the BWKuEnlsLig bacterial strain. To confirm arabinose-inducible expression of the bacterial NHEJ proteins, cell-free extracts were generated from the wild-type strain (BW35), BWKuLig#2 and BWKuEnlsLig after growth in LB or LB containing l-Arabinose, and western analysis was performed. The wild-type Mt-Ligase D and Mt-Ku were detected with an anti-His tag antibody as these proteins have N-terminal histidine tags, while the KuEnls protein was detected with an anti-EGFP antibody. As can be seen from Figure 1B, Mt-Ligase D is expressed in BWKuLig#2 and BWKuEnlsLig when the bacteria are grown in arabinose. Mt-Ku was only detected in BWKuLig#2, while the KuEnls protein was expressed only in BWKuEnlsLig and was ∼65 kDa as expected.
To determine whether KuEnls could bind DNA termini and function in repair, a plasmid re-circularization assay was performed. BWKuLig#2 or BWKuEnlsLig were grown in LB in the presence or absence of arabinose and transformed with Cla I-linearized pBestluc and pACYC184. Re-circularization of pBestluc results in carbenicillin-resistant (CarbR) bacteria, and pACYC184 encodes chloramphenicol resistance (CmR) and was used to normalize for transformation efficiency. An increase in the CarbR/CmR colonies demonstrates an increase in total repair. Without growth in arabinose, the Mt-Ku, KuEnls and Mt-Ligase D are not expressed, and the CarbR/CmR ratio ± standard error for total repair was 0.00144 ± 0.00021 for BWKuLig#2 and 0.00127 ± 0.00021 for BWKuEnlsLig. Total repair was 267 and 145 times greater in the presence of arabinose for BWKuLig#2 and BWKuEnlsLig, respectively. Since ClaI linearizes pBestluc in the luciferase open reading frame, accurate and inaccurate repair was calculated by determining the number of CarbR colonies expressing active firefly luciferase. The results for total, accurate and inaccurate repair are shown in Figure 1C for bacteria grown in the presence of arabinose. These data indicate that the KuEnls fusion protein can function with Mt-Ligase D to repair DNA. There was an ∼2 times difference between the strains for total and inaccurate repair (Figure 1C). It is possible that the addition of the EGFPnls to the Mt-Ku did compromise its ability to a small extent to communicate with Mt-Ligase D or to homodimerize. However, what is evident from these experiments is that the KuEnls fusion protein can function with Mt-Ligase D to re-circularize plasmid in bacteria, and hence has the ability to bind DSBs.
The KuEnls fusion protein can bind DNA damage in the nucleus of mammalian cells
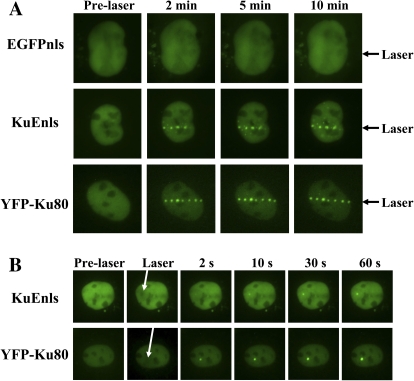
Genomic DNA in mammalian cells is protected by chromatin and has a very different structure from plasmid DNA and the bacterial genome. Although there is evidence of bacterial repair proteins functioning in mammalian cells (31,32), it was necessary to determine whether the bacterial Ku could bind DSBs in nuclear DNA in mammalian cells. The Xrs 6 cell line is deficient in Ku80 (11) and therefore was a good model to test KuEnls binding to damage without the interference of mammalian Ku. Xrs 6 cells were transiently transfected with vectors to express KuEnls, EGFPnls, or YFP-tagged human Ku80 (YFP-Ku80). After 48 h, all three proteins were expressed in the nuclei of the cells (Figure 2). A micropoint ablation laser system was used to generate DNA damage in the nuclei and images were taken at increasing time intervals. The pattern of expression of the control EGFPnls protein did not change after damage induction (Figure 2A). However, the KuEnls (bacterial Ku fusion) and YFP-Ku80 formed foci along the line of the laser damage. Images are shown for up to 10 min post-damage induction (Figure 2A), but subsequent experiments detected KuEnls still bound to the damage in foci after 2 h (D. J. Chen, unpublished data). To determine how fast the KuEnls moved to the damage and formed foci, a point of damage was created with the laser. The YFP-Ku80 formed an intense focus within 2 sec, while the KuEnls was only just detectable at 2 sec post-damage induction (Figure 2B). By 10 sec, the intensity of the KuEnls signal increased and the focus was clearly visible. The intensity of the focus continued to increase at the point of damage throughout the experiment.
Fig. 2.
KuEnls can bind to laser damage in the nuclear DNA of xrs 6 cells. Mammalian expression vectors to express KuEnls or YFP-Ku80 were transiently transfected into xrs 6 cells. The cells were damaged using a pulsed nitrogen laser (365 nm, 10 Hz pulse) and images taken prior to laser damage and up to 10 min post-laser damage in (A) and prior to laser damage and post-laser damage during the first 60 sec in (B). The arrows point to the site of laser damage across the nucleus of the cell in (A) and to the point of laser damage in the nucleus in (B).
Expression of KuEnls in human U2OS cells increases the cell sensitivity to bleomycin sulfate
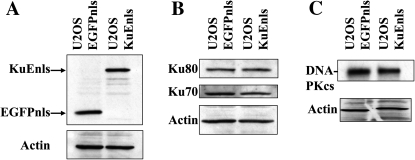
The expression vectors for KuEnls and EGFPnls were individually transfected into the human U2OS cell line, and stable clones isolated and expanded. Western analysis was performed on cell-free extracts from the control cell line (U2OS EGFPnls) and the KuEnls expressing cell line. As can be seen from Figure 3A, KuEnls was expressed at a similar level to the EGFPnls, which is approximately half the size of the KuEnls protein. The expression of Ku70, Ku80 (Figure 3B) and DNA-PKcs (Figure 3C) was also examined and the protein levels of these human NHEJ proteins were comparable in the two cell lines.
Fig. 3.
Expression of KuEnls or EGFPnls in U2OS cells. Cell-free extracts from stably transfected U2OS cells were prepared and analyzed by western analysis for KuEnls and EGFPnls using an anti-EGFP antibody (A). Cell-free extracts were also examined for expression of human Ku70, Ku80 (B) and DNA-PKcs (C).
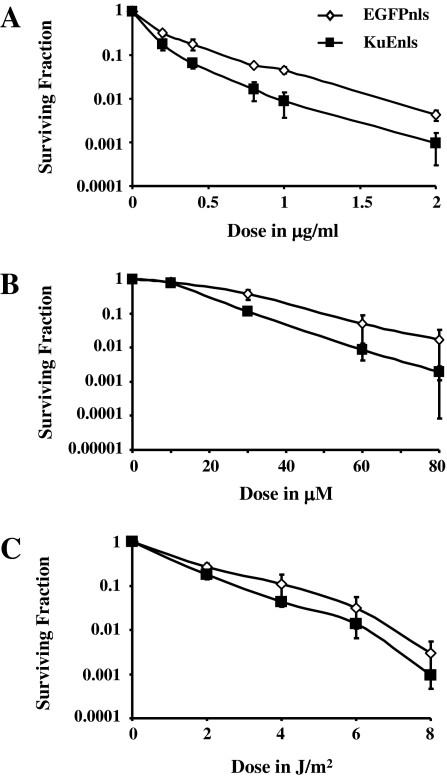
Three DNA damaging agents were selected for survival analyses due to the types of damage they predominantly introduce: bleomycin sulfate introduces DSBs, SSBs and AP sites and is radiomimetic (33), hydrogen peroxide generates SSBs and oxidative base damage, and ultra-violet radiation predominantly introduces cyclobutane pyrimidine dimers and 6-4 photoproducts. At least three experiments were performed for each of the cell lines with each DNA damaging agent, and survival was determined using a colony-forming assay (Figure 4). The expression of KuEnls significantly increased the sensitivity of the U2OS cells to bleomycin sulfate (Figure 4A). Analysis using the linear-quadratic model showed a significant change in the alpha and beta terms for the KuEnls-expressing cell line compared to the EGFPnls control cell line (Table I), and there was a reduction in the dose of bleomycin sulfate required to kill 90% of the cells: 0.3 μg/ml for the KuEnls cell line and 0.6 μg/ml for the EGFPnls control cell line. This is an enhancement ratio of 2. Analysis using the area under the curve also demonstrated a significant sensitization of KuEnls-expressing cells to bleomycin sulfate. Survival analysis following hydrogen peroxide treatment (Figure 4B) did show a significant change in the alpha although not the beta component of the survival curve for KuEnls cells compared to EGFPnls cells (Table I), and the sensitization was marginal when analyzed using the method of area under the curve. The KuEnls protein therefore only slightly increased the sensitivity of the U2OS cells to hydrogen peroxide. Interestingly, there was no change in the sensitivity of the KuEnls cell line compared to the EGFPnls control cell line following treatment with UV radiation (Figure 4C and Table I).
Fig. 4.
Survival analysis after DNA damaging agents. U2OS cells stably expressing EGFPnls or KuEnls were treated with bleomycin sulfate (A), hydrogen peroxide (B) or ultra-violet radiation (C) and a colony-forming assay used to assess the level of survival. At least three experiments were examined for each damaging agent with each cell line, and the mean surviving fraction and standard deviation plotted for each treatment dose.
Table I.
Analysis of survival data
| Treatment | Cell line | Linear-quadratic analysis | Area under the curve ± SD | |
| Alpha ± SD | Beta ± SD | |||
| Bleomycin sulfate | EGFPnls | 4.072 ± 0.348 | −0.682 ± 0.190 | 0.27 ± 0.03 |
| KuEnls | 6.754* ± 1.150 | −1.601* ± 0.370 | 0.17* ± 0.02 | |
| Hydrogen peroxide | EGFPnls | 0.0211 ± 0.012 | 0.0005 ± 0.0003 | 28.08 ± 6.31 |
| KuEnls | 0.0668* ± 0.0098 | 0.0002 ± 0.0001 | 19.74 ± 1.06 | |
| UV | EGFPnls | 0.468 ± 0.309 | 0.0352 ± 0.0318 | 1.82 ± 0.27 |
| KuEnls | 0.862 ± 0.166 | −0.021 ± 0.0367 | 1.46 ± 0.08 | |
The data for the survival studies were analyzed using the linear-quadratic model and using the area under the curve to determine whether there were statistical differences between the EGFPnls control and the KuEnls expressing cell line. SD = standard deviation.
*indicates P < 0.05 compared to the EGFPnls stably-transfected cell line.
Expression of KuEnls does not substantially alter the growth rate of the U2OS cells
Bleomycin is more effective at killing actively dividing cells (34). It was therefore necessary to determine whether stable expression of KuEnls altered the growth of the cells. We performed growth analyses by monitoring the change in cell number as a function of time. For each time point in the experiment, three samples of cells were counted using a hemocytometer, and experiments were performed in triplicate for each cell line. The exponential phase of growth was used to calculate the doubling time of the cells. U2OS cell lines expressing EGFPnls and KuEnls were found to have similar doubling times (Table II), indicating that the KuEnls protein did not significantly disrupt the growth of the U2OS cells.
Table II.
Growth rate
| U2OS cell line | Doubling time (h) ± SD |
| KuEnls | 23.7 ± 3.4 |
| EGFPnls | 25.3 ± 0.8 |
Triplicate experiments measuring the change in cell number as a function of time were performed for each cell line. The exponential phase was determined to occur between 48 and 120 h after initiation of the growth and this time period was used to determine the doubling time. The average and standard deviation (SD) are shown.
* indicates P < 0.05 compared to the EGFPnls stably-transfected cell line.
Mycobacterium tuberculosis Ku does not interact with DNA-PKcs in vitro
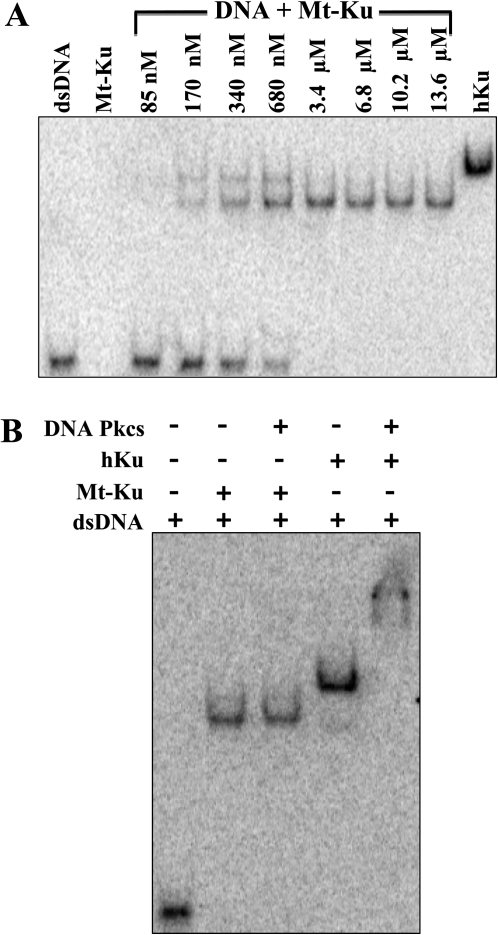
Binding studies were performed with Mt-Ku in vitro with a 32P-labeled 53 mer double-stranded DNA probe. As can be seen from Figure 5A, 640 nM human Ku and 3.4 μM Mt-Ku were required to completely bind the substrate. At lower concentrations of Mt-Ku, two bands are visible, which could represent the binding of two dimers to the DNA. A 33 mer was previously shown to be the minimum DNA size required for Mt-Ku binding, and two bands were detected when a 66 mer was used for binding studies (18).
Fig. 5.
Mt-Ku can bind DNA in vitro but does not bind DNA-PKcs. Linear double-stranded DNA (53 mer) was 32P-labeled on the 3′ end and incubated with increasing amounts of Mt-Ku or 640 nM human Ku (A). To determine whether Mt-Ku interacted with DNA-PKcs when bound to DNA, DNA-binding reactions were prepared with 3.4 μM Mt-Ku or 40 nM human Ku in the presence and absence of 120 nM DNA-PKcs (B). Reactions were incubated at room temperature for 30 min prior to electrophoresis through a 4% polyacrylamide Tris-glycine native gel. Images were analyzed using a phosphoimager.
Binding reactions were also examined that contained 120 nM human DNA-PKcs with Mt-Ku or human Ku and the DNA probe (Figure 5B). There was no change in the electrophoretic mobility of the DNA bound by Mt-Ku when DNA-PKcs was present in the binding reaction. However, the mobility of the DNA incubated with human Ku was substantially reduced when DNA-PKcs was added. This indicates that in vitro DNA-PKcs binds human Ku bound to DNA but not Mt-Ku.
Mycobacterium tuberculosis Ku does not bind to human Ku in vitro
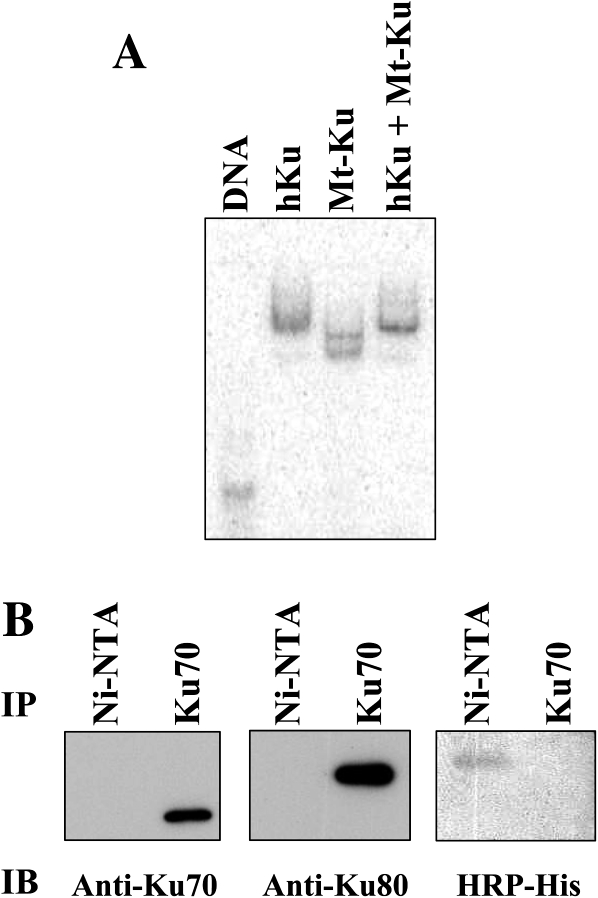
Binding reactions were prepared containing the 32P-labeled 53 mer DNA probe and human Ku, Mt-Ku or human Ku and Mt-Ku. When both types of Ku protein were present, the electrophoretic mobility of the 53 mer was identical to that of the human Ku (Figure 6A). This indicates that human Ku preferentially bound the DNA in vitro, and it is unlikely that a combination of the two proteins were bound to the same DNA molecule. To rule out the possibility that the two proteins interact, pure human Ku and pure Mt-Ku were mixed together and immunoprecipitation was performed with either an anti-Ku70 monoclonal antibody or Ni-NTA beads. Nickel binds the his-tag on the N-terminus of the Mt-Ku protein. After immunoprecipitation, the proteins were analysed by western analysis for the presence of human Ku70, human Ku80 or the his-tagged Mt-Ku. From Figure 6B, it is evident that immunoprecipitation with the anti-Ku70 antibody detected only Ku70 and Ku80. The only protein detected with the Ni-NTA beads was the his-tagged Mt-Ku. This indicates that in vitro the Mt-Ku did not interact with human Ku, and hence in cells it is unlikely that Mt-Ku could form dimers with either Ku70 or Ku80.
Fig. 6.
Mt-Ku does not interact with human Ku. Linear double-stranded DNA (53 mer) was 32P-labeled on the 3′ end and incubated with human Ku (40 nM) or Mt-Ku (3.4 μM) or human Ku and Mt-Ku. After 30 min at room temperature, samples were subjected to electrophoresis through a 4% polyacrylamide Tris-glycine native gel and the image was analyzed using a phosphoimager (A). To examine the interaction of Mt-Ku and human Ku in vitro, 10 μg of each pure protein was mixed and precipitated with either Ni-NTA beads or anti-Ku70 monoclonal antibody, followed by protein G-agarose. After 5 h of gentle agitation at 4°C, samples were washed and then boiled to release the bound proteins. The proteins were separated by SDS-PAGE and western analysis performed (B).
Discussion
This work has developed a green fluorescent fusion protein of the M. tuberculosis Ku (KuEnls) that is targeted to the nucleus of mammalian cells using the nuclear localization signal peptide from human Ku70. Using our E. coli model of bacterial NHEJ, we demonstrated that the KuEnls protein can function with Mt-Ligase D to re-circularize plasmid DNA and so has the ability to bind DSBs. We confirmed that the bacterial Ku fusion protein could not only bind plasmid DNA termini but also damage generated using a laser in the nucleus of Ku80-deficient mammalian cells. Since there were no foci formed by a control green fluorescent molecule that also contained the Ku70 nuclear localization signal (EGFPnls), foci formation by KuEnls was due to the bacterial Ku protein. The micropoint laser ablation system has previously been shown to generate DSBs in genomic DNA (25). This is therefore the first demonstration that the bacterial Ku protein from M. tuberculosis has the ability to bind DSBs in DNA that is bound to chromatin.
In comparison to a human YFP-Ku80 protein, the KuEnls protein was slower at forming foci at the laser damage. This is likely due to the binding affinity of Mt-Ku since in vitro experiments (Figure 6 and ref. 18) do indicate that human Ku binds DNA termini more avidly than bacterial Mt-Ku. To effectively sensitize cells to DNA damage, it will be necessary for the KuEnls protein to bind DSBs prior to human Ku. To test the biological activity of the bacterial Ku, stably transfected U2OS cell lines expressing either KuEnls or EGFPnls were generated. The EGFPnls cell line was the control cell line. We determined that expression of the bacterial Ku fusion protein resulted in sensitization of the cells to bleomycin sulfate, which is known to generate DSBs. Therefore, even in the presence of human Ku, the bacterial Ku had a biological impact on survival from a DSB-generating agent, which suggests some bacterial Ku protein was able to bind the DSBs before the human Ku. A ratio of the dose required to reduce cell survival to SF = 0.1 for the control cell line/KuEnls-expressing cell line was 2. This is comparable to previous studies examining the radiosensitizing effect of antisense RNA against Ku86 (ratio = 1.8) and a virus expressing a dominant negative human Ku70 (1.3–1.8) in human cells (35). Bleomycin is more effective at killing actively dividing cells (34). Since the doubling times were similar for the EGFPnls and KuEnls cell lines, the sensitization of the U2OS cells to bleomycin by KuEnls was not due to changes in growth rate. Survival studies using hydrogen peroxide and UV demonstrated that KuEnls resulted in little or no change to these damaging agents. A lack of substantial sensitization to these agents again suggests that KuEnls is acting by binding to DSBs, as hydrogen peroxide predominantly generates SSBs and oxidative base damage, while UV radiation generates cyclobutane pyrimidine dimers and 6-4 photoproduct.
A sensitization to bleomycin sulfate could have occurred by an alteration in human NHEJ repair protein expression, by KuEnls binding to human Ku70/80 and acting as a dominant negative or by binding the DSBs and blocking DSB repair by human DSB repair pathways. Western analysis demonstrated that expression of the KuEnls protein in U2OS cells did not alter expression of human Ku70, Ku80 and DNA-Pkcs, and work in vitro demonstrated that purified Mt-Ku did not bind to human Ku or to DNA-Pkcs. Therefore, the mechanism of action of KuEnls is different from the radiosensitizing N-terminal deleted Ku70 fragment, which acts as a dominant negative preventing human Ku from binding to DSBs (36). It is therefore likely that KuEnls binds to DSBs generated by bleomycin sulfate but due to a lack of communication with DNA-PKcs prevents further repair by NHEJ. Extended studies using laser micro-irradiation do indicate that the KuEnls protein remains bound to the DNA in foci for up to 2 h after damage initiation.
Cancer treatments work by generating DNA damage in tumor cells resulting in cell death, while limiting the damage in the surrounding normal tissue. DSBs are potentially the most lethal lesions produced and NHEJ is an important repair pathway involved in the survival of cells. Our work suggests that the bacterial Ku protein could be developed for use as a complimentary treatment with existing radiotherapy and chemotherapy regimens to enhance tumor cell death. Small molecule inhibitors have been generated for ATM, DNA-PKcs, PARP, DNA ligases and the MRN complex (for review, see refs. 14,15). These target specific signaling and repair pathways and will be useful in enhancing tumor sensitivity for subsets of tumors that have abnormal levels of certain DSB repair pathways (15): some cancer cells have abnormally high alternative NHEJ (37) or can be deficient in homologous recombination (38), and tumors defective in Fanconi anemia repair pathways are particularly sensitive to ATM inhibition (39). The epidermal growth factor receptor (EGFR) has also been found to promote DSB repair through classical NHEJ (for review, see ref. 40) and inhibitors of EGFR also increase cellular radiosensitivity of a variety of tumor types. It is possible, however, that the inhibition of one DSB repair pathway during treatment could result in the increased activity of an alternative DSB repair mechanism. Therefore, developing a molecule to block all types of DSB repair may increase the effectiveness of standard therapies.
Work in yeast indicates that the choice of DSB repair pathway may be at the level of DNA resection at the termini of the DSB. Cell cycle control of homologous recombination has been demonstrated to be by the phosphorylation of the Sae2 nuclease by cyclin-dependent kinases (41). In mammalian cells, the decision as to which DSB repair pathway proceeds may be determined by the end resection of the DNA termini: if end resection occurs then homology-directed repair or alternative end-joining can be used, but if human Ku binds then extensive resection cannot occur (7) and classical NHEJ repairs the DSB. Therefore, the development of a molecule to bind and block the termini of DSBs may prevent all types of DSB repair. It is possible that the bacterial Ku could be such a protein. We provide evidence that the bacterial Ku does sensitize human cells to bleomycin sulfate, does bind DSBs in human chromatin and does not communicate with the classical human NHEJ proteins. It is possible that homologous recombination as well as NHEJ could be inhibited by KuEnls as binding to DNA termini could prevent DNA end resection. Although it is beyond the scope of this study to examine in more detail how KuEnls is disrupting DSB repair, future studies are planned to examine how/whether the Mt-Ku inhibits specifically NHEJ and homologous recombination.
Funding
National Cancer Institute at the National Institutes of Health (grant numbers CA85693 and CA85693-9S1 for L.H. and CA50519 and CA134991 for D.J.C.).
References
- 1.Teoule R. Radiation induced DNA damage and its repair. Int. J. Radiat. Biol. 1987;51:573–589. doi: 10.1080/09553008414552111. [DOI] [PubMed] [Google Scholar]
- 2.Sage E, Harrison L. Clustered DNA lesion repair in eukaryotes: relevance to mutagenesis and cell survival. Mutat. Res. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Natarajan AT, Obe G, van Zeeland AA, Palitti F, Meijers M, Verdegaal-Immerzeel EAM. Molecular mechanisms involved in the production of chromosomal aberrations. II. Utilization of Neurospora endonuclease for the study of aberration production by X-rays in G1 and G2 stages of the cell cycle. Mutation Res. 1980;69:293–305. doi: 10.1016/0027-5107(80)90094-9. [DOI] [PubMed] [Google Scholar]
- 4.Bryant PE. Enzymatic restriction of mammalian cell DNA using Pvu II and Bam H1: evidence for the double strand break origin of chromosomal aberrations. Int. J. Radiat. Biol. 1984;46:57–65. doi: 10.1080/09553008414551061. [DOI] [PubMed] [Google Scholar]
- 5.Bryant PE, Riches RC, Terry SYA. Mechanisms of the formation of radiation-induced chromosomal aberrations. Mutat. Res. 2010;701:23–26. doi: 10.1016/j.mrgentox.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat. Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Heyer W-D, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4: DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeggo PA, Kemp LM. X-ray sensitive mutants of a Chinese hamster ovary cell line: isolation and cross-sensitivity to other DNA damaging agents. Mutat. Res. 1983;112:313–327. doi: 10.1016/0167-8817(83)90026-3. [DOI] [PubMed] [Google Scholar]
- 11.Taccioli GE, Gottlieb TM, Blunt T, et al. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 12.Al-Minawi AZ, Lee Y-F, Hakansson D, et al. The ERCC1/XPF endonuclease is required for completion of homologous recombination at DNA replication forks stalled by inter-strand cross-links. Nucleic Acids Res. 2009;37:6400–6413. doi: 10.1093/nar/gkp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinert T, Kaochar S, Jones H, Paek A, Clark AJ. The replication fork’s five degrees of freedom, their failure and genome rearrangements. Curr. Opin. Cell Biol. 2009;21:778–784. doi: 10.1016/j.ceb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Bolderson E, Richard DJ, Zhou BB, Khana KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin. Cancer Res. 2009;15:6314–6320. doi: 10.1158/1078-0432.CCR-09-0096. [DOI] [PubMed] [Google Scholar]
- 15.Rassool FV, Tomkinson AE. Targeting abnormal DNA double strand break repair in cancer. Cell. Mol. Life Sci. 2010;67:3699–3710. doi: 10.1007/s00018-010-0493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Della M, Palmbos PL, Tseng HM, et al. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 17.Malyarchuk S, Wright D, Castore R, Klepper E, Weiss B, Doherty AJ, Harrison L. Expression of Mycobacterium tuberculosis Ku and Ligase D in Escherichia coli results in RecA and RecB-independent DNA end-joining at regions of microhomology. DNA Repair (Amst) 2007;6:1413–1424. doi: 10.1016/j.dnarep.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weller GR, Kysela B, Roy R, et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 19.Pitcher RS, Tonkin LM, Green AJ, Doherty AJ. Domain structure of a NHEJ DNA repair ligase from Mycobacterium tuberculosis. J. Mol. Biol. 2005;351:531–544. doi: 10.1016/j.jmb.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 20.Wright D, DeBeaux A, Shi R, Doherty AJ, Harrison L. Characterization of the roles of the catalytic domains of Mycobacterium tuberculosis ligase D in Ku-dependent error-prone DNA end joining. Mutagenesis. 2010;25:473–481. doi: 10.1093/mutage/geq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gell D, Jackson SP. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999;27:3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 23.Hammel M, Yu Y, Mahaney BL, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uematsu N, Weterings E, Yano K, et al. Autophosphorylation of DNA-PKcs regulates its dynamics at DNA double strand breaks. J. Cell Biol. 2007;117:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono M, Tucker PW, Capra JD. Production and characterization of recombinant human Ku antigen. Nucleic Acids Res. 1994;22:3918–3924. doi: 10.1093/nar/22.19.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koike M, Ikuta T, Miyasaka T, Shiomi T. The nuclear localization signal of the human Ku70 is a variant bipartite type recognized by the two components of nuclear pore-targeting complex. Experimental Cell Res. 1999;250:401–413. doi: 10.1006/excr.1999.4507. [DOI] [PubMed] [Google Scholar]
- 29.Koike M, Miyasaka T, Mimori T, Shiomi T. Subcellular localization and protein-protein interaction regions of Ku proteins. Biochem. Biophys. Res. Comm. 1998;252:679–685. doi: 10.1006/bbrc.1998.9368. [DOI] [PubMed] [Google Scholar]
- 30.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus Subtilis. J. Mol. Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 31.Harrison L, Skorvaga M, Cunningham RP, Hendry JH, Margison GP. Transfection of the E. coli nth gene into radiosensitive Chinese Hamster cells: effects on sensitivity to radiation, hydrogen peroxide and bleomycin sulfate. Radiat. Res. 1992;132:30–39. [PubMed] [Google Scholar]
- 32.Harris LC, Margison GP. Expression in mammalian cells of the Escherichia coli O6 alkylguanine-DNA-alkyltransferase gene ogt reduces the toxicity of alkylnitrosoureas. Br. J. Cancer. 1993;67:1196–1202. doi: 10.1038/bjc.1993.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Povirk LF, Houlgrave CW. Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: specific formation and cleavage of closely opposed lesions in complementary strands. Biochem. 1988;27:3850–3857. doi: 10.1021/bi00410a049. [DOI] [PubMed] [Google Scholar]
- 34.Sikic BI. Biochemical and cellular determinants of bleomycin cytotoxicity. Cancer Surv. 1986;5:81–91. [PubMed] [Google Scholar]
- 35.Urano M, He F, Minami A, Ling CC, Li GC. Response to multiple radiation doses of human colorectal carcinoma cells infected with recombinant adenovirus containing dominant-negative Ku70 fragment. Int. J. Radiat. Oncol. Biol. Phys. 2010;77:877–885. doi: 10.1016/j.ijrobp.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He F, Li L, Kim D, Wen B, Deng X, Gutin PH, Ling CC, Li GC. Adenovirus-mediated expression of a dominant negative Ku70 fragment radiosensitizes human tumor cells under aerobic and hypoxic conditions. Cancer Res. 2007;67:634–642. doi: 10.1158/0008-5472.CAN-06-1860. [DOI] [PubMed] [Google Scholar]
- 37.Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIa in chronic myeloid leukemia: consequences for the repair of DNA double strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 39.Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, Moreau LA, Shimamura A, D'Andrea AD. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J. Clin. Invest. 2007;117:1440–1449. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee B, Choy H, Nirodi C, Burma S. Targeting nonhomologous end-joining through epidermal growth factor receptor inhibition: rationale and strategies for radiosensitization. Semin. Radiat. Oncol. 2010;20:250–257. doi: 10.1016/j.semradonc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–693. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]