Abstract
Objective
Positive associations between polymorphisms in the type-three metabotropic glutamate receptor gene (GRM3) and the pathogenesis of schizophrenia as well as response to antipsychotic treatment have been reported. The objective of this study was to determine whether refractory psychiatric symptoms in antipsychotic non-responders are related to polymorphisms in GRM3.
Methods
Ninety-five treatment refractory schizophrenia subjects were enrolled. Prior to a medication switch, global psychopathology and negative symptoms were rated. These subjects were genotyped for seven markers in GRM3. Genotype associations with symptoms were assessed.
Results
Two markers in GRM3 (rs1989796 and rs1476455), were associated with the presence of refractory global symptoms as measured by the Brief Psychiatric Rating Scale (BPRS) total scores. Participants with an rs1476455_CC genotype had significantly higher BPRS scores than A-carriers (55.1±10.4 vs. 48.3±9.2; F=7.6, p=0.0071). Additionally, participants with the rs1989796_CC genotype had significantly higher BPRS scores than T-carriers (50.1±5.7 vs. 55.8±10.5, F=7.1, p=0.0091). No evidence for significant associations with negative symptoms was observed.
Conclusions
Polymorphisms in the GRM3 gene may be associated with refractory global psychosis symptoms but not negative symptoms in persons with schizophrenia.
Keywords: Metabotropic, Glutamate, Antipsychotic, Pharmacogenetics, Schizophrenia
Introduction
Glutamate disposition and signaling in the brain is becoming increasingly recognized as an essential component of the pathogenesis and treatment of schizophrenia (Harrison et al., 2008; Tamminga, 2006). Furthermore, variants in glutamate system genes are associated with cognitive function, medication response, medication dosing, and in some studies with genetic risk for schizophrenia (Bishop et al., 2005; Egan et al., 2004; Fijal et al., 2009; Fujii et al., 2003). Efforts to identify specific genetic variants that predict disease risk, symptom presentation, and medication response in psychiatric disorders have recently begun to focus on the components of the glutamate system and are helping to improve our understanding of the biology of schizophrenia, risk for disease, and targets for new drug treatments.
Many second generation antipsychotics have either direct or indirect effects on the glutamate system that may partially account for their mechanisms of action and effectiveness in treating the cognitive and negative symptoms of schizophrenia (Moghaddam, 2004; Tamminga, 2006). Investigational pharmacological agents for schizophrenia that stimulate selected metabotropic glutamate receptors (mGluRs) as well as agents that modulate N-methyl-D-aspartate (NMDA) receptors have antipsychotic properties in both humans and animal models of disease (Javitt, 2006; Patil et al., 2007). Due to the complex nature of glutamate signaling in the brain, including interactions with dopamine and other neurotransmitter systems, genetic variants in glutamate receptors are likely contributors to the variability seen in symptom presentation and antipsychotic drug response (Bishop et al., 2005).
GRM3 has been localized to chromosome 7q21.1-21.2 and spans 220.1 kb (Scherer et al., 1996). GRM3 codes for the mGluR3 protein which is a G-protein coupled receptor. Along with the structurally similar mGluR2 protein, mGluR3 is localized to the periphery of both pre- and post-synaptic neurons and is essential for optimal signaling of glutamate in the brain (Cartmell and Schoepp, 2000). Initial studies of genetic variation in GRM3 suggest association with schizophrenia in some studies (Egan et al., 2004; Fujii et al., 2003) but not others (Albalushi et al., 2008; Bishop et al., 2007; Jonsson et al., 2009; Marti et al., 2002; Norton et al., 2005; Schwab et al., 2008; Tochigi et al., 2006).
In studies of cognition (Egan et al., 2004) and drug response in schizophrenia (Bishop et al., 2005; Fijal et al., 2009) GRM3 variants have accounted for a significant portion of the variability seen in these outcome measures. The mGluR3 receptor product of this gene has subsequently become one glutamate receptor targeted by developmental antipsychotics (Patil et al., 2007). The heterogeneity of the disease association studies alongside growing evidence for the role of GRM3 in cognition, symptom profile and response to drug treatment further suggests that variants in this gene may influence disease presentation and potentially interact with other genetic (Nicodemus et al., 2007; Tan et al., 2007) and perhaps non-genetic factors as opposed to being solely a disease risk gene.
Collectively, research to date indicates that single nucleotide polymorphisms (SNPs) in the GRM3 gene may be useful to study as genetic markers with potential influence on response to antipsychotics in persons with schizophrenia. In addition to traditional pharmacogenetic analyses of drug response, it is also informative to identify and characterize the biological underpinnings of antipsychotic non-response. Genetic association studies of disease presentation and symptoms that are resistant to drug treatment may provide important information on treatment strategies and drug development efforts for difficult to treat patients. Building on our initial research of treatment response pharmacogenetics and disease association studies of GRM3 in schizophrenia, we investigated the relationship between variants in this gene with symptomatology in a uniquely characterized cohort of treatment refractory patients with the goal of identifying genetic determinants of antipsychotic resistant symptoms that may drive non-response to initial therapies.
Methods
Participants
Ninety-five unrelated subjects meeting DSM-IV criteria for schizophrenia were recruited for this study. Individuals were assessed in this study prior to initiating treatment with a medication regimen for treatment refractory schizophrenia (predominantly clozapine). All subjects gave written informed consent to an IRB-approved protocol. Inclusion criteria consisted of having a DSM-IV diagnosis of schizophrenia, being at least 18 years of age, and refractory to prior antipsychotic treatment according to the operationally defined criteria of Kane et al (Kane et al., 1988) (i.e., persistent psychotic symptoms for at least two years despite adequate separate trials with three antipsychotic drugs from two biochemical classes at doses ≥1000 chlorpromazine equivalents for 6 weeks) (n=93) or inadequate response to at least two prior antipsychotic agents (n=2). Previously utilized second generation antipsychotics were clinically estimated to exceed >1000 (Woods, 2003). Subjects were recruited from inpatient and outpatient clinics in this single site study. Potential subjects were excluded if they had a DSM-IV diagnosis of organic mental disorder, suicidal ideation, other serious medical condition, or had been treated with clozapine previously.
Assessments
Participants were evaluated by a trained rater prior to olanzapine or clozapine initiation with the 18-item Brief Psychiatric Rating Scale (BPRS) with each item rated 1-7, with 1 = not present and 7 = severe. In addition, ratings of negative symptoms were conducted with the Scale for Assessment of Negative Symptoms (SANS), with SANS score defined as the sum of the alogia, anhedonia, avolition, attention, and affective flattening global ratings with each item rated 0-5, with 0 = not present and 5 = severe.
Genotyping
Genomic DNA was isolated from whole blood with the salt precipitation method (Lahiri and Nurnberger, 1991). Genotyping was done with Pyrosequencing™ Technology. Polymerase Chain Reaction (PCR) primers were designed using Oligo 6 (MBI, Cascade, CO, USA). Pyrosequencing primers were designed using Pyrosequencing Primer Design Version 1.01 software (http://www.pyrosequencing.com). Seven SNPs (rs274622, rs724226, rs917071, rs6465084, rs1468412, rs1989796, and rs1476455) in the GRM3 gene (Egan et al., 2004; Fujii et al., 2003) were analyzed for their relationship to BPRS or SANS scores. Genotyping assays for six of the GRM3 variants were done as described previously (Bishop et al., 2005). Assays for rs6465084 were completed with forty-five PCR cycles per reaction in a 50 ul volume with 1.5 mM Mg2+. PCR products were visualized by electrophoresis on 1.5% agarose gels stained with ethidium bromide prior to Pyrosequencing. Forward, reverse, and Pyrosequencing primers for the variants are as follows: rs6465084 (TTGCCTTAATGACACAAAGTTCTC, CCGGTGCTCTTTCCATATTGA, TCCATGAAAAAGGCAC). Genotyping was done blind to symptom ratings. All genotyping assays were validated with direct sequencing.
Statistical Analysis
Allele and genotype frequencies, Hardy Weinberg Equilibrium (HWE), and genotype association tests were conducted with PLINK software (Purcell et al., 2007). Linkage disequilibrium (LD) plots were created with Haploview version 4.2 software (Barrett et al., 2005) (displayed as D’) with solid spine D’>0.8 used to define haplotype blocks (see Figure 1). Genotype associations with BPRS and SANS scores as quantitative traits were conducted with PLINK using the genotypic (2df) association procedure which assesses an additive effect model. The mperm 1000 permutations option was selected to assess significance by controlling for multiple genetic markers and the LD between SNPs. Subsequent one way Analysis of Variance (ANOVA) tests were completed for markers rs1989796, and rs1476455 which were identified as significant from the PLINK procedures to quantify differences across genotype groups. Student's T-tests were performed to test for differences in BPRS and SANS scores between males and females. ANOVA was used to test for differences in SANS and BPRS scores across racial groups. Pearson's correlation coefficient was used to examine relationships between age and SANS/BPRS scores. Power calculations were carried out using Quanto 1.2.4; http://hydra.usc.edu/gxe using our lowest minor allele frequency (0.12 for rs1476455) as a conservative estimate of our ability to detect associations in this sample. At a minor allele frequency 0f 0.12, using the means and standard deviations for BPRS and SANS scores obtained in our study sample of 95 subjects and a two-tailed 0.05 level of significance for an additive effect model, we had 80% power to detect R2 for genotypes as low as 0.08 and a mean difference of difference of 2.65 points on SANS and BPRS scores across genotype groups.
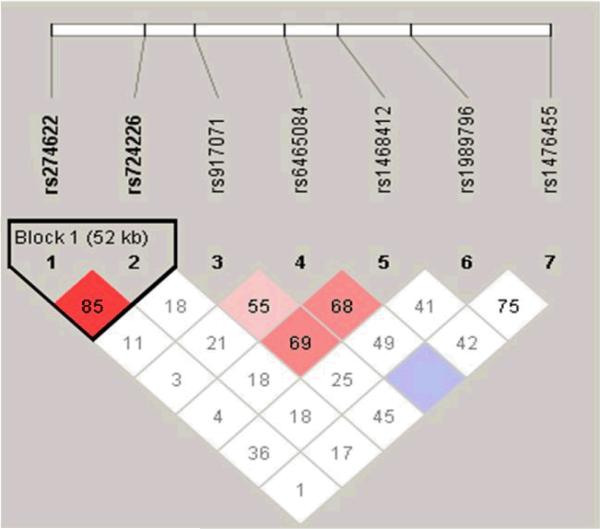
Figure 1. Linkage disequilibrium structure of GRM3.
Linkage disequilibrium for GRM3 SNPs (displayed as D’) with solid spine D’>0.8 used to define haplotype blocks
Policy and ethics
The work described in this article was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans.
Results
Ninety-five subjects with schizophrenia volunteered to participate in this study. Our study population consisted of n=84 (89%) Caucasian, n=8 (8%) African American, n=1 (1%) Hispanic, n=1 (1%) Asian, and n=1 (1%) unknown participants. Subjects were predominantly male n=61 (64%) with a mean age of 37.9±10.2 years. Mean BPRS Total scores were in the moderately severe range (53.5±10.5 points), as were negative symptoms as measured by the SANS (13.9±4.3 points). In this treatment refractory sample, age was not significantly associated with BPRS or SANS scores and scores did not differ across race/ethnic groups (all p's>0.05). SANS scores were significantly higher in female participants (15.1±4.3) vs. males (13.2±4.2) (t=2.1, 1df, p=0.04). BPRS scores did not differ between males and females.
GRM3 Genotypes
Allele and genotype frequencies are presented in Table 1. Genotypes for GRM3 did not deviate from Hardy-Weinberg Equilibrium, with the exception of rs1468412 (p=0.024), which has been previously associated with risk for schizophrenia (Egan et al., 2004). Markers in GRM3 were in moderate linkage disequilibrium (denoted as D’) with one haplotype block consisting of two SNPs with D’>0.80 as indicated in Figure 1.
Table 1.
Characteristics of SNPs assessed for association with refractory symptoms
| SNP | Position | HWEApval | MAF B | Alleles | Genotype Count |
|---|---|---|---|---|---|
| rs274622 | 85917591 | 0.8154 | 0.34 (C) | T:C | 12/41/42 |
| rs724226 | 85970025 | 0.4917 | 0.34 (A) | G:A | 13/39/43 |
| rs917071 | 85998432 | 0.1928 | 0.34 (T) | C:T | 14/36/45 |
| rs6465084 | 86048126 | 0.9842 | 0.28 (G) | A:G | 7/40/48 |
| rs1468412 | 86078102 | 0.0313 | 0.35 (T) | A:T | 17/33/45 |
| rs1989796 | 86118964 | 0.8735 | 0.37 (T) | C:T | 14/43/48 |
| rs1476455 | 86196391 | 0.5059 | 0.12 (A) | C:A | 0/22/73 |
HWE: Hardy Weinberg Equilibrium
MAF: Minor Allele Frequency
Relationship between Symptoms and Genetic Variability
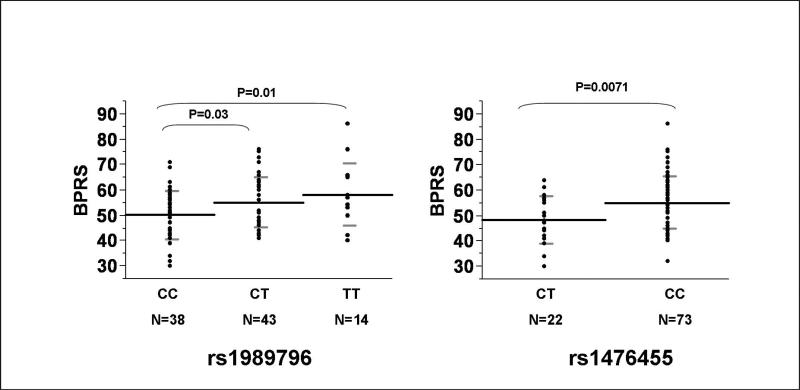
Genotype associations between GRM3 variants and BPRS and SANS scores are summarized in Tables 3 and 4. Two markers (rs1989796 and rs1476455) in the 3’ end of GRM3 were significantly associated with BPRS Total scores, but not SANS scores. BPRS and SANS scores across rs1989796 and rs1476455 genotype groups were then assessed. Mean (SD) BPRS total scores for rs1989796 CC, CT, and TT were 50.1 (10.0), 55.0 (9.9), and 58.2 (12.2) (F=4.07, 2df, p=0.02). Mean (SD) BPRS scores for rs1476455 AC, and CC were 48.3 (9.2) and 55.1 (10.4) (F=7.6, 1df, p=0.007) (see Figure 2). Mean (SD) SANS scores for rs1989796 CC, CT, and TT were 13.4 (4.0), 14.1 (4.6), and 14.5 (4.2) (F=0.44, 2df, p=0.64). Mean (SD) SANS scores for rs1476455 AC, and CC were 12.7 (4.3) and 14.3 (4.3) (F=2.4, 1df, p=0.13). Adjusting these relationships with SANS scores for sex did not alter the significance of these relationships.
Table 3.
Genotype associations between metabotropic glutamate receptor 3 (GRM3) single nucleotide polymorphisms (SNPs) and negative symptom scores as assessed by the Scale for Assessment of Negative Symptoms (SANS)A
| SNP | BETA | SE | R2 | T-score | P-value | Corrected P-value B |
|---|---|---|---|---|---|---|
| rs274622 | -0.65 | 0.64 | 0.011 | -1.01 | 0.32 | 1.0 |
| rs724226 | -0.41 | 0.63 | 0.0045 | -0.64 | 0.52 | 0.90 |
| rs917071 | 0.083 | 0.62 | 1.94E-4 | 0.13 | 0.89 | 0.99 |
| rs6465084 | 0.27 | 0.71 | 0.0016 | 0.38 | 0.70 | 1.0 |
| rs1468412 | -0.14 | 0.59 | 6.30E-4 | -0.24 | 0.81 | 1.0 |
| rs1989796 | 0.59 | 0.64 | 0.0092 | 0.93 | 0.36 | 0.93 |
| rs1476455 | -1.60 | 1.038 | 0.025 | -1.54 | 0.13 | 0.56 |
Additive model
Permuted p-value correcting for multiple comparisons and linkage disequilibrium between SNPs
Figure 2. GRM3 Variants and BPRS Total Scores.
Analysis of variance and student's t-tests were used to compare mean BPRS values ± standard deviation across genotype groups.
Discussion
The principle finding of this study is that global psychosis symptoms as measured by BPRS Total scores were significantly related to two gene variants in the 3’ end of the GRM3 gene. Significant genotype associations between rs1989796 and rs1476455 and BPRS total scores were observed, with potentially clinically meaningful and statistically significant differences across genotype groups. No significant associations between GRM3 variants and negative symptoms were observed in this study sample. To our knowledge this is the first association between GRM3 variants and clinical symptoms in treatment refractory schizophrenia.
Relationship of the glutamate system to schizophrenia and antipsychotic pharmacology
Currently, there are eight known metabotropic glutamate genes (GRMs) coding for metabotropic glutamate receptors (mGluRs) one through eight (GRM1-GRM8). The eight individual mGluRs are subdivided into three groups (Group-I, Group-II, and Group-III) based on amino acid sequence and second messenger signaling similarities (Nakanishi, 1992). GRM1 and GRM5 are the two members of Group I, GRM2 and GRM3 comprise Group II, with the rest falling into Group III. These receptors mediate signal transduction through G-protein second messenger systems which distinguishes them from their ionotropic counterparts, the N-methyl-d-aspartate (NMDA), alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA), and kainate receptors. Metabotropic glutamate receptors seem to “fine tune” glutamate transmission, while ionotropic receptors regulate large scale glutamate fluctuations in the brain (Conn and Pin, 1997).
A number of studies have investigated the relationships between GRM3 with risk for schizophrenia with mixed results. Collectively these data do not provide strong evidence that variants in these genes on their own confer and increased risk for developing this disease. Previous investigations of GRM3 variants as potential predictors of antipsychotic response in schizophrenia (Bishop et al., 2005; Fijal et al., 2009) while not associated with disease risk (Bishop et al., 2007; Marti et al., 2002; Norton et al., 2005) suggests that GRM3 variants are associated with drug response and symptom presentation in rather than the disease. The results obtained from the cohort of treatment refractory persons with schizophrenia studied here is consistent with this possibility.
The primary mechanisms of action for antipsychotics are hypothesized to center on striatal dopamine-2 (D2) and mesolimbic serotonin-2A (5HT2A) receptor antagonism, but an increasing body of animal and human research indicates that these agents may also affect glutamate neurotransmission via regulating gene expression (Girgenti et al., 2010; Molteni et al., 2009) . The relationship of genetic variability in the region of the GRM3 gene with the effects of second generation antipsychotic medications is not surprising. Second generation antipsychotics affect the expression of GRM3 mRNA as well as serum glutamate concentrations. Serum glutamate concentrations increase significantly after subjects are switched from conventional antipsychotic agents to olanzapine (Goff et al., 2002). While not affecting NMDA or AMPA gene expression in rats (Riva et al., 1997; Tascedda et al., 2001), clozapine and olanzapine appear to up-regulate the expression of GRM3 mRNA after chronic administration. This upregulation may explain our earlier results with rs274622 which is a T/C variant residing in the in the “T” of a CCAAT box of the GRM3 promoter region (Corti et al., 2001). This variant may contribute to a state of glutamate hypofunction that is reversed by antipsychotic treatment. In the current study sample of treatment refractory patients, we did not observe any associations between SNPs in the 5’ end of GRM3 with either negative or global symptoms. This is not surprising, as the reasons why an individual is refractory to antipsychotic agents likely involve a number of variables. It is plausible that these SNPs at the 3’ end of the gene are in linkage disequilibrium for unstudied variants in the 3'untranslated region of GRM3, which may be targeted by epigenetic regulatory mechanisms that have increased activity or importance after long term antipsychotic exposure, or which may be general markers for treatment resistance.
These results may also shed light on the heterogeneity seen in studies investigating associations between GRM3 and schizophrenia. At the current time, eleven studies have investigated the association of GRM3 SNPs with schizophrenia, seven in Caucasian samples and four in Asian samples (Albalushi et al., 2008; Bishop et al., 2007; Chen et al., 2005; Egan et al., 2004; Fallin et al., 2005; Fujii et al., 2003; Jonsson et al., 2009; Marti et al., 2002; Norton et al., 2005; Schwab et al., 2008; Tochigi et al., 2006). These studies have resulted in mixed associations between SNPs and/or haplotypes and disease. Additional evidence suggests that GRM3 may interact with the catechol-omethyltransferase (COMT) Val158Met variant to confer risk for schizophrenia (Nicodemus et al., 2007; Tan et al., 2007). Our results may provide additional insight into these heterogeneous findings if there are differential relationships between GRM3 and clinical outcomes for schizophrenia subtypes.
Study limitations
This study describes the association of variation in one candidate gene with symptoms in treatment refractory schizophrenia patients prior to a medication switch. The reasons for non-response to prior antipsychotic treatment are unknown. Further, because of heterogeneity of prior antipsychotic treatment, we were unable to determine the relationships of exposure to specific agents and doses with the outcomes assessed in this study. In addition, the long-standing nature of illness in the subjects enrolled here limited our ability to reliably estimate illness duration. This study sample is predominantly Caucasian, so we were unable to determine unequivocally whether the relationships between GRM3 SNPs and symptom presentation hold across other race and ethnic groups. Of note, our results did not change significantly when non-Caucasian races/ethnicities were excluded from our analyses.
Conclusion
We observed that two variants in the 3’ end of GRM3 were associated with global symptoms of psychosis as assessed by the BPRS but not negative symptoms in this study sample. This suggests that variation in this gene may be related to a mechanism of treatment resistance in a subset of patients. However, it is important to note that not all subjects with these genotypes had higher measures of global psychosis symptoms as measured by the BPRS; rather on average they were higher across selected genotype groups. As previously noted, there may be many reasons why a patient achieves inadequate response to antipsychotic therapy. Dosing, treatment adherence, intolerance to side effects, and agent selection are some modifiable factors that may influence this outcome.
The identification of genetic biomarkers of medication non-response is potentially useful, but the interpretation of this information needs to be considered carefully. However, it is conceivable that the results of this type of study, if replicated, may be useful in agent selection for schizophrenia. It is plausible that the use of treatment strategies reserved for later in treatment, such as clozapine, may be considered earlier on in therapy if we identify markers of non-response to other therapeutic options.
Table 2.
Genotype associations between metabotropic glutamate receptor 3 (GRM3) single nucleotide polymorphisms (SNPs) and total scores of the Brief Psychiatric Rating Scale (BPRS)A
| SNP | BETA | SE | R2 | T-score | P-value | Corrected P-value B |
|---|---|---|---|---|---|---|
| rs274622 | -0.58 | 1.58 | 0.0015 | -0.37 | 0.71 | 1.0 |
| rs724226 | -0.19 | 1.55 | 1.6E-4 | -0.12 | 0.90 | 1.0 |
| rs917071 | -1.93 | 1.50 | 0.018 | -1.29 | 0.20 | 0.69 |
| rs6465084 | -0.67 | 1.73 | 0.0016 | -0.39 | 0.70 | 1.0 |
| rs1468412 | -1.60 | 1.43 | 0.013 | -1.12 | 0.27 | 0.80 |
| rs1989796 | 4.24 | 1.49 | 0.080 | 2.84 | 0.0055 | 0.04 |
| rs1476455 | -6.79 | 2.47 | 0.075 | -2.75 | 0.0071 | 0.04 |
Additive model
Permuted p-value correcting for multiple comparisons and linkage disequilibrium between SNPs
Acknowledgments
Sources of Support and Acknowledgements: This project was supported by grants from the National Institute of Mental Health (K23MH01758, K08MH64158, and K08MH083888), the NIH-NCCR, and the General Clinical Research Center (GCRC) Program (M01-RR-59).
Footnotes
Conflict of Interest Statement:
Dr. Bishop has received research grant support from Ortho-McNeil Janssen and honoraria from Eli Lilly
Dr. Miller has served on data safety monitoring boards for Otsuka Pharmaceuticals and GlaxoSmithKline, and received research grant support in the form of medical supplies from Astra Zeneca, Bristol Myers Squibb, Eli Lilly, Janssen Pharmaceutica and Pfizer.
Dr. Ellingrod has served on an advisory board for Eli Lilly
Mr. Holman has no potential conflicts to disclose
Prior Publication:
Portions of this research were presented as an abstract and poster at the 2009 International Congress on Schizophrenia Research Annual Meeting
References
- Albalushi T, Horiuchi Y, Ishiguro H, et al. Replication study and meta-analysis of the genetic association of GRM3 gene polymorphisms with schizophrenia in a large Japanese case-control population. Am J Med Genet B Neuropsychiatr Genet. 2008;147:392–396. doi: 10.1002/ajmg.b.30610. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Ellingrod VL, Moline J, Miller D. Association between the polymorphic GRM3 gene and negative symptom improvement during olanzapine treatment. Schizophr Res. 2005;77:253–260. doi: 10.1016/j.schres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Wang K, Moline J, Ellingrod VL. Association analysis of the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatr Genet. 2007;17:358. doi: 10.1097/YPG.0b013e3281ac231e. [DOI] [PubMed] [Google Scholar]
- Cartmell J, Schoepp DD. Regulation of neurotransmitter release by metabotropic glutamate receptors. Journal of Neurochemistry. 2000;75:889–907. doi: 10.1046/j.1471-4159.2000.0750889.x. [DOI] [PubMed] [Google Scholar]
- Chen Q, He G, Chen Q, et al. A case-control study of the relationship between the metabotropic glutamate receptor 3 gene and schizophrenia in the Chinese population. Schizophr Res. 2005;73:21–26. doi: 10.1016/j.schres.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annual Review of Pharmacology & Toxicology. 1997;37:205–237. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- Corti C, Xuereb JH, Corsi M, Ferraguti F. Identification and characterization of the promoter region of the GRM3 gene. Biochem Biophys Res Commun. 2001;286:381–387. doi: 10.1006/bbrc.2001.5391. [DOI] [PubMed] [Google Scholar]
- Egan MF, Straub RE, Goldberg TE, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci U S A. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijal BA, Kinon BJ, Kapur S, et al. Candidate-gene association analysis of response to risperidone in African-American and white patients with schizophrenia. Pharmacogenomics J. 2009;9:311–318. doi: 10.1038/tpj.2009.24. [DOI] [PubMed] [Google Scholar]
- Fujii Y, Shibata H, Kikuta R, et al. Positive associations of polymorphisms in the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatr Genet. 2003;13:71–76. doi: 10.1097/01.ypg.0000056682.82896.b0. [DOI] [PubMed] [Google Scholar]
- Girgenti MJ, Nisenbaum LK, Bymaster F, Terwilliger R, Duman RS, Newton SS. Antipsychotic-induced gene regulation in multiple brain regions. J Neurochem. 2010;113:175–187. doi: 10.1111/j.1471-4159.2010.06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff DC, Hennen J, Lyoo IK, et al. Modulation of brain and serum glutamatergic concentrations following a switch from conventional neuroleptics to olanzapine. Biological Psychiatry. 2002;51:493–497. doi: 10.1016/s0006-3223(01)01321-x. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Lyon L, Sartorius LJ, Burnet PW, Lane TA. The group II metabotropic glutamate receptor 3 (mGluR3, mGlu3, GRM3): expression, function and involvement in schizophrenia. J Psychopharmacol. 2008;22:308–322. doi: 10.1177/0269881108089818. [DOI] [PubMed] [Google Scholar]
- Javitt DC. Is the glycine site half saturated or half unsaturated? Effects of glutamatergic drugs in schizophrenia patients. Curr Opin Psychiatry. 2006;19:151–157. doi: 10.1097/01.yco.0000214340.14131.bd. [DOI] [PubMed] [Google Scholar]
- Jonsson EG, Saetre P, Vares M, et al. DTNBP1, NRG1, DAOA, DAO and GRM3 Polymorphisms and Schizophrenia: An Association Study. Neuropsychobiology. 2009;59:142–150. doi: 10.1159/000218076. [DOI] [PubMed] [Google Scholar]
- Kane JM, Honigfeld G, Singer J, Meltzer H. Clozapine in treatment-resistant schizophrenics. Psychopharmacol Bull. 1988;24:62–67. [PubMed] [Google Scholar]
- Lahiri DK, Nurnberger JI., Jr. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991;19:5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti SB, Cichon S, Propping P, Nothen M. Metabotropic glutamate receptor 3 (GRM3) gene variation is not associated with schizophrenia or bipolar affective disorder in the German population. Am J Med Genet. 2002;114:46–50. doi: 10.1002/ajmg.1624. [DOI] [PubMed] [Google Scholar]
- Moghaddam B. Targeting metabotropic glutamate receptors for treatment of the cognitive symptoms of schizophrenia. Psychopharmacology (Berl) 2004;174:39–44. doi: 10.1007/s00213-004-1792-z. [DOI] [PubMed] [Google Scholar]
- Molteni R, Calabrese F, Racagni G, Fumagalli F, Riva MA. Antipsychotic drug actions on gene modulation and signaling mechanisms. Pharmacol Ther. 2009;124:74–85. doi: 10.1016/j.pharmthera.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- Nicodemus KK, Kolachana BS, Vakkalanka R, et al. Evidence for statistical epistasis between catechol-O-methyltransferase (COMT) and polymorphisms in RGS4, G72 (DAOA), GRM3, and DISC1: influence on risk of schizophrenia. Hum Genet. 2007;120:889–906. doi: 10.1007/s00439-006-0257-3. [DOI] [PubMed] [Google Scholar]
- Norton N, Williams HJ, Dwyer S, et al. No evidence for association between polymorphisms in GRM3 and schizophrenia. BMC Psychiatry. 2005;5:23. doi: 10.1186/1471-244X-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil ST, Zhang L, Martenyi F, et al. Activation of mGlu2/3 receptors as a new approach to treat schizophrenia: a randomized Phase 2 clinical trial. Nat Med. 2007;13:1102–1107. doi: 10.1038/nm1632. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva MA, Tascedda F, Lovati E, Racagni G. Regulation of NMDA receptor subunit messenger RNA levels in the rat brain following acute and chronic exposure to antipsychotic drugs. Brain Research Molecular Brain Research. 1997;50:136–142. doi: 10.1016/s0169-328x(97)00175-7. [DOI] [PubMed] [Google Scholar]
- Scherer SW, Duvoisin RM, Kuhn R, Heng HH, Belloni E, Tsui LC. Localization of two metabotropic glutamate receptor genes, GRM3 and GRM8, to human chromosome 7q. Genomics. 1996;31:230–233. doi: 10.1006/geno.1996.0036. [DOI] [PubMed] [Google Scholar]
- Schwab SG, Plummer C, Albus M, et al. DNA sequence variants in the metabotropic glutamate receptor 3 and risk to schizophrenia: an association study. Psychiatr Genet. 2008;18:25–30. doi: 10.1097/YPG.0b013e3282ef48d9. [DOI] [PubMed] [Google Scholar]
- Tamminga CA. The neurobiology of cognition in schizophrenia. J Clin Psychiatry. 2006;67(Suppl 9):9–13. discussion 36-42. [PubMed] [Google Scholar]
- Tan HY, Chen Q, Sust S, et al. Epistasis between catechol-O-methyltransferase and type II metabotropic glutamate receptor 3 genes on working memory brain function. Proc Natl Acad Sci U S A. 2007;104:12536–12541. doi: 10.1073/pnas.0610125104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tascedda F, Blom JM, Brunello N, et al. Modulation of glutamate receptors in response to the novel antipsychotic olanzapine in rats. Biological Psychiatry. 2001;50:117–122. doi: 10.1016/s0006-3223(01)01135-0. [DOI] [PubMed] [Google Scholar]
- Tochigi M, Suga M, Ohashi J, et al. No association between the metabotropic glutamate receptor type 3 gene (GRM3) and schizophrenia in a Japanese population. Schizophr Res. 2006;88:260–264. doi: 10.1016/j.schres.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Woods SW. Chlorpromazine equivalent doses for the newer atypical antipsychotics. J Clin Psychiatry. 2003;64:663–667. doi: 10.4088/jcp.v64n0607. [DOI] [PubMed] [Google Scholar]