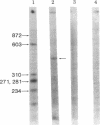
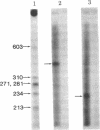
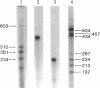
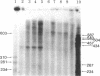
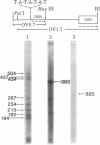
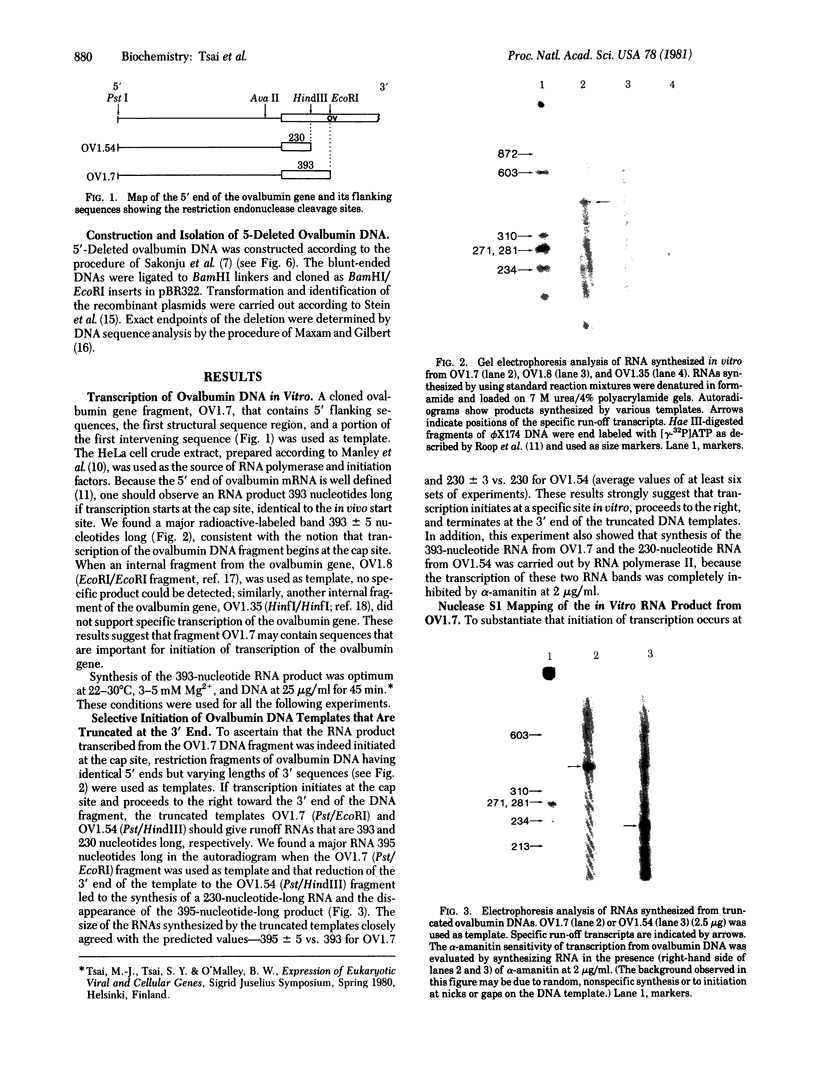
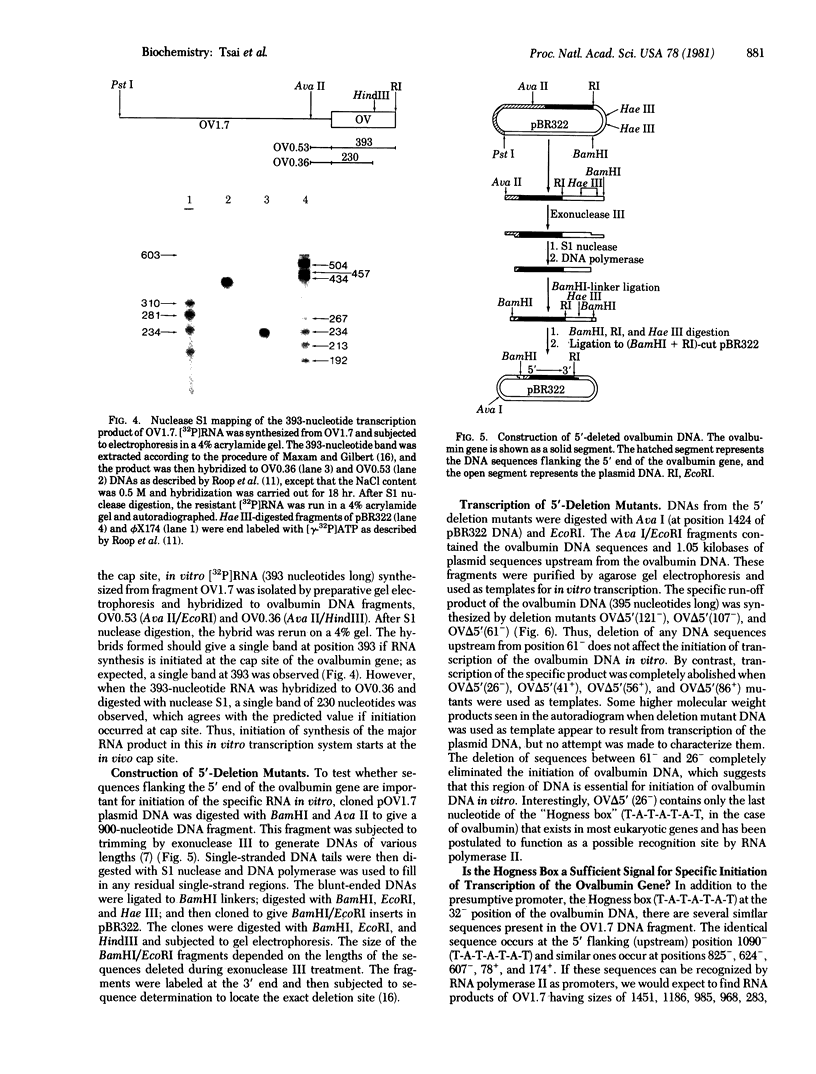
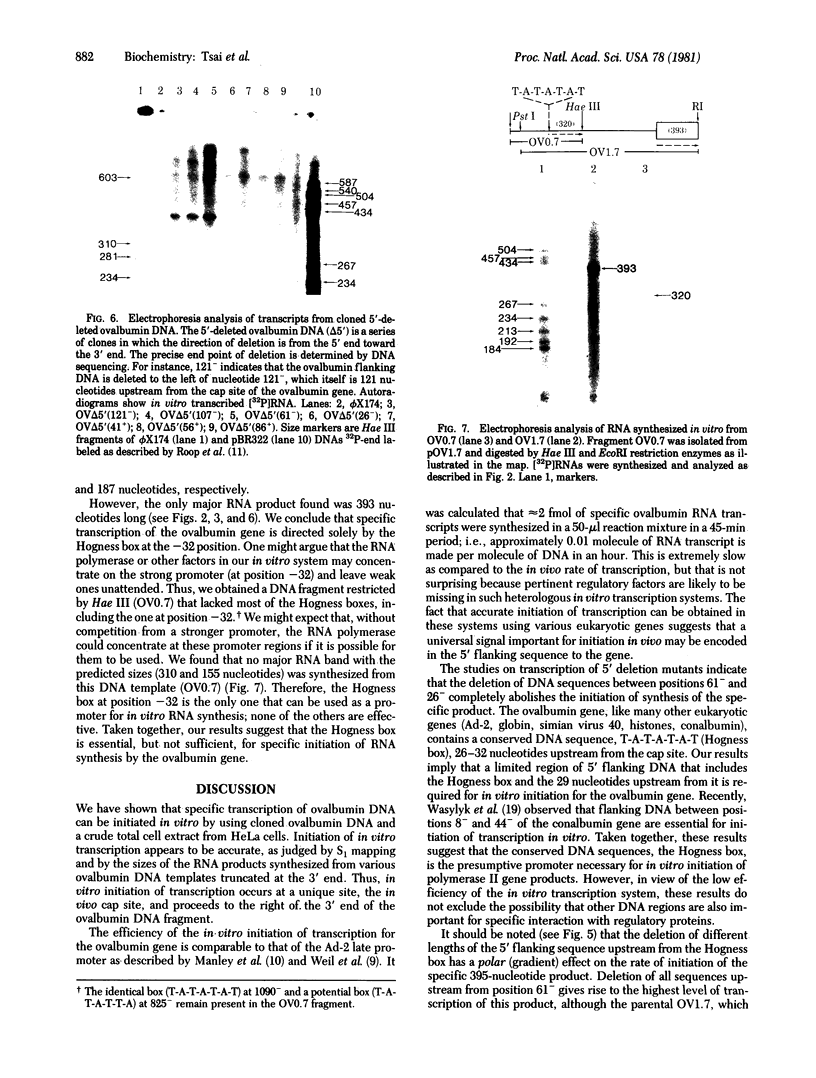
Abstract
An in vitro system [Weil, P. A., Luse, D. S., Segall, J. & Roeder, R. G. (1979) Cell 18, 469-484 and Manley, J. L., Fire, A., Cano, A., Sharp, P. A. & Gefter, M. L., (1980) Proc. Natl. Acad. Sci USA 77, 3855-3859] was adapted for studying initiation of transcription of the ovalbumin gene. The DNA template was a cloned ovalbumin gene fragment that contained 5′ flanking sequences, the first structural sequence region, and a portion of the first intervening sequence. A HeLa cell crude extract was used as the source of RNA polymerase and initiation factors. Correct initiation was judged by the sizes of the transcription products generated from ovalbumin templates truncated at various positions before the 3′ end of the gene. Transcription of the specific product was carried out by RNA polymerase II, as judged from α-amanitin sensitivity. A series of deletion mutants was constructed by trimming 5′ flanking sequences of the ovalbumin DNA template by using exonuclease III. The DNAs generated were then cloned in pBR322 and used as templates to determine which sequences were necessary for initiation of transcription. Specific initiation of the ovalbumin gene was unaffected by deletion of all but 61 nucleotides of the 5′ flanking sequence but completely abolished by deletion of all but 26 nucleotides of 5′ flanking sequence. Thus, a region between 61 and 26 nucleotides upstream from the cap site, which includes the Hogness box (T-A-T-A-T-A-T) at position 32-26, is essential for the correct initiation of the ovalbumin gene. Nevertheless, natural DNA fragments containing false Hogness boxes (T-A-T-A-A-A-A and T-A-T-A-T-A-T) not normally located in the immediate 5′ flanking region of an authentic gene did not serve as promoters for initiation of transcription. These results suggest that the Hogness box is essential, but not sufficient, for specific initiation of RNA synthesis.
Keywords: deletion mutants, Hogness box, eukaryotic promoter
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Woo S. L., Colbert D. A., Lai E. C., Mace M. L., Jr, O'Malley B. W. The ovalbumin gene: cloning and molecular organization of the entire natural gene. Proc Natl Acad Sci U S A. 1979 May;76(5):2253–2257. doi: 10.1073/pnas.76.5.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugaiczyk A., Woo S. L., Lai E. C., Mace M. L., Jr, McReynolds L., O'Malley B. W. The natural ovalbumin gene contains seven intervening sequences. Nature. 1978 Jul 27;274(5669):328–333. doi: 10.1038/274328a0. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. Y., Parker C. S., Roeder R. G. Transcription of cloned Xenopus 5S RNA genes by X. laevis RNA polymerase III in reconstituted systems. Proc Natl Acad Sci U S A. 1979 Jan;76(1):136–140. doi: 10.1073/pnas.76.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roop D. R., Tsai M. J., O'Malley B. W. Definition of the 5' and 3' ends of transcripts of the ovalbumin gene. Cell. 1980 Jan;19(1):63–68. doi: 10.1016/0092-8674(80)90388-8. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Schmidt O., Mao J. I., Silverman S., Hovemann B., Söll D. Specific transcription of eukaryotic tRNA genes in Xenopus germinal vesicle extracts. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4819–4823. doi: 10.1073/pnas.75.10.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. P., Catterall J. F., Woo S. L., Means A. R., O'Malley B. W. Molecular cloning of ovomucoid gene sequences from partially purified ovomucoid messenger RNA. Biochemistry. 1978 Dec 26;17(26):5763–5772. doi: 10.1021/bi00619a025. [DOI] [PubMed] [Google Scholar]
- Tsai S. Y., Roop D. R., Stumph W. E., Tsai M. J., O'Malley B. W. Evidence that deoxyribonucleic acid sequences flanking the ovalbumin gene are not transcribed. Biochemistry. 1980 Apr 29;19(9):1755–1761. doi: 10.1021/bi00550a005. [DOI] [PubMed] [Google Scholar]
- Tsai S. Y., Roop D. R., Tsai M. J., Stein J. P., Means A. R., O'Malley B. W. Effect of estrogen on gene expression in the chick oviduct. Regulation of the ovomucoid gene. Biochemistry. 1978 Dec 26;17(26):5773–5780. doi: 10.1021/bi00619a026. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Kédinger C., Corden J., Brison O., Chambon P. Specific in vitro initiation of transcription on conalbumin and ovalbumin genes and comparison with adenovirus-2 early and late genes. Nature. 1980 Jun 5;285(5764):367–373. doi: 10.1038/285367a0. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Luse D. S., Segall J., Roeder R. G. Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979 Oct;18(2):469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- Wu G. J. Adenovirus DNA-directed transcription of 5.5S RNA in vitro. Proc Natl Acad Sci U S A. 1978 May;75(5):2175–2179. doi: 10.1073/pnas.75.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]