Abstract
Amino acid response (AAR) pathway is activated when cells are deprived of amino acids. In the present study, using the human colon cancer cell line SW480, we observed that DKK1, an antagonist of the Wnt pathway, was significantly induced at the mRNA level after the removal of amino acids from the medium. Addition of the amino alcohol histidinol, which prevents the formation of histidinyl-tRNAHis, also increased DKK1 mRNA to a level similar to that observed when cells were deprived of all amino acids. Transcriptional activity and stability of DKK1 mRNA were both increased in the amino acid-deprived condition. The induction of DKK1 gene expression was confirmed by the increased immunofluorescent staining of the DKK1 protein in the amino acid deprived condition. Although Chromatin Immunoprecipitation assays showed increased RNA Polymerase II binding at the DKK1 promoter in amino acid-limited conditions, ATF4 binding to the promoter is absent. Luciferase reporter assays did not detect any functional AARE within the DKK1 gene structure. Knockdown of ATF4 by siRNA did not affect the increase of DKK1 mRNA during amino acid limitation. Inhibition of ERK phosphorylation abolished the induction of DKK1. Our study revealed that DKK1 is a novel target gene in the response to amino acid deficiency and that the expression of DKK1 is up-regulated through an ATF4-independent and an ERK-dependent pathway.
Keywords: amino acid deficiency; ATF4, DKK1; phosphorylated ERK; transcription; Wnt
1. Introduction
The availability of amino acids is critical to the maintenance of body and organ protein homeostasis [1]. At the cellular level, AAR pathways are activated in response to amino acid deficiency. The GCN2/ATF4 pathway is one of the well-documented AAR pathways involved in the regulation of gene expression in response to amino acid availability [2]. In this pathway, amino acid limitation induces the phosphorylation of eukaryotic initiation factor α (eIFα), which leads to a general decrease in translation of mRNA but an increase of specific amino acid responsive genes such as activating transcription factor 4 (ATF4) [3]. Elevated ATF4 activates many downstream genes including asparagine synthetase (ASNS) [4], ATF3 [5] and system A amino acid transporter 2 (SNAT2) [6]. Emerging evidence shows that the activation of the GCN2/ATF4 pathway plays critical role in tumor cell survival [7, 8].
Mitogen-activated protein kinase (MAPK) pathways are signaling cascades by which cells respond to extracellular stimuli. One of the three major MAPK pathways, the ERK pathway, is initiated by the activation of Raf and subsequent MEK and ERK kinases [9]. A recent study demonstrated that MEK/ERK signaling is required for phosphorylation of eIFα in human hepatoma cells by amino acid limitation [10].
The Wnt signaling pathway plays an important role in development and carcinogenesis [11]. Wnt is a combination of two homologous gene families, the wingless gene functioning during Drosophila embryogenesis [12], and the int genes identified as the vertebrate counterpart [13]. The canonical Wnt pathway in mammalian cells is activated when Wnt proteins bind to cell-surface receptors of the Frizzled family, thus activating the Dishevelled proteins, which then block the β-catenin degradation complex (GSK3β/APC/AXIN). This results in the translocation of β-catenin from the cytosol to the nucleus and the initiation of the downstream transcription [14]. Aberrantly activated Wnt signaling was observed in over 90% of colorectal cancers [15]. DKK1, a Wnt antagonist, blocks the Wnt/β-catenin pathway through competitive binding to the Wnt co-receptor LPR5/6 and the transmembrane protein Kremen1/2 [16–19]. Consistent with the state of Wnt signaling, DKK1 expression was down-regulated in human colon tumors [20].
Our laboratory previously reported that the expression of one of the Wnt signals, WNT5a, was reduced during amino acid deprivation in human colon cancer cell line SW480 [21]. This was accompanied with the reduction of the intracellular Wnt signal β-catenin. In the present study, our results showed the up-regulation of one of the antagonists, DKK1, of the Wnt signaling pathway by amino acid deprivation. Therefore the Wnt signaling pathway may be responsive to environmental signals through the amino acid response pathway. The increased expression of DKK1 is mediated in an ATF4-independent and MAPK/ERK-dependent manner.
2. Materials and Methods
2.1. Cell culture and amino acid deprivation treatments
Human colon adenocarcinoma cell line SW480 was obtained from American Type Culture Collection (Manassas, VA) and was cultured in standard Minimum Essential Medium (MEM) supplemented with 10% (v/v) fetal bovine serum (Mediatech; Herndon, VA) and 100 IU/mL Penicillin, and 100 μg/mL Streptomycin (Mediatech; Herndon, VA). Cells were maintained at 37 °C in 95% humidity and 5% CO2. For amino acid deprivation treatment, cells were first seeded at a density of 1.5 × 106 cells per 60-mm dish, grown for 24 h, and then given fresh media 14 h before treatments. Amino acid deprivation treatment was performed by incubating the cells in MEM or in amino acid-free MEM (MEM-AA, Cell Media Facility at the University of Illinois at Urbana-Champaign, Urbana, IL) supplemented by 10% (v/v) dialyzed fetal bovine serum (Sigma-Aldrich; St. Louis, MO). For single amino acid deprivation treatment, the above-mentioned regular MEM was made without adding the amino acid histidine (MEM-His). When inhibitors were used, an aliquot of either the stock solution or the solvent as the control was added directly to the cells in fresh media. Histidinol (HisOH) and Actinomycin D (ActD) were purchased from Sigma-Aldrich while U0126 was purchased from Cell Signaling Technology (Danvers, MA).
2.2. Quantitative real-time RT PCR
Total RNA was isolated using TRI Reagents (Sigma-Aldrich). cDNA was generated using high-capacity cDNA kit (Applied Biosystems; Foster City, CA). Real-time PCR was performed in a 20 μL reaction volume containing 12.5 μL POWER SYBR green master mix (2×, Applied Biosystems), 5 μmol/L of each primer, and 25 ng cDNA template by cycling at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 sec and 60 °C for 1 min on a 7300 real-time PCR system (Applied Biosystems). After amplification, melting curves were acquired stepwise from 55 to 95 °C to ensure that a single product was amplified in the reaction. The primers, synthesized by Integrated DNA Technologies, are as follows: DKK1, sense 5′- GATCATAGCACCTTGGATGGG -3′, antisense 5′-GGCACAGTCTGATGACCGG -3'; ASNS, sense, 5′- GCAGCTGAAAGAAGCCCAAGT-3′, antisense 5′-TGTCTTCCATGCCAATTGCA-3′; ATF3, sense 5′- GCTGTCACCACGTGCAGTATCTCA -3′, antisense 5′- CTGTTCCTCCTCTTGCTGACAAGC -3′; ATF4, sense 5′- TGAAGGAGTTCGACTTGGATGCC -3′, antisense 5′- CAGAAGGTCATCTGGCATGGTTTC -3′; p21, sense 5′- CCGCGACTGTGATGCGCTAATG -3′, antisense 5′- CTCGGTGACAAAGTCGAAGTTC -3′; L7a, sense 5′- TTTGGCATTGGACAGGACATCC -3′, antisense 5′- AGCGGGGCCATTTCACAAAG -3′. L7a was used as an internal control to normalize raw data.
2.3. Western blotting
Cells were lysed with 1× Laemmli Buffer [62.5mmol/L Tris-HCl, pH6.8, 2% SDS, 10% Glycerol v/v, 0.01% Bromophenol blue, 5% 2- mercaptoethanol, 1× protease inhibitors (Roche, Indianapolis, IN) and 1× phosphatase inhibitors (Sigma-Aldrich)]. Protein concentration was quantified by Lowry protein assay. Equal amounts of samples were size-fractionated on a 12% Tris-HCl polyacrylamide gel and transferred to a PVDF membrane (Bio-Rad, Hercules, CA). PVDF membranes were then incubated with blocking solution [Tris-buffered saline/Tween (TBST), 20 mmol/L Tris-HCl, pH 7.6, 137 mmol/L NaCl, and 0.1% v/v Tween-20] containing 5% (w/v) Carnation nonfat dry milk for 1 h at room temperature. Primary antibodies were diluted in blocking solution according to manufacturers' instructions and incubated for 3 h at room temperature. Subsequently, the membranes were washed with TBST 5 times for 5 min each time. HRP-conjugated secondary antibody was diluted 1:10,000 in blocking solution containing 1% (w/v) Carnation nonfat dry milk and incubated for 1 h at room temperature. After 5 washes of 5 min each with TBST, the membranes were exposed to the enhanced chemiluminescence reagent SuperSignal West Dura (Pierce; Rockford, IL). Signals were detected and quantified using ChemiDoc XRS imaging system (Bio-Rad). For primary antibodies, ATF4 (sc-200) and phosphorylated ERK (p-ERK, sc-7383) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and phosphorylated eIF2α (p-eIF2α, #9721) was purchased from Cell Signaling Technology (Danvers, MA). Secondary antibodies were purchased from Bio-Rad. For normalization of loading and blotting, a non-specific band from ATF4 antibody was used. Data were presented as ratio to the non-specific band.
2.4. Chromatin immunoprecipitation assay
For ChIP analysis, 1.5 × 107 cells were seeded per 150-mm dish. Amino acid deprivation treatment was performed as above. Cells from three 150-mm dishes were used for each treatment and later pooled together as one sample. At 8 h after treatment, protein and DNA were cross-linked by adding formaldehyde directly to the culture medium to a final concentration of 1%. Cross-linking was stopped 10 min later by adding 2 mol/L glycine to a final concentration of 0.125 mol/L. Cross-linked chromatin was solubilized by sonication using a Sonic Dismembrator (Model 100, Fisher Scientific Co.) for five bursts of 40 s at 50% of maximum power output with 2 min cooling on ice between each burst. The chromatin immunoprecipitation procedure used was previously described [22]. Briefly, solubilized chromatin from three dishes of cells was diluted 1:5 to 1:10 with dilution buffer and each portion was incubated with 2 μg of antibody. Normal rabbit IgG was used as a negative control for the immunoprecipitation procedure. Antibodies against ATF4 (sc-200) and Pol II (sc-899) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Acetyl-histone H3 (06-599) and acetyl-histone H4 (06-866) were from Upstate Biotechnology (www.millipore.com). The antibody-bound complex was precipitated by Protein A-Sepharose beads (Zymed). The DNA fragments in the immunoprecipitated complex were released by reversing the cross-linking at 65 °C for 5 h and purified using a QIAprep Spin Miniprep Kit (Qiagen). Purified DNA was analyzed by quantitative real time PCR with primers covering the DKK1 proximal promoter region −95 to −28 (sense, 5'-GGCTTTGTTGTCTCCCTCCCAAG-3' and antisense, 5'-CCACCGCGGCTGCCTTTATA-3'), and the ASNS proximal promoter region −87 to −22 (sense, 5'- TGGTTGGTCCTCGCAGGCAT -3' and antisense, 5'- CGCTTATACCGACCTGGCTCCT -3').
2.5. Transcriptional rate analysis
To measure the transcriptional activity of the DKK1 gene, the method of Sandoval et al. [23] was used, which relies on ChIP analysis to examine Poll II binding at the coding region distal to the promoter. Primers within a downstream region away from the transcription start site were used to monitor the transcription rate according to the quantity of DNA bound to Pol II. A higher transcription rate is represented as more Pol II binding at the given time point. Primers used to measure the transcriptional activity were: DKK1 +710 to +809 (sense, 5'-ATCTGTCTCGCCTGCAGGAAGC-3' and antisense, 5'-AAGGGAGCTTTCAGGACTCACCAT-3').
2.6. Short interfering RNA (siRNA) assay
The siRNA for control Hypoxanthine-guanine phosphoribosyltransferase and ATF4 were purchased from IDT DNA technologies (www.idtdna.com) and the DharmaFECT-4 transfection reagent was purchased from Dharmacon, Inc. (Lafayette, CO). Cells were seeded into 6-well plates at a density of 3× 105 cells/well in MEM and grown for 16 h. Transfection was performed according to the instructions from Dharmacon, using 3 μL of DharmaFECT-4 and 20 nmol/L per well final siRNA concentration. Cells were treated with transfection reagent for 48 h, then rinsed with PBS, given fresh MEM, and cultured for another 12 h. The medium was then removed and replaced with control MEM or amino acid free MEM. Total RNA and protein extracts were isolated at 8 h after the treatment and analyzed by quantitative real-time RT-PCR and immunoblotting, respectively.
2.7. Construction of DKK1 promoter and enhancer sequences for reporter assay
The DKK1 gene sequence was obtained from The Ensembl Project using the accession number ENSG00000107984. Partially truncated promoter regions were cloned into the pGL3-basic vector (Promega; Madison, WI) as following: P1 (−960/ +23), P2 (−264/+2173), P3 (−349/ +23), P4 (−733/ +23), and P5 (−555/ +23). Enhancer regions covering predicted amino acids response elements (AARE) were cloned into the pGL3-promoter vector (Promega) as following: E1 (+1109/ +1305), E2 (+1210/ +1406), E3 (+1860/ +2060), E4 (+2036/ +2253), E5 (−3000/ −2701), and E6 (+2915/ +3101). The human ASNS promoter region (−115/ +1) was used as a positive control. The resulting clones were confirmed by DNA sequencing at the DNA sequencing facility at the University of Illinois at Urbana-Champaign, Urbana, IL. Plasmids were harvested and purified using Wizard SV plasmid midi-prep kit (Promega).
2.8. Luciferase reporter assay
Cells were plated at a density of 1 × 105 cells / well in 24-well plates (Costar; Cambridge, MA) 24 h before transfection. One μg plasmid and 5 ng pRL-SV40 were co-transfected into each well using Superfect transfection reagent (Qiagen; Valencia, CA) following manufacturer's instructions. Amino acid deprivation treatment was started at least 16 h after transfection and lasted for 8 h. Cells were then lysed with PLB buffer and subjected to one round of freeze-thaw. Luciferase activity was measured using Dual-Luciferase Assay (Promega) using a luminometer (Fermomaster FB12, Zylux Corporation; Huntsville, AL). For the ATF4 over-expression experiment, reporter-gene plasmid was co-transfected with plasmid containing ATF4 cDNA sequence or empty vector itself following the same procedure as mentioned above.
2.9. Immunofluorescent staining
Cells were plated at a density of 0.3 × 106 cells / well of 6-well plates with glass cover slips in the wells. Subsequent treatments were performed following the above-mentioned procedures. At the end of each treatment, cells were washed with 1× PBS for 3 times and fixed by 4% Paraformaldehyde treatment for 30 min on the cover slips, followed by 0.5% Triton X-100 for 15 min for permeabilization. Image-iT FX signal enhancer (Invitrogen) was used to block the samples for 30 min and anti-DKK1 antibody (Abcam 61304, 1:250) was applied to the cover slips for 2 hours at 37 °C. A goat anti-rabbit antibody with Alexa Fluor 647 (1:200 dilution, Invitrogen) was applied to the cover slip for 45 min in a dark chamber. The samples were counterstained with Hoechst (Invitrogen) for 15 min and washed off by 1xPBS. Cover slips were mounted onto microscope slides using Prolong Gold antifade reagent (Invitrogen). Pictures were taken using the Zeiss Axiovert 200M microscope (Zeiss).
2.10. Statistical analysis
Standard student's t test was performed for each experiment. Each data point represents the mean of at least three independent experiments. Error bars represent calculated standard deviations from the mean. P values were calculated using student's t-test and asterisks indicate P <0.05.
3. RESULTS
3.1. DKK1 mRNA level was increased in SW480 after amino acid deprivation
We have shown previously that amino acid deprivation in human colon cancer cell line SW480 caused reduction of Wnt signaling through the down-regulation of WNT5a [21]. In the present study, we investigated the expression of DKK1, an antagonist of the Wnt pathway. Using real time PCR, we observed that the DKK1 mRNA level was significantly increased in amino acid-deprived SW480 colon cancer cells (Fig. 1a). The induction peaked at approximately 8 h and then decreased almost to the initial level within 24 h. To investigate the contribution from either transcriptional up-regulation or increase of mRNA stability by amino acid limitation, we monitored the transcription rate by following the binding of RNA Polymerase II within the DKK1 coding region [23]. There was a 3-fold increase of transcriptional activity in amino acid-deprived cells (Fig. 1b). For mRNA stability assay, SW480 was deprived from total amino acids for 8 h and then treated with MEM, MEM-AA, or MEM-AA plus the p-ERK inhibitor, U0126, with all media containing 5 μmol/L Actinomycin D. The addition of Actinomycin D blocked further transcription; thereby allowing the observation of changes in mRNA stability. In the amino acid-deprived condition, the half-life of DKK1 mRNA increased to 2.25 h compared to 1 h in MEM (Fig. 1c). More importantly, the addition of the phosphor-ERK inhibitor U0126 in the amino acid deprived media did not change the half life of the DKK1 mRNA, indicating that phosphorylation of ERK may not be involved in the regulation of mRNA stability of DKK1.
Fig. 1. DKK1 gene expression was induced by amino acid deprivation.
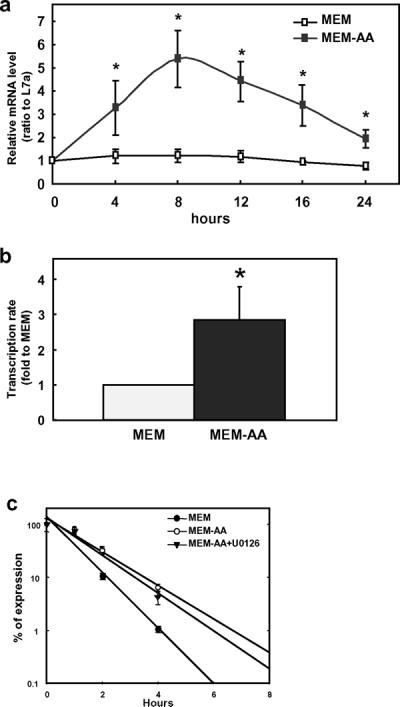
a) Time course of the total amino acid deprivation treatment. SW480 was grown in complete medium (MEM) and amino acid deficiency medium (MEM-AA) for 0–24 h. Relative mRNA level was determined by quantitative real time RT-PCR. Data points were normalized by internal control L7a and to time zero. b) Transcription rate of DKK1 gene during amino acid deprivation. ChIP analysis was performed to examine Pol II binding at the coding region distal to the promoter. Data were presented as ratio to MEM. c) mRNA stability of DKK1 following amino acid deprivation. Each data point represents the mean ± S. D of three independent experiments. Asterisks (*) represent statistical significance compared to values in MEM (P < 0.05).
3.2. DKK1 expression was increased by histidinol treatment to the similar level as the amino acid deprivation
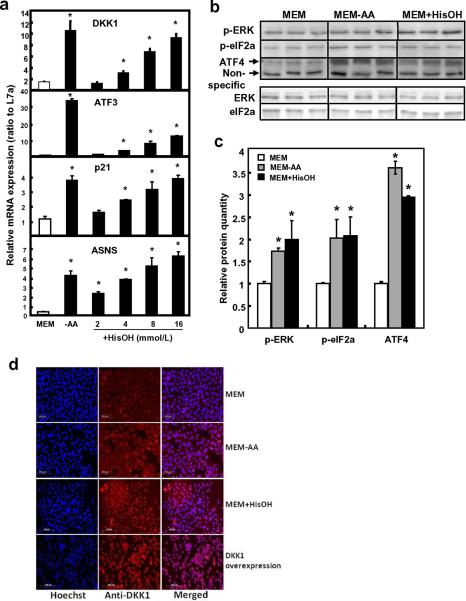
Amino alcohol histidinol (HisOH) prevents the formation of histidinyl-tRNAHis and could be used to test the effect from histidinyl-tRNAHis blockade. Results showed that serial concentrations of HisOH also increased DKK1 mRNA to a level similar to that observed when cells were amino acid-deprived (Fig. 2a), illustrating that tRNA charging is involved. Typical AAR pathway responsive genes, such as ASNS, ATF3 and p21waf1, were also activated in a similar manner (Fig. 2a). Likewise, protein expression of ATF4, p-eIF2α and p-ERK was induced under amino acid deprivation and by the addition of 16 mmol/L of HisOH (Fig. 2b, 2c). The induction of DKK1 gene expression was confirmed by increased immunofluorescent staining of the DKK1 protein in both the amino acid deprived condition and the MEM+HisOH condition where histidine availability was blocked (Fig. 2d). A parallel treatment where DKK1 protein was overexpressed by in vitro transfection was used as a positive control to show the increased staining for the DKK1 protein by immunohistochemistry (Fig. 2d).
Fig. 2. Expression of mRNA and protein in SW480 after histidinol treatment.
a) mRNA level was quantified in human colon cancer cell line SW480 after histidinol (HisOH) treatment. SW480 cells were grown in MEM, MEM without amino acid, and MEM containing 2, 4, 8, or 16 μmol/L histidinol for 8 h. Relative mRNA level was determined by quantitative real time RT-PCR. Data were normalized by the internal control L7a. b) A representative blot from Western blotting analysis for ATF4, p-eIF2α and p-ERK protein expression. c) Protein level was quantified in SW480 by Western blotting with 16 mmol/L HisOH treatment. A non-specific band was used as the loading control to normalize the raw data. Each bar represents the mean ± S. D. of three independent experiments. Asterisks (*) represent statistical significance compared to values in MEM (P < 0.05). d) The MEM-, MEM-AA-, and MEM+HisOH-treated cells were fixed on glass cover slips and subsequently used for immunofluorescent staining. DKK1 protein was analyzed after immunofluorescent staining using an antibody against DKK1 protein and an Alexa Fluor 647-labeled secondary antibody (red). The cover slips were also counterstained with Hoechst 33342 fluorescent staining for nucleus (blue). The two pictures were overlaid to show DKK1 staining on top of the nuclear staining (merged). Cells treated with in vitro transfection of DKK1-containing plasmid was used as a positive control for the staining of DKK1 protein.
3.3. The effect from HisOH treatment on the induction of DKK1 was not due to osmolarity change
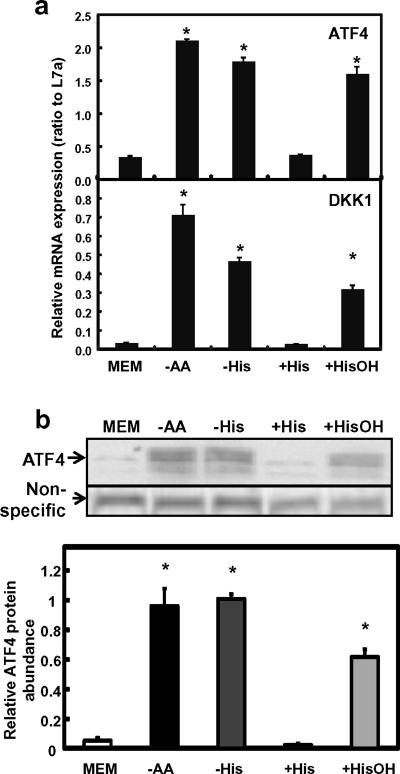
To confirm the effect from HisOH treatment, SW480 cells were treated with complete MEM, MEM without any amino acids (MEM-AA), MEM without histidine (MEM-His), MEM with 16 mmol/L histidine (MEM+His), or MEM with 16 mmol/L HisOH (MEM+HisOH, Fig. 3). MEM+His treatment was included in this set of the experiments to eliminate potential effect from the potential change in osmolarity when 16 mmol/L of HisOH was added in the MEM+HisOH treatment. Results showed that MEM-AA, MEM-His, and MEM+HisOH treatments were all effective in inducing the amino acid response (Fig. 3). Specifically, mRNA level of both DKK1 and ATF4 was upregulated similarly under either total amino acid starvation or histidine deprivation (by -His or +HisOH, Fig. 3a). ATF4 protein level increased in the three amino acid-deprived conditions as well (Fig. 3b). In contrast to the MEM+HisOH, no induction on either DKK1 or ATF4 expression was observed when the same concentration of histidine was used in the treatment media, eliminating the possibility of an osmolaric shock.
Fig. 3. DKK1 expression by total or single amino acid deprivation.
SW480 was treated with either MEM (complete), -AA (total amino acid deprivation), and -His or +HisOH (single amino acid limitation) for 8 h before samples were collected and analyzed. MEM+His was used as a negative control to amino acid starvation treatment (+His). a) mRNA expression of ATF4 and DKK1. Real time RT-PCR was performed for gene expression. L7a was used as the internal control and to normalize the data. Each bar represents the mean ± S. D. of three samples. Asterisks (*) represent statistical significance compared to values in MEM (P < 0.05). b) ATF4 protein expression analyzed by Western blotting. A non-specific band was used as the loading control to normalize the raw data. Each bar represents the mean ± S. D. of three independent samples. Asterisks (*) represent statistical significance compared to values in MEM (P < 0.05).
3.4. The induction of DKK1 mRNA by amino acid deprivation involved phosphorylation of ERK
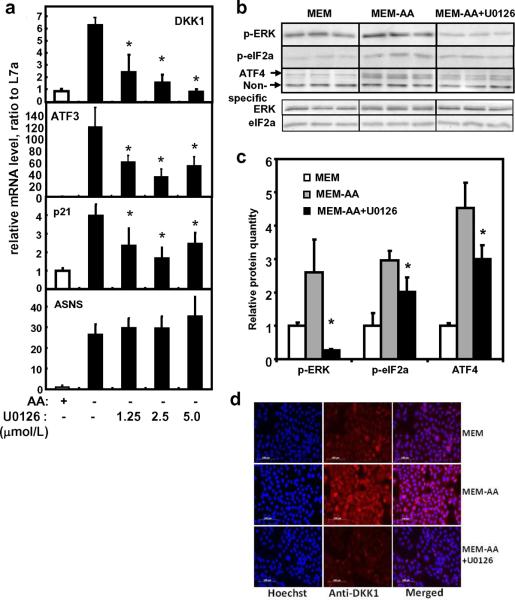
To examine the possible involvement of ERK signaling pathway in the induction of DKK1, the MEK specific inhibitor U0126 was used. Blocking of phosphorylation of ERK with U0126 at 5 μmol/L totally abolished the induction of DKK1 (Fig. 4a), indicating the dependence on the MEK/ERK pathway for DKK1 expression. On the other hand, p-eIF2α, ATF4 and the expression of target genes ATF3 and p21 were partially decreased (Fig. 4a, b, and c). Interestingly, the positive control for amino acid responsive pathway, ASNS mRNA expression, was not affected by U0126 treatment (Fig. 4a). The dependence of DKK1 gene expression on the phosphorylation of ERK was further confirmed using immunofluorescent staining of the DKK1 protein (Fig. 4d). Immunofluorescent staining clearly showed the induction of DKK1 protein by amino acid deprivation. The addition of U0126 blocked the induction of DKK1 protein and decreased the level to what is observed in the amino acid-containing condition (Fig. 4d).
Fig. 4. Expression of mRNA and protein in SW480 after U0126 treatment.
a) mRNA expression after U0126 treatment. SW480 cells were treated with either control DMSO (MEM), MEM-AA, or MEM-AA containing 1.25, 2.5, or 5 μmol/L of the MEK1/2 inhibitor U0126 for 8 h. Relative mRNA level was determined by quantitative real time RT-PCR and normalized to DMSO control. b) Western blot analysis after U0126 treatment. ATF4, p-eIF2α, and p-ERK proteins were analyzed by Western blot analysis in SW480 after U0126 treatment. Total ERK and total eIF2α were also analyzed to ensure that the changes observed for p-ERK and p- eIF2α are specific for phosphorylated forms. c) Protein abundance after U0126 treatment. Protein level of ATF4, p-eIF2α, and p-ERK was quantified after Western blot analysis in SW480 cells treated with U0126. A non-specific band was used as the loading control to normalize the raw data. Each data point represents the mean ± S. D. of at least three independent samples. For data from the U0126 treated samples, asterisks (*) represent statistical significance compared to values in amino acid deprived condition (MEM-AA, P < 0.05). d) DKK1 protein expression. The MEM-, MEM-, and MEM-AA+U0126-treated cells were fixed on glass cover slips and subsequently used for immunofluorescent staining. DKK1 protein was analyzed after immunofluorescent staining using an antibody against DKK1 protein and an Alexa Fluor 647-labeled secondary antibody (red). The cover slips were also counterstained with Hoechst 33342 fluorescent staining for nucleus (blue). The two pictures were overlaid to show DKK1 staining on top of the nuclear staining (merged).
3.5. DKK1 gene lacked a functional responsive element to amino acid deprivation
To identify a functional responsive element to amino acid limitation, we performed reporter gene assays. Using Vector NTI (Invitrogen), we analyzed a region of the DKK1 gene sequence consisting of the upstream 3 kb to the downstream 3 kb. The software predicted 5 putative AARE-like elements according to the consensus AAREs (NSRE-1: TGATGAAAC; NSRE-2: RTTKCATCA) [24]. We individually cloned 5 truncated promoter fragments of the DKK1 gene and cloned them into the pGL3-basic luciferase reporter vector and 6 DNA fragments containing the putative AARE-like elements into the pGL3-promoter vector. The constructs are tested for luciferase reporter activity. Results showed that none of the promoter regions nor any of the other predicted enhancer fragments contained a functional AARE (see supplementary file). Furthermore, when ATF4 was co-expressed with reporter plasmids containing a DKK1 promoter region or a putative AARE, the luciferase assay showed that this exogenous, over-expressed ATF4 could not induce DKK1 expression (see supplemental file). Luciferase reporter plasmid containing ASNS (−115 to +1) promoter fragment was used as a positive control and it showed the expected, reported response to the amino acid starvation (supplemental file) [22, 24].
3.6. Transcription of DKK1 was increased without ATF4 binding at the promoter
To define the possible role of transcription factor binding and chromatin modification in amino acid regulated control of DKK1 expression, we investigated the binding of ATF4 and Pol II as well as local intensity of acetyl-histone H3 and acetyl-histone H4 at the proximal promoter region of DKK1 and ASNS using chromatin immunoprecipitation. The results showed that Pol II binding to the promoters of both ASNS and DKK1 was increased under amino acid deprived condition, indicating that both ASNS and DKK1 were transcriptionally activated (Fig. 5). However, in contrast to the increased ATF4 binding to the ASNS promoter under amino acid deprived condition, ATF4 binding to the DKK1 promoter was similar to the binding level of the negative control IgG in either MEM or MEM-AA condition, which indicated that the ATF4 binding to the DKK1 promoter is not present. There was no significant difference in acetyl-histone H3 or acetyl-histone H4 level at the promoter of DKK1 in amino acid free condition compared to complete MEM. On the other hand, both acetylated histones were increased at the ASNS promoter region by amino acid deprivation (Fig. 5).
Fig. 5. Protein association at the promoter region of DKK1.
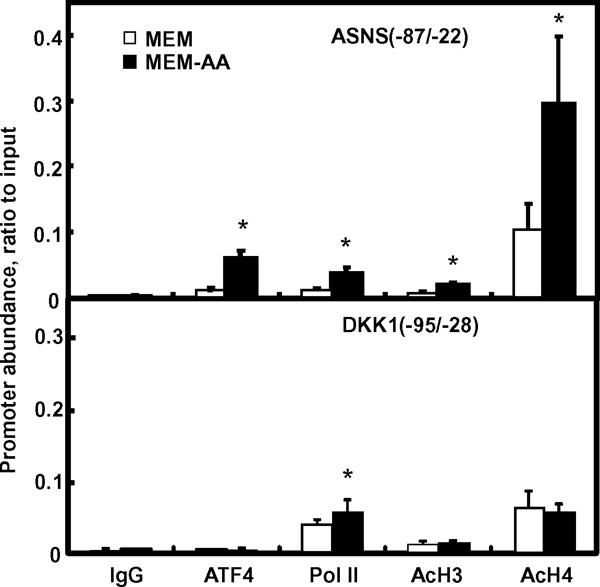
SW480 cells were grown in MEM or MEM-AA for 8 h. Samples were collected for chromatin immunoprecipitation assay. Primers located at the proximal promoter region of either DKK1 (−95/−28) or ASNS (−87/−22) were used in the real time PCR to quantify the association of promoter DNA to ATF4, Pol II, acetyl-histone H3, and acetyl-histone H4. Normal rabbit IgG was used as a negative control (IgG). Each bar represents the mean ± S. D. from three independent experiments. Asterisks (*) indicate statistical significance compared to values in MEM (P < 0.05).
3.7. Induction of DKK1 mRNA expression was independent of the expression of ATF4
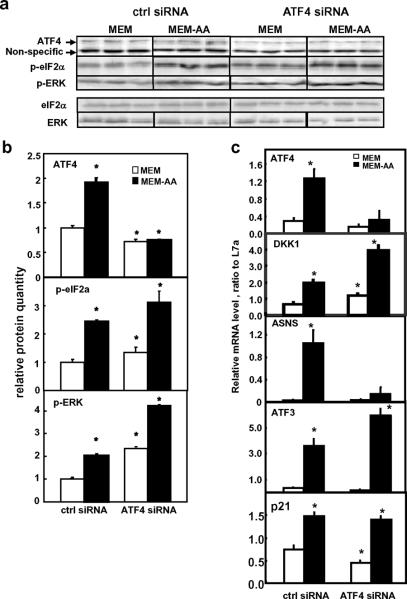
To further confirm the independence of DKK1 expression on ATF4, we investigated the expression of DKK1 when ATF4 level were knocked down by an ATF4-specific siRNA in both MEM and MEM-AA conditions. During amino acid deprivation, both ATF4 mRNA and protein were decreased by the ATF4 siRNA to a level below the well-nourished control siRNA-treated cells (Fig. 6, ctrl siRNA in MEM). Control siRNA treatment did not change the ATF4 induction by amino acid deprivation. The induction of the ATF4 responsive-, AAR-target gene ASNS was almost completely blocked by the ATF4 siRNA treatment (Fig. 6c). However, during amino acid limitation, the silencing of ATF4 did not affect the elevated DKK1, ATF3, and p21 mRNA expression (Fig. 6c) and even resulted in an increase of DKK1 mRNA. At the same time, both p-eIF2α and p-ERK remained elevated when ATF4 was blocked by siRNA (Fig. 6a and b).
Fig. 6. Effects of ATF4 siRNA transfection in SW480.
SW480 cells were transfected with control (ctrl siRNA) and ATF4 siRNA for 48 h. Culture media were changed to fresh MEM 12 h before treatment for 8 h in either MEM or MEM-AA. a) Representative blots of Western blotting analysis for ATF4, p-eIF2α and p-ERK protein expression. Total eIF2α and total ERK were examined as negative control. b) Quantification of ATF4, p-eIF2α, and p-ERK protein levels. Relative protein level was presented after normalization to the non-specific band observed. c) Relative mRNA level was determined by quantitative real time RT-PCR and normalized to the internal control L7a. Each data point represents the mean ± S. D. Asterisks (*) represent statistical significance compared to values in control siRNA in MEM (P < 0.05).
4. DISCUSSION
The present study demonstrates the link between the amino acid response pathways and the Wnt signaling pathway. We showed that DKK1, an antagonist of the Wnt pathway, is unique in its regulation, in that its expression was up-regulated by amino acid deprivation in the human colon cancer cell line SW480. The induced expression was partly due to mRNA stabilization and partly due to transcriptional activation. The transcriptional control of DKK1 expression is mediated through an ATF4-independent and MAPK/ERK-dependent manner. Histone acetylation at the promoter region under the amino acid deprived condition also contributes to the induction of the gene expression for DKK1.
AAR pathways are known for regulating specific cellular responses when there is limitation of amino acid availability. Our previous report has shown that Wnt5a, another Wnt signaling related gene, was down-regulated in the SW480 colon cancer cell line by amino acid deprivation [21]. Nuclear β-catenin, the major intracellular factor of canonical Wnt signaling, was also decreased, which is consistent with the down-regulation of the whole Wnt signaling pathway following amino acid deprivation [21]. In the present study, we observed that an antagonist of the Wnt pathway, DKK1, was up-regulated under the amino acid deprived conditions. Therefore, it is possible that the down-regulation of Wnt signaling was also a result of the up-regulation of DKK1. DKK1 has wide and complex effects on cell progression and differentiation [25–28]. The induced DKK1 by amino acid limitation is likely a link between the Wnt signaling in cancer development in response to nutritional stresses.
Amino acid response pathway has been shown to affect gene expression at multiple stages of gene expression [3]. Specifically amino acid limitation has been shown to regulate gene expression of the transcription factor ATF3 by ways of transcriptional control and mRNA stability [5, 29, 30]. These studies illustrated the amino acid-induced transcriptional upregulation of ATF3 which is mediated through the AARE by ATF4 [5]. At the same time, ATF3 expression is induced by amino acid limitation through stabilization of its mRNA by increased interaction with the mRNA-binding protein HuR1 [30]. We showed that DKK1 expression is responsive at both transcriptional level and mRNA stability, further confirmed the multiplicity of amino acid control of gene expression. The involvement of mRNA stabilizing machinery in the regulation of DKK1 during amino acid limitation is one of the critical components for amino acid regulation that warrants further investigation.
We hypothesized that the amino acid response of DKK1 transcription was mediated through the classic GCN2/ATF4 pathway, as previously reported for a number of amino acid responsive genes, including SNAT2 system A amino acid transporter [10] and ASNS [8]. We therefore targeted our study at identifying an AARE on the DKK1 genomic sequence through reporter gene analysis. However, we did not identify any amino acid responsive elements at either proximal and distal promoter regions or downstream regions as in SNAT2 and C/EBPbeta. It is entirely possible that we have not yet included the specific region related to the amino acid deficiency condition. On the other hand Palii et al [31] reported that total amino acid deprivation (KRB based medium) did induce a portion of transcription of the SNAT2 gene that is AARE-independent. Similarly in the present study, the single amino acid deprivation, either by histidine elimination or by the addition of histidinol, provides the similar induction of DKK1 transcription as the total amino acid limitation. However when the entire DKK1 gene was specifically mapped for a potential AARE using overexpression of ATF4, we still could not identify any element that is responsive to ATF4. In future research, we may need to focus the search of an amino acid responsive element on the newly proposed C/EBP-ATF response element (CARE) [4]. On the other hand, based on the results from both over-expression and knockdown of ATF4 and from the ChIP assay, it is highly likely that the increased transcription of DKK1 was regulated by a GCN2/ATF4-independent pathway that requires activation of p-ERK. Therefore, the DKK1 gene may not contain an AARE that is responsive to ATF4-dependent induction. As proposed recently, multiple pathways exist in response to amino acid deficiency [32]. The pathways proposed include the GCN2/ATF4 pathway, the signaling pathway leading to ATF2 activation, and the GCN2-independent pathway. From the evidence presented here, we propose that the SW480 cell line is utilizing the ATF4-independent pathway. It has been reported that, in HepG2 cells, blockage of ERK phosphorylation totally eliminated the phosphorylation of eIF2α and induction of ATF4 [10]. This response indicated the interdependence in HepG2 cells between ERK signaling and GCN2/ATF4 for amino acid responsiveness. In the present study in SW480 cells, blockage of ERK phosphorylation did not completely eliminate the induction of ATF4 and p-eIF2α but the induction of DKK1 gene expression, on the other hand, is completely eliminated. This indicates the dependence of DKK1 induction on the MEK/ERK signaling through phosphorylation of ERK by amino acid deprivation. On the other hand, inhibition of ATF4 expression did not affect the phosphorylation of ERK and eIF2α. And the induction of DKK1 gene expression remains unaffected, as long as there is signaling from phosphorylated ERK. We tried going back up the ATF4 signaling pathway and tested GCN2 dependence. When GCN2 mRNA is knocked down by siRNA, we did not observe any reduction in DKK1 mRNA level induced by amino acid deprivation (data not shown). Since the induction of ASNS was also only partially reduced in this experiment, we need to utilize a better system to knock down GCN2 level for future investigation (data not shown). Further studies are needed then to decipher the ATF4-independent pathway to illustrate the transcription factor that is responsive to the amino acid regulation and that relays the activating signal from the phosphorylation of ERK.
The histone modifications at the promoter regions have been related to critical regulations of mammalian gene expression. Specifically, amino acid deprivation has been shown to induce acetylation of histone H3 and H4 on the ASNS promoter [22]. For ASNS gene, the temporal mapping of the amino acid response showed that the acetylation of both histone H3 and H4 coincided with the increased binding of ATF4 at ASNS promoter when there was the histidine deprivation [22]. This is also confirmed in the current study when the total amino acids were removed. For another amino acid responsive gene CHOP, a single amino acid limitation (-Leu) induced an increased acetylation of histone H4 and H2B but not H3 [33]. These amino acid-induced histone modifications are shown to be dependent on ATF2 binding at the promoter of CHOP gene [33]. In future studies, it is therefore important to identify the transcription factor that is responsible for the induction of DKK1 transcription and to examine the needs for a regionally modified histone profile.
In summary, our present study identifies DKK1 as a gene with a novel response to amino acid deprivation in a colon cancer cell line. The transcriptional control of DKK1 by amino acid deprivation in this study defines a novel amino acid responsive pathway that is independent of ATF4 expression and dependent on the phosphorylation of ERK. Further experiments are needed to characterize the cascades of this signaling pathway. Future studies will also investigate the interactions between the Wnt signaling pathway and the amino acid responsive pathways, as this may be a critical component during colon cancer development.
Supplementary Material
Research Highlights
The Wnt signaling pathway was affected by amino acid deprivation through the induction of DKK1
Transcriptional regulation of DKK1 by amino acid deficiency was by the phosphorylation of ERK
Knockdown of ATF4 does not inhibit the induction of DKK1 mRNA
The study demonstrated the interactions among Wnt, AAR, and MAPK/ERK signaling pathways in a colon cancer cell line
DKK1 is regulated by a novel amino acid response pathway that is independent of the master regulator ATF4.
ACKNOWLEDGEMENTS
The authors would like to thank the members of the Pan and Chen laboratories for advice and assistance. The authors acknowledge Ms. Min Huang for the technical assistance she offered during the study. This work is sponsored by the University of Illinois startup fund to HC. The authors would like to thank Anita Snyder (www.anitaksnyder.com) for professional copy editing.
Abbreviations
- AAR
amino acid response
- AARE
amino acids response elements
- ATF
activating transcription factor
- ASNS
asparagines synthetase
- DKK1
dickkopf homolog 1
- eIFα
eukaryotic initiation factor 2 alpha
- ERK
extracellular signal-regulated kinase
- GCN2
general control non-derepressible 2
- HisOH
histidinol
- MAPK
mitogen-activated protein kinase
- p-eIF2α
phosphorylated eukaryotic initiation factor 2 alpha
- p-ERK
phosphorylated extracellular signal-regulated kinase
- Pol II
RNA polymerase II
- Raf
rapidly accelerated fibrosarcoma protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors do not have any conflict of interest.
Reference List
- [1].Fafournoux P, Bruhat A, Jousse C. Amino acid regulation of gene expression. Biochemical Journal. 2000;351:1–12. doi: 10.1042/0264-6021:3510001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sikalidis A, Lee J-I, Stipanuk M. Gene expression and integrated stress response in HepG2/C3A cells cultured in amino acid deficient medium. Amino Acids. 2010:1–13. doi: 10.1007/s00726-010-0571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kilberg MS, Pan YX, Chen H, Leung-Pineda V. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu.Rev.Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. 59–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends in Endocrinology & Metabolism. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pan YX, Chen H, Thiaville MM, Kilberg MS. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochemical Journal. 2007;401:299–307. doi: 10.1042/BJ20061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Palii SS, Thiaville MM, Pan YX, Zhong C, Kilberg MS. Characterization of the amino acid response element within the human sodium-coupled neutral amino acid transporter 2 (SNAT2) System A transporter gene. Biochemical Journal. 2006;395:517–527. doi: 10.1042/BJ20051867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wek RC, Staschke KA. How do tumours adapt to nutrient stress[quest] EMBO J. 2010;29:1946–1947. doi: 10.1038/emboj.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol.Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Thiaville MM, Pan YX, Gjymishka A, Zhong C, Kaufman RJ, Kilberg MS. MEK Signaling Is Required for Phosphorylation of eIF2{alpha} following Amino Acid Limitation of HepG2 Human Hepatoma Cells. Journal of Biological Chemistry. 2008;283:10848–10857. doi: 10.1074/jbc.M708320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Logan CY, Nusse R. THE WNT SIGNALING PATHWAY IN DEVELOPMENT AND DISEASE. Annual Review of Cell and Developmental Biology. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- [12].Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- [13].Nusse R, van OA, Cox D, Fung YK, Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984;307:131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- [14].Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim.Biophys.Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- [15].Lustig B, Behrens J. The Wnt signaling pathway and its role in tumor development. J.Cancer Res.Clin.Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Semenov MV, Tamai K, Brott BK, Kuhl M, Sokol S, He X. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr.Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- [17].Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- [18].Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat.Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- [19].Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A, Niehrs C. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- [20].Gonzalez-Sancho JM, Aguilera O, Garcia JM, Pendas-Franco N, Pena C, Cal S, Garcia de HA, Bonilla F, Munoz A. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene. 2005;24:1098–1103. doi: 10.1038/sj.onc.1208303. [DOI] [PubMed] [Google Scholar]
- [21].Wang Z, Chen H. Amino acid limitation induces down-regulation of WNT5a at transcriptional level. Biochem.Biophys.Res.Commun. 2009;378:789–794. doi: 10.1016/j.bbrc.2008.11.124. [DOI] [PubMed] [Google Scholar]
- [22].Chen H, Pan YX, Dudenhausen EE, Kilberg MS. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. Journal of Biological Chemistry. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- [23].Sandoval J, Rodriguez JL, Tur G, Serviddio G, Pereda J, Boukaba A, Sastre J, Torres L, Franco L, Lopez-Rodas G. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004;32:e88. doi: 10.1093/nar/gnh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. Journal of Biological Chemistry. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- [25].Gregory CA, Singh H, Perry AS, Prockop DJ. The Wnt signaling inhibitor dickkopf-1 is required for reentry into the cell cycle of human adult stem cells from bone marrow. J.Biol.Chem. 2003;278:28067–28078. doi: 10.1074/jbc.M300373200. [DOI] [PubMed] [Google Scholar]
- [26].Morvan F, Boulukos K, Clement-Lacroix P, Roman RS, Suc-Royer I, Vayssiere B, Ammann P, Martin P, Pinho S, Pognonec P, Mollat P, Niehrs C, Baron R, Rawadi G. Deletion of a single allele of the Dkk1 gene leads to an increase in bone formation and bone mass. J.Bone Miner.Res. 2006;21:934–945. doi: 10.1359/jbmr.060311. [DOI] [PubMed] [Google Scholar]
- [27].Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, Shaughnessy JD., Jr. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N.Engl.J.Med. 2003;349:2483–2494. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- [28].Christodoulides C, Laudes M, Cawthorn WP, Schinner S, Soos M, O'Rahilly S, Sethi JK, Vidal-Puig A. The Wnt antagonist Dickkopf-1 and its receptors are coordinately regulated during early human adipogenesis. J.Cell Sci. 2006;119:2613–2620. doi: 10.1242/jcs.02975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pan Y, Chen H, Siu F, Kilberg MS. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple activating transcription factor-3 mRNA species that, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. Journal of Biological Chemistry. 2003;278:38402–38412. doi: 10.1074/jbc.M304574200. [DOI] [PubMed] [Google Scholar]
- [30].Pan YX, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3'-untranslated region regulates its amino acid limitation-induced stabilization. Journal of Biological Chemistry. 2005;280:34609–34616. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Palii S, Kays C, Deval C, Bruhat A, Fafournoux P, Kilberg M. Specificity of amino acid regulated gene expression: analysis of genes subjected to either complete or single amino acid deprivation. Amino Acids. 2009;37:79–88. doi: 10.1007/s00726-008-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bruhat A, Cherasse Y, Chaveroux C, Maurin AC, Jousse C, Fafournoux P. Amino acids as regulators of gene expression in mammals: molecular mechanisms. Biofactors. 2009;35:249–257. doi: 10.1002/biof.40. [DOI] [PubMed] [Google Scholar]
- [33].Bruhat A, Chrasse Y, Maurin A-C, Breitwieser W, Parry L, Deval C, Jones N, Jousse C, Fafournoux P. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic acids research. 2007;35:1312–1321. doi: 10.1093/nar/gkm038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.