Abstract
Inherent difficulties with blocking many desirable targets using conventional approaches have prompted many to consider using RNA interference (RNAi) as a therapeutic approach. Although exploitation of RNAi has immense potential as a cancer therapeutic, many physiological obstacles stand in the way of successful and efficient delivery. This Review explores current challenges to the development of synthetic RNAi-based therapies and considers new approaches to circumvent biological barriers, to avoid intolerable side effects and to achieve controlled and sustained release.
Since the discovery of RNA interference (RNAi), there has been an explosion of interest and knowledge in using this technology for clinical applications1. Although small-molecule inhibitors and monoclonal antibodies have led to many successful therapies for cancer2,3, many important cancer therapy targets are difficult to inhibit using these strategies. For example, a small-molecule inhibitor of a specific kinase will not affect its kinase-independent oncogenic functions and so will not restrict the entire function of the protein4-6. Most small-molecule inhibitors are also not specific with regard to target modulation, which can introduce undesirable toxicity. In the case of monoclonal antibodies, the protein might simply be inaccessible if it is not present on the cell surface or in circulation. The use of RNAi in the clinic is attractive as it can circumvent many of these problems and its potential for use as a therapeutic has recently been bolstered by a report of systemic small interfering RNA (siRNA) delivery into human tumours7. This study highlights how a molecule, the M2 subunit of ribonucleotide reductase (RRM2), which is difficult to inhibit using conventional approaches, can be targeted using siRNA. Although highly attractive as a therapeutic approach, several hurdles must be overcome to successfully introduce RNAi-based therapies into the clinic. Some of these include efficient and safe systemic delivery, avoidance of undesirable off-target effects, and the development of methods for assessing systemic biodistribution and subcellular localization. In addition, methods for crossing compartmental boundaries and avoiding intracellular trapping are needed. In this Review we discuss the existing challenges and future directions for developing RNAi as a clinical modality for cancer therapy.
Mechanisms of RNAi
The mechanism by which RNAi inhibits the conversion of mRNA into protein has been reviewed elsewhere8-10. Briefly, double-stranded RNA (dsRNA) is recognized by an RNase type III enzyme, Dicer, and cleaved into small fragments 21–23 base pairs in length11,12. The dsRNA has sequences that form a sense (passenger) strand and an antisense (guide) strand with respect to the target mRNA (FIG. 1). The dsRNA fragment binds to a protein complex called RNA-induced silencing complex (RISC), and the passenger strand of dsRNA is cleaved and discarded while the guide strand is directed to the 3′ untranslated region (UTR) of the complementary target mRNA13. When dsRNA is exogenously introduced, referred to as siRNA, a cleavage enzyme within RISC (argonaute 2) degrades the target mRNA, thereby preventing translation14,15. Endogenous non-coding RNAs (ncRNAs), such as microRNAs (miRNAs), also exist in cells and are pre-processed by a nuclear RNase III (Drosha) before export into the cytoplasm by nuclear transport receptor complex, exportin 5–RanGTP16,17. Owing to imperfect matching with 3′ UTRs, miRNAs in some instances do not lead to the cleavage of mRNA with the RISC but instead result in translational suppression18. As our understanding of the role that ncRNAs have in the pathogenesis of cancer has expanded, it has become clear that our ability to harness their potential as an anticancer therapeutic is a formidable task19.
Figure 1. Mechanism of RNAi following intracellular dsRNA delivery.
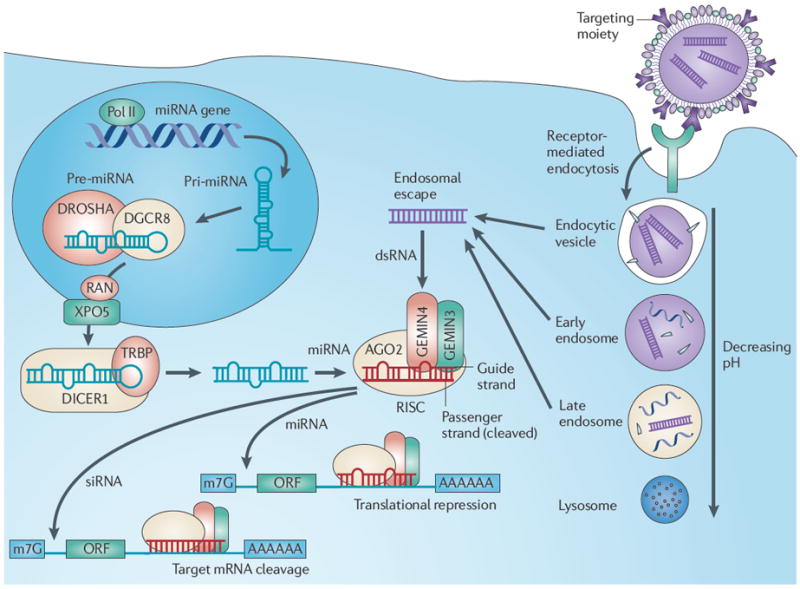
This demonstrates the intracellular fate of a nanoparticle following receptor-mediated endocytosis. Prior to degradation, the delivered double-stranded RNA (dsRNA) (whether microRNA (miRNA) or synthetic small interfering RNA (siRNA)) must escape the increasingly acidic endosome. After endosomal escape, the dsRNA bypasses Dicer and incorporates into the RNA-induced silencing complex (RISC). This leads to RNA interference (RNAi) by either translational repression or mRNA cleavage. AGO2, argonaute 2; ORF, open reading frame; Pol II, RNA polymerase.
Current challenges in RNAi delivery
There are several challenges that currently limit the use of RNAi in the clinic. Methods that overcome these are being developed and are discussed below (FIG. 2; TABLE 1).
Figure 2. Overcoming the biological barriers of RNAi delivery.
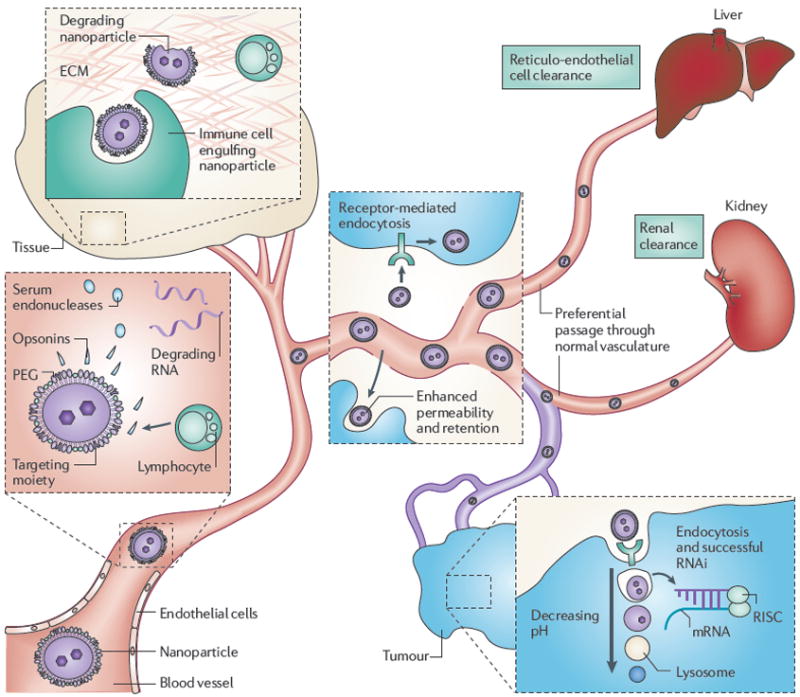
Representation of the fate of a liposomal nanoparticle from the point of intravenous delivery to RNA-induced silencing complex (RISC) incorporation. Intravascular barriers include opsonins that mediate engulfment by macrophages and serum endonucleases that degrade any naked double-stranded RNAs (dsRNAs). Extravascular nanoparticles encounter degradation by immune cells and the extracellular matrix (ECM). Particles preferentially traffic to tumours by the enhanced permeability and retention effect and receptor-mediated endocytosis. Once intracellular, for successful RNA interference (RNAi) naked dsRNAs must undergo endosomal escape for RISC incorporation before acidification by lysosomes. PEG, polyethelene glycol.
Table 1.
Clinical delivery of RNAi: challenges and solutions
| Challenges | Solutions |
|---|---|
| Biological barriers | |
| Intravascular degradation | Chemical modification and nanoparticle encapsulation |
| Tissue penetrance | Exploit abnormal angiogenesis and EPR effect |
| Intracellular delivery | Exploit receptor-mediated endocytosis |
| Intracellular trafficking | Incorporate biomaterials that enhance endosomal escape |
| Extracellular matrix | Optimize physico-chemical properties to escape immune and stromal interactions |
| Toxicities of RNAi | |
| Immune response to RNAi | Sequence optimization and oligonucleotide modifications |
| Carrier toxicity | Use of biodegradable, biocompatible and non-immunogenic materials |
| Off-target effects | Sequence optimization and oligonucleotide modifications |
| Oversaturation of RISC | Annotation of ‘dose’ effects on direct and indirect gene regulation |
| Tissue specificity | |
| Healthy versus cancer | Study of long-term RNAi side effects and exploit cancer features for targeting |
| Predictable delivery | Create carrier libraries annotating delivery efficacy based on healthy tissue and tumour type, and in silico modelling |
| Monitoring delivery | |
| Pharmacokinetics | Use of PEGylation or logic-embedded vectors |
| Pharmacodynamics | Use of bioluminescent imaging and novel biomarkers of delivery and response |
| Resistance | Study of altered processing machinery and SNP variants affecting RNAi |
EPR, enhanced permeability and retention; PEG, polyethylene glycol; RISC, RNA-induced silencing complex; RNAi, RNA interference; SNP, single nucleotide polymorphism.
Intravascular degradation
Naked siRNA is unstable in circulation owing to serum RNase A-type nucleases and rapid renal clearance, leading to degradation and a short half-life20 (FIG. 2). Some investigators have turned towards the chemical modification of the sugars, the backbone or the bases of oligoribonucleotides for stabilization21,22. However, the hydrophobic cell membranes create a challenge for the intracellular delivery of negatively charged polymers. Additionally, once siRNAs are intracellular they only transiently cause gene silencing, as the concentration of the siRNAs decreases with each cell division23. Nanoparticle carriers have the potential to overcome the challenges of intravascular degradation, and can provide safe and efficient delivery of synthetic dsRNAs. On entering the bloodstream, nanoparticles encounter a complex environment of plasma proteins and immune cells. Nanoparticle uptake by immune cells in circulation, such as monocytes, leukocytes, platelets and dendritic cells, occurs through various pathways and is facilitated by the adsorption of opsonins to the surface of the particle24 (FIG. 2). Additionally, physical and chemical properties of the nanoparticle surface, such as size and surface charge, can lead to haemolysis, thrombogenicity and complement activation, resulting in altered biodistribution and potential toxicity24.
Tissue penetrance and intracellular delivery
For safe and effective delivery of RNAi to the mRNA target of interest, many variables must be negotiated. Although nanoparticles by definition range in size from 1 nm to 1,000 nm in size, it has become clear that intravenously injected particles > 100 nm in diameter are likely to be trapped by the reticuloendothelial system (RES) in the liver, spleen, lung and bone marrow, leading to degradation by activated monocytes and macrophages (FIG. 2). Compared with normal capillaries, tumour vessels are irregularly shaped, defective, leaky and have varying widths25. Higher interstitial fluid pressure in solid tumours can also pose challenges for the diffusion of nanoparticles26 (FIG. 2). Potential strategies to overcome these barriers include using nanoparticles with diameters larger than the fenestrations of normal vessels (from a few nanometers to 150 nm); injecting vectors with varying radii; and normalizing tumour vasculature, such as with anti-angiogenic agents26,27. Overall, nanoparticles of less than 100 nm in diameter are thought to be optimal for intra-tumoural delivery and for avoiding RES clearance. Another important factor for increased capillary leak is the enhanced permeability and retention (EPR) effect, which can be exploited and enhanced for improved nanoparticle delivery28,29 (FIG. 2).
Intracellular trafficking
Many nanoparticles enter cells through endocytosis30. Once endocytosed, intracellular trafficking begins in early endosomal vesicles (FIG. 2). These subsequently fuse with sorting endosomes, which transfer their contents to late endosomes. These late endosomes are acidified (pH ~ 5–6) by membrane-bound proton-pump ATPases. The late endosomal content is relocated to lysosomes, which are further acidified (pH ~4.5) and contain nucleases that degrade RNA31. Thus, if endosomal escape does not occur before these later phases, RNAi will not occur. Some mechanisms to enhance endosomal escape include the use of fusogenic lipids, fusogenic peptides, photosensitive molecules, pH-sensitive lipoplexes and pH-sensitive polyplexes31. Fusogenic lipids promote endosomal release by increasing the interactions between the liposomal and endosomal membranes. The pH-sensitive materials function by a proton ‘sponge’ effect; as pH inside the endosome drops, protonation on the delivery material allows for an influx of chloride ions and subsequent osmosis, resulting in endosomal rupture and the release of its contents31.
The fate of RNAi in biological fluids
Although intracellular barriers to RNAi delivery are being explored in depth, extracellular factors in the microenvironment are less well studied and could be more important. For successful delivery, nanoparticles must extravasate and move through the complex extracellular matrix (ECM) to reach the cancer cells (FIG. 2). The desmoplasia associated with pancreatic cancer is a good example of the difficulties that a complex tumour microenvironment presents in achieving therapeutic drug concentrations32. The biological, chemical and physical properties of the ECM can result in the unpackaging of nanoparticles and the premature release of their contents33. In vitro studies are typically carried out under static conditions that are not reflective of the dynamic forces that are found in vivo. Three-dimensional models can be useful to recapitulate ECM barriers to nanoparticle delivery34. The immune system is also an important extracellular barrier. Phagocytosis by macrophages, such as Kupffer cells in the liver or dendritic cells in the lymph nodes, occurs more frequently with particles bearing cationic or anionic charges than with neutral nanoparticles24 (FIG. 2). In addition to phagocytosis by scavenger macrophages, neutrophils can function as extracellular traps for nanoparticles35,36.
Immune-mediated toxicities
Unknown or unexpected toxicities that are related to systemic RNAi delivery must be anticipated. The innate immune response to siRNA is highly contextualized and divided into two groups: Toll-like receptor (TLR)-mediated or non-TLR-mediated groups. TLR3, TLR7 and TLR8 are activated when engaged with nucleic acids within endosomal and lysosomal compartments. Ligands that activate TLR7 and TRL8 include duplex siRNA, or its corresponding single strand, although this is highly dependent on nucleotide sequence. For example, uridine- and guanosine-rich sequences with either UG dinucleotide or 5′-UGU-3′ motifs potently activate TLR7 and TLR8, and avoidance of such sequences decreases this response37. In addition, AU-rich sequences have been shown to preferentially activate TLR8 (REF. 38). TLR3 is only activated by duplex siRNA, and whether the sequence is important is not yet known39. Following endocytosis from the cell surface, TLR-mediated activation requires endosomal acidification and maturation. TLR7 is predominately expressed in plasmacytoid dendritic cells and B cells, TLR8 in myeloid dendritic cells, monocytes and macrophages, and TLR3 in mature myeloid dendritic cells40. TLR3 is also expressed in lung, aorta, dermis, choroidal and umbilical vein endothelial cells. TLR7 and TLR8 signal through the adaptor protein MYD88, which forms a signalling complex with interleukin-1 receptor-associated kinase 1 (IRAK1), IRAK4 and tumour necrosis factor receptor-associated factor 6 (TRAF6). This leads to the nuclear translocation of nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), with downstream activation of interferon-α (IFNα) and inflammatory cytokines40. Additionally, a unique off-target effect of naked siRNA binding to cell-surface TLR3 has been shown to cause anti-angiogenic effects through the activation of IFNγ and interleukin-12 (IL-12)41. The use of nanoparticles can overcome these effects by shielding the dsRNAs from cell surface interaction.
The non-TLR-mediated innate immune response predominately occurs through the cytoplasmic RNA sensors retinoic acid-inducible gene 1 (RIG1; also known as DDX58) and dsRNA-binding protein kinase (PKR). On dsRNA binding, RIG1 binds to IFNβ-promoter stimulator protein 1, which in turn activates interferon regulatory factor 3 (IRF3) and NF-κB, causing production of IFNβ and other inflammatory mediators42. PKR is a serine/threonine kinase that is expressed in most mammalian cells and that leads to the phosphorylation of eIF2α and inhibitor of NF-κB proteins followed by NF-κB activation. This non-TLR-mediated pathway can occur in a wide variety of mammalian cells, but the TLR pathway is limited to haematopoietic cells. As nanoparticle targeting improves, innate immune response mechanisms could be better predicted on the basis of the tissue of interest. Some of the chemical modifications to the sugars, backbone or bases of dsRNA (such as modifications to the 2′-OH group in the ribose sugar backbone, which include 2′-OMe, 2′-F and 2′-deoxy (2′-H) or locked nucleic acid (LNA) modifications) that result in improved intravascular stabilization have also been able to evade innate immune recognition while retaining RNAi activity23,40.
Nanoparticle-mediated toxicities
Although different types of delivery systems have shown efficacy in intra-tumoural delivery of siRNA, nanoparticle-related toxicity can also pose potential limitations. An ideal system would deliver a dsRNA payload while itself being bio-compatible, biodegradable, mostly inert to the immune system and having predictable systemic clearance. Liposomes are phospholipid structures that are assembled when water is added to a dried lipid film, and can range in size from submicrometers to several micrometers in diameter. These can incorporate water-soluble or water-insoluble drugs, therapeutic proteins, DNA and siRNA into their hydrophobic or hydrophilic compartments. Several liposomal-based therapeutics have been approved for various applications43. Cationic lipids such as 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethyl-ammonium methyl sulphate (DOTMA) represent an attractive media for delivery owing to their electrostatic interaction with negatively charged nucleic acids forming lipoplexes. Although useful for in vitro experiments, these lipids have been shown to invoke a robust type I and type II interferon response44. Cationic lipid-containing liposomes can cause gene expression changes independently of their RNAi payload, raising concerns about safety45. Additionally, cationic liposomes have shown dose-dependent toxicity and pulmonary inflammation that arises as a result of reactive oxygen intermediates46. As this effect was found to be charge related46, neutral nanoliposomes such as 1,2-dioleoyl-snglycero-3-phosphatidylcholine (DOPC)47 might avoid such toxicities. These nanoliposomes are tenfold more effective in tissue delivery than DOTAP and have no identified toxicities to date48,49. Negatively charged liposomes can induce an immune response and be subject to electrostatic repulsion by negatively charged cell membranes50.
Other lipid and cholesterol formulations, such as polyelectrolyte complex (PEC), along with polyethylenimine (PEI), form micelles that can efficiently deliver siRNA without a detectable IFNα response51. The conjugation of siRNAs to high-density lipoprotein (HDL) or low-density lipoprotein (LDL) as nanovehicles has also been shown to allow effective delivery to select tissues. HDL-bound siRNAs are predominately taken up by the liver, adrenals, ovaries and kidneys by high-affinity binding to scavenger receptor class B, type I, and LDL-bound siRNAs predominately enter the liver by LDL receptor expression52. A fairly new class of lipid-like compounds that is composed of amino-alkylacrylate and amino-acrylamide materials has been termed lipidoids; and these lipidoids have proved safe and well-tolerated in mice and cynomolgus primates53,54. These types of approaches could allow for the identification of not only well-tolerated delivery vehicles, but also the identification of compounds that require less substrate and payload, resulting in an improved therapeutic index.
The use of synthetic and natural polymer carrier systems has been extensively studied. Similar to lipids, their cationic substrate forms electrostatic interactions and allows the incorporation of negatively charged RNA. PEI, dendrimers and natural polymers, including chitosan, cyclodextrins and collagen, are among the more promising synthetic polymers that are currently in development. Although PEI has shown high transfection efficiency and the ability to buffer the acidic endosomal environment by the proton sponge effect, some intracellular and extracellular cytotoxicity has been observed, and chemical modifications might be necessary before achieving an acceptable toxicity profile for clinical use55. Dendrimers consist of a central core molecule from which multiple branching arms stem. Early generation dendrimers, such as polyamidoamine (PAMAM), were cytotoxic owing to cell membrane disruption caused by nanohole formation, membrane thinning and erosion56. The biocompatibility of these agents has been improved through surface modificiations56.
Chitosan is a deacetylated derivative of chitin, an abundant mucopolysaccharide in crustaceans and insects. Its low cost, biodegradability, absence of immunogenicity and high delivery efficacy make it a suitable delivery carrier. Thus far, in vivo toxicology studies in mice have shown chitosan to be well tolerated57. Tumour-specific delivery of chitosan particles with Arg-Gly-Asp peptide-labelling has recently been demonstrated58, as has the ability of these particles to target tumour-associated endothelial cells59. Cyclodextrins are cyclic oligomers of glucose with an amphipathic structure. The first targeted siRNA therapy in non-human primates was with a cyclodextrin polycation harbouring a transferrin ligand. This agent only caused increases in IL-6 and IFN levels, and hepatic and renal dysfunction, when administered at high doses60. This led to the first Phase I trial of a targeted, nanoparticle-delivered siRNA therapy in humans7. Atelocollagen, a highly purified type I collagen that is free of immunogenic telopeptides, has a varied clinical utility, such as the promotion of wound healing and vessel bio-prosthesis. When complexed with siRNA to form nanoparticles it has high delivery efficiency with little immunogenicity61.
Non-immune off-target effects
One of the bigger hurdles that needs to be cleared to ensure specific transcript silencing is non-immune off-target effects of RNAi. Although siRNA was initially thought to be target gene specific, it soon became clear that cross-hybridization with unintended transcripts that contained partial identity to the siRNA sequence could result in gene silencing (sometimes with as few as 6–8 contiguous nucleotides)62. Just as miRNAs usually lead to translational suppression owing to imperfect matching with 3′ UTRs18, siRNA sequences at positions 2–7 of the sense strand best predict off-target translational suppression, suggesting an miRNA-like off-target effect63. Although concomitant silencing of many target genes by a single miRNA is an evolutionarily conserved phenomenon64, potential toxicities of such unintended gene silencing can have important phenotypic effects63. Off-target effects of siRNA treatment can account for observed phenotypic changes independently of the target gene of interest65. Another unique property of 2′-O-methylation of the lead siRNA strand, other than attenuated intravascular degradation22 and immune stimulation40, is a substantial tapering of the observed off-target effects without losing silencing of perfectly complementary targets65. As RNAi therapeutics become a clinical reality, the long-term implications of these unintended off-target effects will need to be better understood and controlled.
Oversaturation of an RNAi-silencing complex
On successful intracellular delivery of dsRNA into the cytoplasm, some important barriers can still impede targeted gene silencing. We have shown that cancers can have altered levels of Drosha and Dicer, affecting the processing of precursors of siRNA such as short hairpin RNAs66. Low expression levels of Dicer in ovarian, lung and breast cancers correlated with a worse prognosis and poor treatment response66. Additionally, other investigators have shown that the high-level expression of short hairpin RNAs in the livers of mice overwhelmed the hepatic RNAi processing machinery, causing hepatic failure and death67. Others have shown that the RISC can be oversaturated by competing siRNAs68. Some studies suggest that the exogenous introduction of either siRNA or miRNA can lead to competition with endogenous miRNAs for RISC, leading to derepression of miRNA-regulated genes69. Thus, as target genes are silenced, derepression of miRNA-regulated genes results in their re-expression, which could have consequences in the tissues of interest. However, whether systemic delivery of dsRNA will be capable of RISC oversaturation is currently unknown.
Limited ability to ‘hit the mark’
Although the goal of harnessing RNAi is to attack cancer, some delivery into healthy tissue is unavoidable. Most in vitro and in vivo experiments rarely exceed a few weeks duration. However, cancer therapeutics must be effective for much longer periods of time: in some cases, years. Although many features of cancer allow for novel and specific targeting, consideration should be given to the unknown long-term consequences of RNAi delivery to healthy tissues, such as hepatic, renal or immune-mediated sequelae.
Once a guide strand is loaded into RISC, the complex is stabilized and able to recycle after multiple rounds of mRNA cleavage70. This implies a need for predictable degradation of the guide sequence. However, normal tissues are likely to handle systemically introduced RNAi differently from cancerous tissues71. For example, intracellular levels of enhanced RNAi-1 nuclease can substantially alter the duration of gene silencing, which can vary from a few days to several months depending on tissue type and expression levels72. Cancer cells can have a shorter duration of gene silencing owing to intracellular dilution of the RNAi molecules with each cell division, whereas gene silencing in slowly dividing or senescent cells generally lasts much longer73. Now that targeted nanoparticles delivering RNAi are entering the clinic7, it will be important to study the effects of long-term gene silencing in healthy tissues.
Exploiting the features of cancer
Tremendous progress has been made in exploiting the biological and physical features of cancer to enhance nanoparticle delivery. The ability of a nanoparticle to negotiate all known biological barriers, recognize cancer compared with healthy tissue and deliver therapeutic RNAi represent key determinants of therapeutic index. First-generation nanoparticles rely on the EPR effect for passive diffusion into the tumour microenvironment28. As the EPR effect is due to enlarged fenestrations in tumour-associated neovasculature, there is greater nanoparticle accumulation in tumours compared with healthy tissues with normal vasculature25,26 (FIG. 2). The main subclasses that reach tumour cells through this mechanism include liposomes, polymers and dendrimers; however, liposomal drug formulations have the longest track record in the clinic. Liposomal doxorubicin entered the cancer clinic in 1995, spawning multiple subsequent liposomal formulations for delivering cytotoxic chemotherapies43. Polymer and dendrimer drug conjugates have followed suit, and many different formulations are currently under clinical investigation74. Although several liposomal50,75 and polymeric57 nanoparticles can be effective delivery vehicles for in vivo RNAi, a more tumour-specific approach is desired.
Although second-generation nanoparticles can also arrive in tumours through the EPR effect, surface modifications allow a direct interface with cancer cells. By exploiting a particular epitope, receptor or other unique features of the cancer cell that are absent on healthy tissues, a more sophisticated mode of delivery by receptor-mediated endocytosis is realized30 (FIG. 2). Attaching antibodies to the nanoparticle surface allows for improved surface-specific delivery76, but these large moieties can increase cross-sectional area and make navigation across biological barriers more difficult and counterproductive77. Aptamer–nanoparticle conjugates have recently been shown to specifically recognize several different cancer types with high efficiency, creating a new method of targeting cell-surface proteins78. Through the use of natural ligands, such as folic acid79 or transferrin7,60, a ‘trojan horse’-type mechanism allows for more directed delivery in cancers overexpressing these receptors. The search for greater specificity for cancer using phage display biopanning approaches recently led to the discovery of a radiation-induced cell surface receptor (GRP78) that binds the ligand GIRLRG. Nanoparticles conjugated with GIRLRG were able to specifically target and deliver paclitaxel to radiation-treated cancer cells both in vitro and in vivo80. Further refinement of these targeting techniques, without interfering with nanoparticle arrival or capacity to carry a dsRNA payload, should bring this technology a step closer to the clinic.
Third-generation nanoparticles, such as logic-embedded vectors (LEVs), may have the ability to confront biological barriers in a time- and sequence-dependent manner, resulting in therapeutic release at the desired lesion77. These multi-component carriers rely on the material properties of the carriers and their payload43. We recently reported the first in vivo multistage siRNA delivery system for sustained gene silencing81. We used systemically delivered first-stage microparticle carriers that consisted of mesoporous silicon, which enabled the sustained release of second-stage neutral nanoliposomes (DOPC). After a single intravenous administration, the siRNA-targeted gene of interest, EphA2 (which encodes ephrin type A receptor 2), was inhibited for 3 weeks causing significant antitumour effects in two orthotopic models of ovarian cancer without observable toxicity81. Although this approach needs further development, it may open the door further for RNAi-based cancer therapeutics.
Systemic versus local delivery
This Review focuses on the systemic delivery of RNAi, but some studies have shown that direct intratumoural injection of nanoparticles containing siRNA can effectively result in gene silencing and tumour response51. Such an approach may offer higher bioavailability and reduced toxicity from systemic delivery, but many tumours cannot be reached by local delivery. Additionally, local delivery is not a viable option for treating micrometastatic disease. Therefore, although local delivery of RNAi could represent a treatment option for small, easily accessible primary tumours, such as melanoma, breast cancer or cervical cancer, it will probably have a limited role in clinical oncology.
Mathematical and computer modelling
The ability to predict and control the location of RNAi following systemic administration could help to maximize clinical efficacy. Mathematical and computer modelling has begun to shed light on how small modifications in nanoparticles can make dramatic differences to successful RNAi delivery. This has important implications for individualized treatment regimens that are affected by tumour histopathology, location, vasculature, microenvironment and host differences. Design maps provide preliminary multi-dimensional reference tables that show how alterations in size, shape, charge and chemical composition dictate the fate of nanoparticles82. These models take into account how the adhesive and endocytic performance of a nanoparticle is altered with changes in geometric (particle radius, for example), biophysical (such as degree of receptor-mediated endocytosis based on ligand-to-receptor surface density ratio) and biological (ligand–receptor binding affinity, for example) variables under different biological conditions30,82. Given the almost infinite possible nanoparticle combinations83, multiscale in silico modelling is being developed to predict how various properties such as tumour growth, normal and tumour-associated vasculature, extracellular matrix, pressure distributions and response to treatment will affect the efficacy of delivery84. These reference tables and models will establish a framework for predictable RNAi delivery in the highly plastic and sophisticated environment of malignancy.
Monitoring delivery and therapeutic response
The importance of pharmacokinetics and pharmacodynamics
In simple terms, pharmacokinetics is what the body does to a drug, and pharmacodynamics is what the drug does to the body. Therapeutic index of a drug refers to the ratio of therapeutic effect to toxicity. Thus, the ultimate success of nanoparticle-delivered RNAi will depend on the ability to negotiate biological barriers over a specific duration, selectively ‘hit the mark’, degrade predictably, be well tolerated and provide a high therapeutic index. Although naked siRNA can be rapidly cleared by the kidneys20, nanoparticles can be cleared quickly through opsonization by highly concentrated phagocytic cells in the RES85. This RES clearance seems to be predominately mediated by the phagocytic recognition of specific proteins that are absorbed on the surface of the nanoparticles in circulation. The use of polyethylene glycol (PEG) drug conjugates has proved to be a safe and effective strategy in prolonging the half-life of many US Food and Drug Administration-approved therapeutics. Addition of PEG to nanoparticles substantially mitigates RES uptake because its inherent hydrophilicity and steric repulsion effects reduce phagocyte interactions and complement activation86. Although chain length, shape and density of PEG on nanoparticles help to dictate circulation half-life, it is important to emphasize that PEG adds considerable cross-sectional diameter and decreases the ability to add targeting moieties, thus decreasing EPR effect and targeting ability, respectively77,86. As previously described, newer techniques such as the use of LEVs might allow the staged release of secondary particles81. This technique allows for targeting moieties to be used without the bulkiness of many conjugated PEG particles. An important element of RNAi delivery that must be considered is patient convenience and compliance. As many intravenous cancer therapies are delivered at weekly intervals in an outpatient clinic, successful accrual to clinical trials and patient adherence to treatment schedules will be important considerations in the design concept. Additionally, dose-finding Phase I/II trials will need to rely on models that simultaneously consider both protein expression modulation (efficacy) and dose-specific toxicity. An adaptive Bayesian model termed ‘EffTox’ is an example of how the probability of treatment efficacy and toxicity can be co-primary end points to determine Phase II dosing87.
Biomarkers to monitor nanoparticle delivery and response to RNAi are currently underdeveloped. A recently completed first-in-human Phase I clinical trial used post-treatment biopsies to confirm intratumoural nanoparticle delivery and PCR of cleavage fragments to confirm effective RNAi7. Imaging techniques using quantum dots88 incorporated into nanoparticles show promise for high quality in vivo imaging and provide a viable means for long-term tracking of nanoparticle delivery89. However, quantum dots can be toxic, and techniques to optimize their size and structure will determine their use in the clinic90. Various other promising modes of in vivo molecular imaging are being developed and are currently in the early stages of clinical assessment91. Proof of intratumoural delivery still does not confirm endosomal escape, RISC incorporation and successful RNAi. Therefore, there is an urgent need to discover and develop non-invasive biological surrogates of RNAi. Despite the circulating RNases, investigators have been able to capture and amplify circulating mRNAs in cancer patients92,93. Thus, one possible biomarker is circulating mRNA products following RISC cleavage, which could be detected with PCR amplification. Circulating mRNAs are thought to derive from nucleated apoptotic cells, tumour necrosis and nucleic acid shedding by metastases and micrometastases94. If validated, these would represent an easy, inexpensive and non-invasive biomarker of RNAi. Another strategy would be to analyse protein expression changes following RNAi-based therapy. Recently, investigators generated isotope tags for relative and absolute quantification (iTRAQ)-labelled peptides from cell lysates following siRNA treatment and verified protein silencing using liquid chromatography–mass spectrometry (LC–MS)/MS95.
Mechanisms of resistance
As RNAi enters the clinic, given the heterogeneity of tumours, the development of resistance seems inevitable. Additionally, some cancers could have an inherent resistance to some types of RNAi owing to factors such as ethnicity, somatic mutations, altered RNAi processing machinery and germline polymorphisms. Significant genetic variation across major ethnic groups exists96, which might partly account for variable responses to RNAi. In support of this, a recent report showed that germline single nucleotide polymorphisms (SNPs) within miRNA binding regions at 5′ UTRs, coding sequence and 3′ UTRs of target mRNAs lead to altered binding affinities97. These miRNA–mRNA binding sites occurred at previously annotated SNPs that confer breast cancer susceptibility and lead to altered protein expression97. This has important therapeutic implications as it suggests that the delivered miRNAs and siRNAs that target binding regions containing such SNPs could have altered efficacy. Alterations in the RNAi processing machinery such as Dicer, Drosha and RISC complex proteins will need to be further studied and accounted for in RNAi therapeutics66,98. Although exogenous co-expression of the argonaute 2 protein in lung cancer cell lines enhanced RISC cleavage99, cancers may adapt to decrease such proteins and effectively inhibit gene silencing. In anticipation of clinically useful RNAi, such resistance mechanisms in cancer will need to be understood.
Beyond the barriers
For more than a decade, there has been tremendous growth in our understanding of RNAi. The potent ability that small dsRNAs have in gene silencing makes them desirable as novel cancer therapeutics, but many biological barriers exist from drug infusion to target mRNA silencing. However, as shown by the first Phase I trial using targeted nanoparticles for RNAi delivery, rapid discoveries are leading us beyond these barriers. We believe, in due time, that the great translational potential of RNAi will continue to navigate such barriers and find its place in the oncology clinic.
At a glance.
Many cancer targets are difficult to block with conventional therapies. Although RNA interference (RNAi) as a therapeutic approach is appealing, many challenges to delivery must be overcome.
Nanoparticles hold promise for the safe and effective intracellular delivery of RNAi-based molecules.
Physiological barriers and systemic toxicity of nanoparticle-based carrier systems create multiple challenges to bringing RNAi-based therapeutics to the clinic.
Nanoparticles can be used to help avoid immune-mediated responses to systemic RNAi-based therapy.
Solutions to improving tumour specificity and the ability to monitor and control short-term and long-term RNAi-based therapies are crucial next steps before clinical use.
As the technology for delivery improves, so we will also need to improve our understanding of the heterogeneity of RNAi processing in different cancer types.
Various resistance mechanisms to RNAi-based therapies must be anticipated.
Acknowledgments
Portions of this work were supported by the NIH (CA110793, CA109298, P50 CA083639, P50 CA098258, CA128797, RC2GM092599 and U54 CA151668), the Ovarian Cancer Research Fund, Inc. (Program Project Development Grant), the DOD (OC073399, W81XWH-10-1-0158 and BC085265), the Zarrow Foundation, the Laura and John Arnold Foundation and the Betty Anne Asche Murray Distinguished Professorship.
Glossary
- RNA interference (RNAi)
Refers to the mechanism of potent and specific gene-silencing caused by double-stranded RNA (dsRNA) produced by endogenous (miRNA) or exogenous (siRNA) sources.
- Opsonins
Plasma proteins that act as binding enhancers for phagocytosis.
- Reticuloendothelial system (RES)
System composed of scavenging monocytes and macrophages located in the reticular connective tissue (notably the liver, spleen, lung and marrow).
- Enhanced permeability and retention (EPR)
The property by which certain sizes of molecules tend to accumulate and remain in tumour tissue more than in normal tissues.
- Endocytosis
The active uptake of molecules into a cell by clathrin-dependent and clathrin-independent receptor-mediated endocytosis, pinocytosis and phagocytosis.
- Fusogenic lipids
Lipoplexes (containing cationic lipids and nucleic acids) that adopt an inverted hexagonal phase and fuse with anionic membranes resulting in endosomal release.
- Fusogenic peptides
Peptides that have cell penetrating properties (such as being highly hydrophobic), which cause cell membrane destabilization and intra-cytoplasmic release.
- pH-sensitive lipoplexes
Liposomes that hydrolyse and trigger release of contents owing to subtle drops in pH, such as with endosomal fusion, allowing for endosomal escape.
- pH-sensitive polyplexes
Polyplexes (cationic polymer complexed with nucleic acid) that act as proton ‘sponges’, preventing acidification after endosomal fusion and allowing influx of counter ions, osmotic swelling and endosome rupture.
- Aptamer
A DNA or RNA oligonucleotide sequence with a high-affinity binding for specific proteins.
- Logic-embedded vector
Vehicles that work in a time-sequential manner to cross biological barriers.
- Mesoporous silicon
A biodegradable and biocompatible material made from non-oxidized silicon.
- Polyethylene glycol (PEG)
A synthetic polymer that is non-toxic, non-immunogenic and highly water soluble.
- Quantum dots
Colloidal semiconductor nanocrystals with optical and electronic properties superior to conventional organic fluorophores that can be used for imaging purposes.
Footnotes
Competing interests statement The authors declare no competing financial interests.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-strandedRNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 3.Smith I, et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: a randomised controlled trial. Lancet. 2007;369:29–36. doi: 10.1016/S0140-6736(07)60028-2. [DOI] [PubMed] [Google Scholar]
- 4.Kikani CK, Dong LQ, Liu F. “New”-clear functions of PDK1: beyond a master kinase in the cytosol? J Cell Biochem. 2005;96:1157–1162. doi: 10.1002/jcb.20651. [DOI] [PubMed] [Google Scholar]
- 5.Lim ST, Mikolon D, Stupack DG, Schlaepfer DD. FERM control of FAK function: implications for cancer therapy. Cell Cycle. 2008;7:2306–2314. doi: 10.4161/cc.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weihua Z, et al. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956.. This is the first report of using targeted nanoparticles for siRNA delivery in humans.
- 8.Sashital DG, Doudna JA. Structural insights into RNA interference. Curr Opin Struct Biol. 2010;20:90–97. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nature Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 10.Heo I, Kim VN. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell. 2009;139:28–31. doi: 10.1016/j.cell.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 12.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 13.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:565–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 15.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 17.Bohnsack MT, Czaplinski K, Gorlich D. Exportin-5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu C, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabbri M, Calin GA. Beyond genomics: interpreting the 93% of the human genome that does not encode proteins. Curr Opin Drug Discov Devel. 13:350–358. [PubMed] [Google Scholar]
- 20.Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nature Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 22.Czauderna F, et al. Structural variations and stabilizing modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 24.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skinner SA, Tutton PJ, O’Brien PE. Microvascular architecture of experimental colon tumors in the rat. Cancer Res. 1990;50:2411–2417. [PubMed] [Google Scholar]
- 26.Decuzzi P, Causa F, Ferrari M, Netti PA. The effective dispersion of nanovectors within the tumor microvasculature. Ann Biomed Eng. 2006;34:633–641. doi: 10.1007/s10439-005-9072-6. [DOI] [PubMed] [Google Scholar]
- 27.Tong RT, et al. Vascular normalization by vascular endothelial growth factor receptor 2 blockade induces a pressure gradient across the vasculature and improves drug penetration in tumors. Cancer Res. 2004;64:3731–3736. doi: 10.1158/0008-5472.CAN-04-0074. [DOI] [PubMed] [Google Scholar]
- 28.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–6392.. This work describes the first observation of the EPR effect in the tumour microenvironment.
- 29.Maeda H. Tumor-selective delivery of macromolecular drugs via the EPR effect: background and future prospects. Bioconjug Chem. 2010;21:797–802. doi: 10.1021/bc100070g. [DOI] [PubMed] [Google Scholar]
- 30.Decuzzi P, Ferrari M. The receptor-mediated endocytosis of nonspherical particles. Biophys J. 2008;94:3790–3797. doi: 10.1529/biophysj.107.120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 32.Kleeff J, et al. Pancreatic cancer microenvironment. Int J Cancer. 2007;121:699–705. doi: 10.1002/ijc.22871. [DOI] [PubMed] [Google Scholar]
- 33.Burke RS, Pun SH. Extracellular barriers to in Vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjug Chem. 2008;19:693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- 34.Goodman TT, Ng CP, Pun SH. 3-D tissue culture systems for the evaluation and optimization of nanoparticle-based drug carriers. Bioconjug Chem. 2008;19:1951–1959. doi: 10.1021/bc800233a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1477–1478. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 36.Bartneck M, Keul HA, Zwadlo-Klarwasser G, Groll J. Phagocytosis independent extracellular nanoparticle clearance by human immune cells. Nano Lett. 2010;10:59–63. doi: 10.1021/nl902830x. [DOI] [PubMed] [Google Scholar]
- 37.Judge AD, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature Biotech. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 38.Forsbach A, et al. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J Immunol. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 39.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature Rev Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 40.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–101. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 41.Kleinman ME, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765.. This work shows that TLR3 activation by naked siRNAs can cause an anti-angiogenic off-target effect independently of gene silencing.
- 42.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 43.Sakamoto JH, et al. Enabling individualized therapy through nanotechnology. Pharmacol Res. 2010:1–31. doi: 10.1016/j.phrs.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma Z, et al. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem Biophys Res Commun. 2005;330:755–759. doi: 10.1016/j.bbrc.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 45.Omidi Y, Barar J, Akhtar S. Toxicogenomics of cationic lipid-based vectors for gene therapy: impact of microarray technology. Curr Drug Deliv. 2005;2:429–441. doi: 10.2174/156720105774370249. [DOI] [PubMed] [Google Scholar]
- 46.Dokka S, Toledo D, Shi X, Castranova V, Rojanasakul Y. Oxygen radical-mediated pulmonary toxicity induced by some cationic liposomes. Pharm Res. 2000;17:521–525. doi: 10.1023/a:1007504613351. [DOI] [PubMed] [Google Scholar]
- 47.Gutierrez-Puente Y, et al. Cellular pharmacology of P-ethoxy antisense oligonucleotides targeted to Bcl-2 in a follicular lymphoma cell line. Leuk Lymphoma. 2003;44:1979–1985. doi: 10.1080/1042819031000099733. [DOI] [PubMed] [Google Scholar]
- 48.Landen CN, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 49.Ahmed AA, et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell. 2010;18:109–121. doi: 10.1016/j.ccr.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J Intern Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J Control Release. 2008;129:107–116. doi: 10.1016/j.jconrel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature Biotech. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 53.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nature Biotech. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love KT, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv Drug Deliv Rev. 2006;58:1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 56.Jain K, Kesharwani P, Gupta U, Jain NK. Dendrimer toxicity: let’s meet the challenge. Int J Pharm. 2010;394:122–142. doi: 10.1016/j.ijpharm.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 57.Pille JY, et al. Intravenous delivery of anti-RhoA small interfering RNA Loaded in nanoparticles of chitosan in mice: safety and efficacy in xenografted aggressive breast cancer. Hum Gene Ther. 2006;17:1019–1026. doi: 10.1089/hum.2006.17.1019. [DOI] [PubMed] [Google Scholar]
- 58.Han HD, et al. Targeted gene silencing using RGD-labeled chitosan nanoparticles. Clin Cancer Res. 2010;16:3910–3922. doi: 10.1158/1078-0432.CCR-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu C, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heidel JD, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci USA. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeshita F, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci USA. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotech. 2003;21:635–637. doi: 10.1038/nbt831.. This study demonstrated that siRNAs can cause miRNA-like translational suppression owing to imperfect complementarity in the seed sequence.
- 63.Jackson AL, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 65.Jackson AL, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merritt WM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785.. This study shows changes in Dicer and Drosha levels in cancer have effects on gene silencing.
- 67.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 68.Castanotto D, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khan AA, et al. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nature Biotech. 2009;27:549–555. doi: 10.1038/nbt.1543.. This paper demonstrates how exogenous introduction of RNAi can compete with endogenous miRNAs, causing derepression of miRNA-regulated genes.
- 70.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nature Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 71.Raemdonck K, Vandenbroucke RE, Demeester J, Sanders NN, De Smedt SC. Maintaining the silence: reflections on long-term RNAi. Drug Discov Today. 2008;13:917–931. doi: 10.1016/j.drudis.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takabatake Y, et al. Chemically modified siRNA prolonged RNA interference in renal disease. Biochem Biophys Res Commun. 2007;363:432–437. doi: 10.1016/j.bbrc.2007.08.189. [DOI] [PubMed] [Google Scholar]
- 73.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;31:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duncan R. The dawning era of polymer therapeutics. Nature Rev Drug Discov. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 75.Shahzad MMK, et al. Dual targeting of EphA2 and FAK in ovarian carcinoma. Cancer Biol Ther. 2009;8:1027–1034. doi: 10.4161/cbt.8.11.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hogrefe RI, et al. Chemically modified short interfering hybrids (siHYBRIDS): nanoimmunoliposome delivery in vitro and in vivo for RNAi of HER-2. Nucleosides Nucleotides Nucleic Acids. 2006;25:889–907. doi: 10.1080/15257770600793885. [DOI] [PubMed] [Google Scholar]
- 77.Ferrari M. Frontiers in cancer nanomedicine: directing mass transport through biological barriers. Trends Biotech. 2010;28:181–188. doi: 10.1016/j.tibtech.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Estévez MC, et al. Nanoparticle-aptamer conjugates for cancer cell targeting and detection. Methods Mol Biol. 2010;624:235–248. doi: 10.1007/978-1-60761-609-2_16. [DOI] [PubMed] [Google Scholar]
- 79.Kim SH, Mok H, Jeong JH, Kim SW, Park TG. Comparative evaluation of target-specific GFP gene silencing efficiencies for antisense ODN, synthetic siRNA, and siRNA plasmid complexed with PEI-PEG-FOL conjugate. Bioconjug Chem. 2006;17:241–244. doi: 10.1021/bc050289f. [DOI] [PubMed] [Google Scholar]
- 80.Passarella RJ, et al. Targeted nanoparticles that deliver a sustained, specific release of paclitaxel to irradiated tumors. Cancer Res. 2010;70:4550–4559. doi: 10.1158/0008-5472.CAN-10-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanaka T, et al. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Res. 2010;70:3687–3696. doi: 10.1158/0008-5472.CAN-09-3931.. This work demonstrates the first in vivo validation of multi-staged siRNA delivery for sustained gene silencing.
- 82.Decuzzi P, Ferrari M. Design maps for nanoparticles targeting the diseased microvasculature. Biomaterials. 2008;29:377–384. doi: 10.1016/j.biomaterials.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 83.Ferrari M. The mathematical engines of nanomedicine. Small. 2008;4:20–25. doi: 10.1002/smll.200701144. [DOI] [PubMed] [Google Scholar]
- 84.Sanga S, et al. Mathematical modeling of cancer progression and response to chemotherapy. Expert Rev Anticancer Ther. 2006;6:1361–1376. doi: 10.1586/14737140.6.10.1361. [DOI] [PubMed] [Google Scholar]
- 85.Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 86.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thall PF, Cook JD. Dose-finding based on efficacy-toxicity trade-offs. Biometrics. 2004;60:684–693. doi: 10.1111/j.0006-341X.2004.00218.x. [DOI] [PubMed] [Google Scholar]
- 88.Bruchez MJ, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 89.Tan WB, Jiang S, Zhang Y. Quantum-dot based nanoparticles for targeted silencing of HER2/neu gene via RNA interference. Biomaterials. 2007;28:1565–1571. doi: 10.1016/j.biomaterials.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 90.Tavares AJ, Chong L, Petryayeva E, Algar WR, Krull UJ. Quantum dots as contrast agents for in vivo tumor imaging: progress and issues. Anal Bioanal Chem. 2010 Jul 25; doi: 10.1007/s00216-010-4010-3. [DOI] [PubMed] [Google Scholar]
- 91.Hong H, Zhang Y, Cai W. In vivo imaging of RNA interference. J Nucl Med. 2010;51:169–172. doi: 10.2967/jnumed.109.066878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen XQ, et al. Telomerase RNA as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6:3823–3826. [PubMed] [Google Scholar]
- 93.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger RNA in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5:1961–1965. [PubMed] [Google Scholar]
- 94.Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids – a promising, non-invasive tool for early detection of several human diseases. FEBS J. 2007;581:795–799. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 95.Abdrakhmanova A, et al. RNAi and iTRAQ reagents united: targeted quantitation of siRNA-mediated protein silencing in human cells. Anal Bioanal Chem. 2009;395:773–785. doi: 10.1007/s00216-009-3028-x. [DOI] [PubMed] [Google Scholar]
- 96.Spielman RS, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nature Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nicoloso MS, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Merritt WM, Bar-Eli M, Sood AK. The dicey role of Dicer: implications for RNAi therapy. Cancer Res. 2010;70:2571–2574. doi: 10.1158/0008-5472.CAN-09-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Diederichs S, et al. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci USA. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]


