Abstract
Astrocyte heterogeneity remains largely unknown in the CNS due to lack of specific astroglial markers. In this study, molecular identity of in vivo astrocytes was characterized in BAC ALDH1L1 and BAC GLT1 eGFP promoter reporter transgenic mice. ALDH1L1 promoter is selectively activated in adult cortical and spinal cord astrocytes, indicated by the overlap of eGFP expression with ALDH1L1 and GFAP, but not with NeuN, APC, Olig2, IbaI, PDGFRα immunoreactivity in BAC ALDH1L1 eGFP reporter mice. Interestingly, ALDH1L1 expression levels (protein, mRNA, and promoter activity) in spinal cord were selectively decreased during postnatal maturation. In contrast, its expression was up-regulated in reactive astrocytes in both acute neural injury and chronic neurodegenerative (G93A mutant SOD1) conditions, similar to GFAP, but opposite of GLT1. ALDH1L1+ and GLT1+ cells isolated through fluorescence activated cell sorting (FACS) from BAC ALDH1L1 and BAC GLT1 eGFP mice share a highly similar gene expression profile, suggesting ALDH1L1 and GLT1 are co-expressed in the same population of astrocytes. This observation was further supported by overlap of the eGFP driven by the ALDH1L1 genomic promoter and the tdTomato driven by a 8.3kb EAAT2 promoter fragment in astrocytes of BAC ALDH1L1 eGFP X EAAT2-tdTomato mice. These studies support ALDH1L1 as a general CNS astroglial marker and investigated astrocyte heterogeneity in the CNS by comparing the molecular identity of the ALDH1L1+ and GLT1+ astrocytes from astroglial reporter mice. These astroglial reporter mice provide useful in vivo tools for the molecular analysis of astrocytes in physiological and pathological conditions.
Keywords: astroglia, BAC, ALDH1L1, GLT1, GFAP, oligodendroglia, ALS
INTRODUCTION
Astrocytes are the most abundant cells in the CNS, having the unique anatomical localization that bridges the blood vessel and neuronal synapses through their highly ramified processes. This unique anatomical localization allows astrocytes not only provide essential metabolic support to neurons, but also participate in the information processing by active modulation of neuronal/synaptic activity in the CNS (Halassa et al. 2007). In pathophysiological conditions, astrocytes undergo dramatic morphological and molecular changes that can cause potentially both beneficial and detrimental effects, depending upon the exact pathological context (Sofroniew 2009).
Although astrocytes are essential in normal CNS function and are involved in most neuropathological conditions, their molecular identity remains largely uncharacterized, partially due to insufficient mature astrocyte specific markers, preventing investigation of astrocyte heterogeneity and detailed understanding of their functional interaction with local neighboring cells in the CNS. A number of proteins unique to astroglia have been identified including Glutamine Synthetase (GS), Excitatory Amino Acid Transporter 2 (EAAT2, rodent analog GLT1), Aquaporin 4, Connexin 43 (ctx43) (Kimelberg 2009). These proteins are primarily distributed at the membrane of astroglial processes, and thus it becomes impractical to clearly identify individual astrocytes by their immunostaining. Instead, immunostaining of glial fibrillary acidic protein (GFAP), an intermediate filament has been conventionally used to identify astrocytes in the CNS, though it only labels a limited number of astrocytes and their extensive processes in the CNS (Kimelberg 2009).
Recent studies on astroglial gene expression profiles have started to characterize the molecular identity of astrocytes in vivo(Lovatt et al. 2007) and identified aldehyde dehydrogenase 1 family, member L1 (ALDH1L1) as the new astroglial marker that also selectively labels cortical astrocytes in vivo (Cahoy et al. 2008). ALDH1L1, also known as 10-formyltetrahydrofolate dehydrogenase (FDH), is a folate enzyme that converts 10-formyltetrahydrofolate (10-formyl-THF) to tetrahydrofolate, playing an important role in many reactions like de novo nucleotide biosynthesis and the regeneration of methionine, thus having a major impact on cell division and growth (Krupenko 2009). Unique functions of ALDH1L1 in the CNS have not been reported except a potential link to neural tube defects during early CNS development (Anthony and Heintz 2007). Alterations of ALDH1L1 in reactive astrocytes associated with neural injury or neurodegenerative disease are unknown.
A transgenic mouse that overexpress a fluorescence reporter driven by a cell-specific promoter, usually with the use of a bacteria artificial chromosome (BAC) clone has been demonstrated to be an effective way to selectively label CNS cells (Heintz 2001). In the current study, expression specificity of ALDH1L1 was examined in BAC ALDH1L1 eGFP reporter mice(Doyle et al. 2008; Heiman et al. 2008) and compared against other known astroglial markers. Molecular property of ALDH1+ and GLT1+ astrocytes were also directly compared by employing transgenic reporter mice and whole genome transcriptome analysis. The expression change of ALDH1L1 in astrocytes in neurodegenerative/neural injury conditions is also characterized.
MATERIALS AND METHODS
Animals
Wild type mice (C57/B6), SOD1 G93A transgenic mice, BAC GLT1 eGFP (previously generated in our lab) (Regan et al. 2007), BAC NG2 DsRed (kind gift of Dr. Dwight Bergles, Dept. of Neuroscience, Johns Hopkins University), BAC ALDH1L1 eGFP mice (GENSAT project), GFAP eGFP mice (Jackson laboratory) were used for in vivo experiments. The care and treatment of animals in all procedures strictly followed the NIH Guide for the Care and Use of Laboratory Animals and the Guidelines for the Use of Animals in Neuroscience Research and the Johns Hopkins University IACUC. Mice were housed at standard temperature (21°C) and in a light controlled environment with ad libitum access to the food and water. BAC ALDH1L1 eGFP (and GFAP eGFP) mice were crossed with SOD1 G93A mice to obtain double transgenic mice. Littermates were used as control. Mice were sacrificed at designated time point following breeding or experiment. A 30-gauge needle was used to induce an acute lesion in the striatum.
Generation of the EAAT2-tdTomato transgenic mice
EAAT2-tdTomato transgenic mouse was generated by inserting the tdTomato reporter downstream of a 8.3kb EAAT2 promoter fragment. Multiple (4) founders were established following pronuclear injection at the transgenic core laboratory of Johns Hopkins University. The mice will be available upon request.
Primary neuron and astrocyte cultures
Procedures used for culturing primary neuron and astrocytes were described before (Yang et al. 2009). Briefly, cortical astrocyte cultures were prepared from P2-4 wild type mouse pups and grown in DMEM supplemented with 10% heat-inactivated FBS and 1% of penicillin/streptomycin. Astrocytes were ready for experiments once confluent (5-7 DIV). No dbcAMP was added into the culture medium. For neuronal cultures, cortex was dissected out from E14-16 mouse embryos of wild type mice, cortical neurons were cultured in Neurobasal medium supplemented with N27, penicillin/streptomycin, and glutamine.
Immunohistochemistry and Microscopy
For immunohistochemistry, mice were perfused with 4% paraformaldehyde and brain/spinal cord tissue were collected and cryoprotected in 30% sucrose. Tissue sections (20μm) were then prepared by cryostat sectioning and were treated with blocking buffer (0.4% BSA, 5% goat-serum, and 0.2% Triton-X 100 in PBS) for 30-60 min at room temperature. Primary antibodies for ALDH1L1 (NeuroMab), GFAP (Chemicon), APC (cc1 clone, Calbiochem), PDGFRα (BD Pharmingen), Iba1 (Wako), Olig2 (Millipore), and NeuN (Millipore) were incubated overnight at 4 °C in blocking buffer. After rinse in PBS, anti-mouse Alexa 555 conjugated antibody was added for 90min at room temperature. Cells labelled with either immunostain or transgenic reporters were manually counted in Axiovision from digital images taken by fluorescent microscopy. For some single cell analyses, mean fluorescence intensity (MFI) was determined. Typically, MFI measurements were based upon at least 4000 cells/sample with a minimum of 3 mice for each experiment. Data was plotted in PRISM (GraphPad Software, Inc). One-way ANOVA (Bonferroni posthoc analysis) and Student’s t-test were used for statistical analysis where appropriate (see figure legend for details).
Preparation of cell suspension and FAC sorting of astrocytes
Brains or spinal cords from BAC GLT1 eGFP or BAC ALDH1L1 eGFP transgenic reporter mice (age from P5-P130) were used. Mice were anesthetized with pentobarbital (50 mg/kg, i.p.), perfused with cold Hanks buffer (Invitrogen, Carlsbad, CA), and decapitated. The brain was immediately dissected in cold Hanks buffer containing glutamate receptor antagonists, 3 mM DNQX and 100 mM APV (Sigma, St. Louis, MO), and cut into small pieces. Cell suspension was prepared as described in the neural tissue dissociation kit (Miltenyi biotech, Auburn, CA). Briefly, small pieces of tissue were treated with papain enzymatic mix (37C, 15min) and then digested with DNase I (37C, 10min), followed by careful trituration. Cell mixtures were then filtered through a cell strainer (40-70mm) and resuspended in cold HBSS solution (5-10 × 106 cells/ml) for FACS. The whole cell suspension procedure was completed in 1-2 h. Cells were sorted by using MoFlo MLS high speed cell sorter (Beckman coulter) with Summit version 4.3 software. Propidium iodide (PI) and eGFP were all excited by a 488 nm laser, and emissions were collected by 575/26 nm and 530/30 nm discrimination filters, respectively. The signals were manually compensated, and cells were sorted into cold HBSS. The whole procedure for cell suspension preparation and FAC sorting process was completed within 2-3h.
RNA isolation and QRT-PCR
Total RNA was prepared from brain or spinal cord tissue by using Absolutely RNA Miniprep Kit from Stratagene (Stratagene, La Jolla, CA). Total RNA were then converted to cDNA by using high archive cDNA synthesis kit (Applied Biosystems, Foster city, CA). The relative abundance of ALDH1L1 mRNA was determined by using a TaqMan pre-made ALDH1L1 probes (Applied Biosystems, Foster city, CA). Ribosomal 18s rRNA was used as endogenous control for the normalization of RNA quantity.
Western blot analysis
Primary cultures or tissue were lysed in a buffer containing 62.5 mM Tris, 2% SDS, 10% sucrose, and protease inhibitor cocktail (Roche). Homogenates were briefly sonicated and were incubated at 37C for 30min. The supernatants were separated by 4-12% gradient PAGE gel and transferred to PVDF membrane. Blots were placed in blocking solution (5% non-fat milk in TBST) for 1 h at room temperature and then incubated with ALDH1L1 antibody (Neuromab) or β-actin antibody (Sigma) diluted in TBST overnight at 4C. After extensive washing, the blots were incubated with horseradish peroxidase conjugated antibodies (Sigma) for 1h and washed again. Immunoreactive bands were detected using the ECL procedure.
Microarray and data analysis
Total RNA was prepared from FAC sorted astrocytes. Total RNA was lineally amplified and labeled in a Nugene protocol. Sample labeling and hybridization with Mouse Exon 1.0 ST chips (Affymetrix) were performed in the Johns Hopkins University microarray facility. After hybridization, hybridization signals were acquired and normalized with the use of RMAexpress (http://rmaexpress.bmbolstad.com/). Data was further filtered to remove genes with signals less than 80 in all the samples to reduce the noise. Differential gene expression between the different conditions was assessed by statistical linear model analysis using the bioconductor package limma. The moderated t-statistic p-values derived from the limma analysis above were further adjusted for multiple testing by Benjamini and Hochberg’s method to control false discovery rate (FDR). The FDR cutoff of <10% was used to obtain the list of differentially expressed genes.
RESULTS
ALDH1L1 expression is selectively decreased in adult spinal cord astrocytes through transcriptional inactivation of the ALDH1L1 promoter
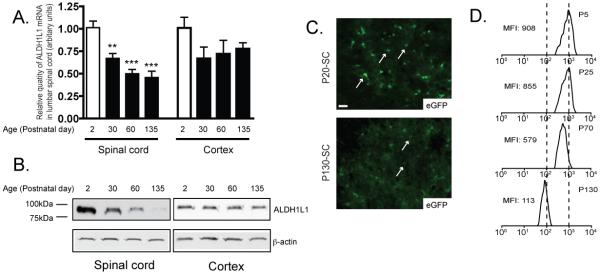
ALDH1L1 expression has been suggested as being astroglia specific by immunostaining in young rodent tissue (Neymeyer et al. 1997). In the current study, ALDH1L1 mRNA levels in cortex and spinal cord were first examined at different postnatal age (P2, P30, P60, and P135). The mRNA levels of ALDH1L1 in cortex are only slightly decreased as the cortex matures. In contrast, the mRNA level in spinal cord is significantly decreased (Figure 1A). Interestingly, ALDH1L1 protein levels also show significant age-dependent decrease in the spinal cord, with the highest expression at the age of P2. To investigate whether the ALDH1L1 promoter becomes inactivated in spinal cord of mature mice, a BAC ALDH1L1 eGFP transgenic mouse line that overexpress eGFP driven by its full genomic promoter was employed for in situ assessment of the ALDH1L1 promoter activity by the eGFP intensity at the single cell level (Doyle et al. 2008; Heiman et al. 2008). As shown in Figure 1C, eGFP expression is dramatically lower in astrocytes (indicated by white arrows) of the ventral horn of lumbar cord at the age of P135 than that of P20. Similar decrease was also found in other regions of spinal cord. Quantitative analysis through FACS of at least 4000 spinal cord eGFP+ cells per spinal cord showed a reduction of ~8 fold in the mean fluorescence intensity (MFI) from P5 to P135, clearly indicating that ALDH1L1 promoter becomes inactivated in the spinal cord as the spinal cord matures, which is responsible for the loss of ALDH1L1 mRNA and protein loss. The loss of ALDH1L1 expression appears to be specific in the spinal cord, as its expression (mRNA, protein, and promoter activity) is not altered in other CNS regions.
Figure 1.
ALDH1L1 expression is selectively decreased in adult spinal cord astrocytes through transcriptional inactivation of the ALDH1L1 promoter A, Selective decrease of ALDH1L1 mRNA levels in spinal cord during postnatal development. N=5, one-way ANOVA with Bonferroni posthoc analysis, **P<0.01, *** P<0.001; mean ± SEM B, Selective decrease of ALDH1L1 protein levels in spinal cord during postnatal development. N=5 C, Loss of ALDH1L1 promoter activity indicated by eGFP fluorescence intensity in astrocytes of lumbar cord of BAC ALDH1L1 eGFP mice. Individual astrocytes are indicated by a white arrow. Scale bar, 20μm D, Quantitative measurement of the eGFP intensity decrease by FACS from spinal cord cell preparation of heterozygous BAC ALDH1L1 eGFP mice during postnatal development. MFI: mean fluorescence intensity. MFI was measured based upon at least 4000 cells/sample with a minimum of 3 mice for each experiment.
ALDH1L1 promoter is selectively activated in astrocytes in BAC ALDH1L1 eGFP reporter mice
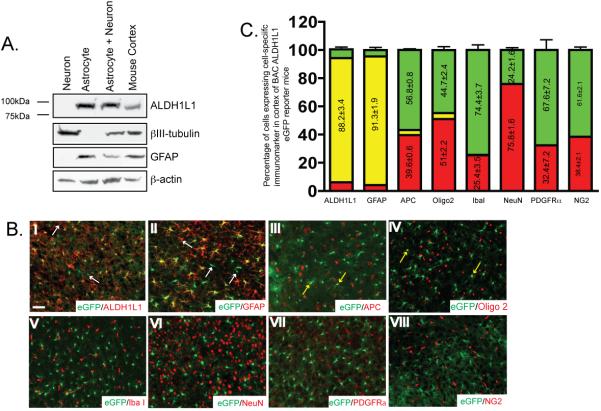
A recent gene profile study from CNS cells identified ALDH1L1 as an astroglial specific marker in the CNS (Cahoy et al. 2008). We first examined ALDH1L1 expression in cultured primary neurons and astrocytes. As shown in Figure 2A, ALDH1L1 protein is selectively expressed only in cultured astrocytes, not in cultures neurons. Relative purity of cultures was confirmed by known astroglial and neuronal marker GFAP and βIII-tubulin, respectively. To fully characterize the expression pattern of ALDH1L1 in the CNS cells, cell type specific markers were immunostained on the brain and spinal cord sections of BAC ALDH1L1 eGFP mice. Transgenic mice that overexpress eGFP driven by a specific Bacteria Artificial Chromosome (BAC) clone, including ALDH1L1 (http://www.gensat.org), has been previously generated for cell type specific and predominant cell body labeling, as a result of cytoplasmic eGFP expression. Co-localization of eGFP expression with different cell type makers, including astroglial marker ALDH1L1, GFAP, oligodendrocyte lineage marker Olig2 and APC (cc1 clone), neuronal marker NeuN, NG2 cell marker PDGFRα, and microglial marker Iba1 was examined in brain and spinal cord sections of BAC ALDH1L1 mice at P25, P60, and P130. Representative images of immunostaining with individual cell specific marker in cortex sections (P20) were shown in Figure 2B. Co-localization of ALDH1L1 and GFAP immunoreactivity was largely found in eGFP+ cells (Figure 2B I-II). ALDH1L1 appeared to selectively label astroglia branches than the cell body (Supplementary Figure 1B). Occasional co-localization of eGFP expression with APC or Olig2 immunoreactivity was also observed (yellow arrows in Figure 2B III-IV). Quantitative analysis of the co-localization of eGFP expression with different cell specific markers showed that eGFP+ cells overlapped well with ALDH1L1 immunoreactivity (88.2±3.4%), and are predominantly GFAP+ (91.3±1.9%) in the area when GFAP has abundant immunoreactivity. A small portion of eGFP+ cells are APC+ (~3.6%) or Olig2+ (~4.3%). Overlap of eGFP with other cell type markers was essentially undetected. Magnified images with ALDH1L1, GFAP, APC, and Olig2 staining are shown in the Supplementary Figure 1. In addition, no apparent overlap of eGFP driven by the ALDH1L1 promoter and the DsRed driven by the NG2 promoter was found in the BAC ALDH1L1 eGFP X BAC NG2 DsRed mice (Figure 2B VIII). The predominant co-localization pattern of eGFP expression with GFAP immunoreactivity but not other cell type markers are generally preserved in both brain and spinal cord tissue (Supplementary Figure 1C) of BAC ALDH1L1 eGFP mice at different age.
Figure 2.
ALDH1L1 promoter is selectively activated in astrocytes in BAC ALDH1L1 eGFP reporter mice A, Selective expression of ALDH1L1 protein in primary astrocyte cultures. B, Expression of eGFP reporter is selectively overlapped with ALDH1L1 and GFAP immunoreactivity but not with other major CNS cell type markers in BAC ALDH1L1 eGFP mice. Representative images from P25 cortex sections were shown. Scale bar, 50μm C, Quantitative analysis of the eGFP expression in major CNS cell types in BAC ALDH1L1 eGFP mice. Following immunostaining, Immunopositive cells and eGFP+ cells were counted from 3-5 fields/slide for 3-4 slides per region of interest for 3 mice.
Molecular comparisons of ALDH1L1+ and GLT1+ cells in astroglial reporter mice
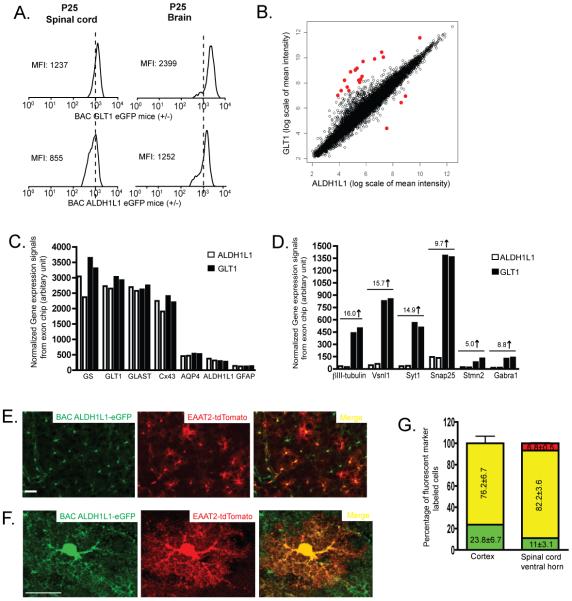
The predominant astroglial expression of eGFP in BAC ALDH1L1 eGFP mice provides a new tool, together with the previously characterized astroglial BAC GLT1 eGFP reporter mice (Regan et al. 2007), to compare the molecular identity of astrocytes in vivo. The overall eGFP expression intensity in BAC ALDH1L1 eGFP and BAC GLT1 eGFP was first compared through FACS and found that BAC GLT1 eGFP has a much higher mean flourescence intensity (MFI) than that in BAC ALDH1L1 in both spinal cord and brain at the age of P25 (Figure 3A), suggesting that GLT1 promoter may have a considerable higher activity than that of ALDH1L1 promoter in the CNS. ALDH1L1+ cells and GLT1+ cells were further isolated by FACS from brain of these two reporter mice at P30 and were subject to the whole genome expression profile analysis. As shown in Figure 3B, vast majority of genes have similar expression levels in both ALDH1L1+ and GLT1+ cells, suggesting that the molecular identity of ALDH1L1+ and GLT1+ cells are highly similar. The number of astrocyte-enriched genes that showed differential expression levels (2 fold) between ALDH1L1+ and GLT1+ is very limited (less than 20 out of the 15,000 genes) and is summarized in the Supplementary Table 1. In particular, both ALDH1L1+ and GLT1+ cells have a highly similar expression pattern of known astroglial genes(Lovatt et al. 2007) (Figure 3C), as well as the astrocyte-enriched genes from previous database (Supplementary Figure 2), confirming that both ALDH1L1+ and GLT1+ cells share very similar astroglial identity. Genes that are co-expressed in ALDH1L1+ and GLT1+ cells were summarized in Supplementary Table 2. In addition, the expression levels of GLT1 mRNA is ~5 fold higher than that of ALDH1L1 mRNA, also indicating that GLT1 promoter activity is higher than that of ALDH1L1 (Figure 3C). On the other hand, GLT1+ but not ALDH1L1+ cells also express a subset of neuronal genes (Figure 3D), which is consistent to the previous observation that GLT1 promoter is also active in a small subset of hippocampal CA3 neurons (Regan et al. 2007). In contrast, ALDH1L1 promoter activity is restricted to astrocytes. The whole genome gene expression analysis also indicates that neuronal and oligodendrocytic genes are not apparently expressed in ALDH1L1+ cells.
Figure 3.
Molecular comparisons of ALDH1L1+ and GLT1+ cells in astroglial reporter mice A, Quantitative comparison of the eGFP intensity by fluoscence activated cell analysis from brain and spinal cord cell preparation of BAC GLT1 eGFP and BAC ALDH1L1 eGFP mice at the age of P25. B, Scatter plot of whole genome gene expression profile in ALDH1L1+ and GLT1+ cells. Normalized expression signals from two duplicated exon arrays were averaged and presented in log2 scale. Most differentially expressed genes were highlighted in red. C, Highly similar expression levels of known astroglial genes in ALDH1L1+ and GLT1+ cells D, Selective expression of a subset of neuronal genes in GLT1+ but not in ALDH1L1+ cells. □: ALDH1L1+ cells; ■: GLT1+ cells E, Co-expression of eGFP and tdTomato reporters in protoplasmic cortical astrocytes in BAC ALDH1L1 eGFP X EAAT2-tdTomato mice. Scale bar, 50μm F, Magnified view of the co-expression of eGFP and tdTomato reporter in the same protoplasmic cortical astrocyte in BAC ALDH1L1 eGFP X EAAT2-tdTomato mice. Scale bar, 20μm G, Quantitative analysis of the co-expression of eGFP and tdTomato reporters in spinal cord and cortex of BAC ALDH1L1 eGFP X EAAT2-tdTomato mice. eGFP+ and tdTomato+ cells were counted from images of brain and spinal cord sections of BAC ALDH1L1 eGFP X EAAT2-tdTomato mice (3-5 fields/slide for 3-4 slides per region of interest for 3 mice).
Previously generated astroglial reporter mice often employ eGFP as the fluorescent reporter, making it infeasible to examine the co-expression of different astroglial markers in the same astrocyte in vivo. Here we generated a new astroglial reporter mouse line that overexpress tdTomato fluorescence reporter driven by a 8.3kb EAAT2 promoter fragment. This EAAT2-tdTomato transgenic mouse line showed selective and strong expression of tdTomato in many but not all astrocytes by crossing with the BAC GLT1 eGFP mice (~80%, data not shown). Quantitative analysis showed that 76.2±6.7% of cortical astrocytes co-express both tdTomato and eGFP reporters and 23.8±6.7% of cortical astrocytes only express eGFP (Figure 3G), suggesting that ALDH1L1 and GLT1 are commonly co-expressed in the same astrocyte in the cortex. It was also observed, though very rarely, that some cells only express tdTomato, not eGFP in cortex (<0.5%). Direct immunostaining of ALDH1L1 in the cortex of BAC GLT1 eGFP mice also showed a high percentage of overlap (>90%, Supplementary Figure 2). In spinal cord, tdTomato and eGFP reporters are also co-expressed in the majority of the cells (82.2±3.6%) with a small percentage of cells only expressing tdTomato or eGFP (6.8±0.5% and 11±3.1%, respectively). Expression of tdTomato is not restricted to the cell body, but also localized to the distal fine processes of astrocytes, visualized as morphology typical of protoplasmic astrocytes (Figure 3F).
Expression change of ALDH1L1 in spinal cord astrocytes in SOD1 G93A transgenic mice
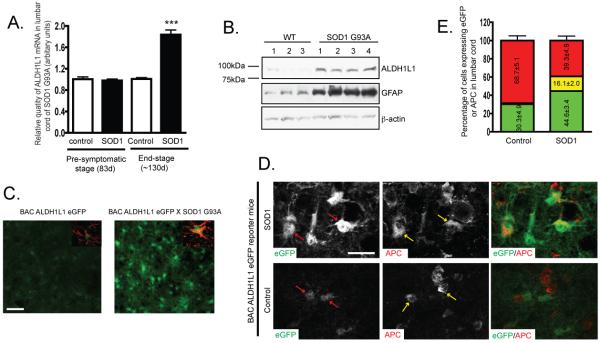
Astroglial genes undergo dramatic expression changes in pathological conditions, such as the strong up-regulation of GFAP in reactive astrocytes in neurological disease or injury. As a newly identified astroglial specific gene, the expression change of ALDH1L1 in both acute lesion and chronic neurodegenerative conditions has not been explored. Total RNA was collected from lumbar cord of SOD1 G93A mice at both symptomatic and end stages. QRT-PCR results by using specific ALDH1L1 probe found that ALDH1L1 mRNA levels were up-regulated in the end stage but not in the symptomatic stage. Consistent with the mRNA change, ALDH1L1 protein levels were also up-regulated in the end-stage lumbar cord of SOD1 G93A mice. To investigate the ALDH1L1 promoter activity change in the SOD1 G93A mice, BAC ALDH1L1 eGFP mice were crossed with SOD1 G93A mice. Microscopic analysis of the coronal sections of lumbar cord from BAC ALDH1L1 eGFP X SOD1 G93A mice found that ALDH1L1 promoter activity was up-regulated indicated by the dramatically increased eGFP fluorescence intensity in the reactive astrocytes of the ventral horn of lumbar cord. Similar increased eGFP intensity (and GFAP immunoreactivity) was also observed in reactive astrocytes in striatum of BAC ALDH1L1 eGFP 7d following needle-induced acute stab lesion (Supplementary Figure 3). These results suggest that ALDH1L1 can serve as an additional marker for the reactive astrocytes in neurodegenerative/neural injury conditions. Interestingly, extensive overlap (16.1±2.0%) of eGFP expression with APC immunoreactivity (but not with NeuN, Iba1, or PDGFRα) was observed in the lumbar cord section of BAC ALDH1L1 eGFP X SOD1 G93A mice, though only occasional co-localization (1-3%) of eGFP expression and APC immunoreactivity was found in control lumbar cord of BAC ALDH1L1 eGFP mice. Similar observations were found in lumbar cord of GFAP eGFP X SOD1 G93A mice (Supplementary Figure 4). The increased overlap of eGFP expression with APC immunostaining could indicate a possible loss of the mature astrocyte status in this pathological condition.
DISCUSSION
In this study we report on several new tools to investigate astroglial heterogeneity in normal and diseased brain. We found that ALDH1L1 is primarily and selectively expressed in cortical and spinal cord astrocytes. We noted rare expression of Olig2+ or APC+ cells in eGFP+ cells in the BAC ALDH1L1 eGFP transgenic mouse (Figure 2). Although Olig2 is generally considered a marker of oligodendrocyte linage , it is transiently expressed in gray and white matter astrocytes during early postnatal development, playing a crucial role for the proper postnatal development of white matter astrocytes (Marshall et al. 2005). Olig2 can also direct astrocyte formation from Subventricular Zone (SVZ) cells (Cai et al. 2007). The overlap of Olig2/APC and ALDH1L1 in astrocytes observed in the current study may suggest that these cells are a subpopulation of astroglial progenitor cells in postnatal brain. Because the whole genome expression analysis of ALDH1L1+ cells reveal essentially no selective oligodendrocyte gene expression, it is unlikely that the ALDH1L1 promoter is normally activated in oligodendrocytes.
By using these astroglial reporter mice, the molecular identity of astrocytes labeled with different markers, namely, ALDH1L1 or GLT1, was directly compared, as a first attempt to investigate the molecular differences of heterogeneous astrocytes in the CNS. Our results indicated that ALDH1L1 and GLT1 appear to co-express in the same population of astrocytes in both cortex and spinal cord (Figure 3). Surprisingly, ALDH1L1 expression levels are selectively and very significantly decreased in mature spinal cord, posting a potential concern for its use as a marker of spinal cord astrocytes in older rodents, especially in normal physiological conditions (Figure 1). On the other hand, its expression (mRNA, protein, and promoter activity) is up-regulated, similar to the expression changes of GFAP, in acute and chronic pathological conditions (Figure 4). As ALDH1L1 is a key enzyme in folate metabolism which is important in nucleotide biosynthesis and cell division (Krupenko 2009), its expression change in spinal cord during postnatal maturation and pathological conditions may reflect the changing demands for nucleotides in astrocytes and potentially may regulate the astrocyte division and growth in these conditions. Interestingly, we observed the significant overlap of eGFP expression with APC immunoreactivity in the ventral horn of lumbar cord of end-stage BAC ALDH1L1 eGFP X SOD1 G93A and GFAP eGFP X SOD1 G93A mice. Because APC (cc1 clone) specifically labels oligodendrocytes, the overlap may indicate that some reactive astrocytes may lose their fully differentiated status. Conversion of a mature astrocyte status into stem-cell like cells was also found previously in an injury condition by genetic fate mapping and cell-type specific viral targeting technique (Buffo et al. 2008). Further genetic fate mapping experiments will be needed to elucidate the change of mature astrocyte status in SOD1 G93A mice.
Figure 4.
Expression change of ALDH1L1 in spinal cord astrocytes of SOD1 G93A transgenic mice A, QRT-PCR analysis of ALDH1L1 mRNA levels in lumbar cord of SOD1 G93A mice. Total RNA from lumbar spinal cord of SOD1 G93A mice at symptomatic and end stages was prepared (n=5-7) (Student’s t-test, ***P<0.001, mean±SEM). B, Up-regulation of ALDH1L1 protein levels in lumbar cord of end-stage SOD1 G93A mice (n=3-4). C, Increased ALDH1L1 promoter activity indicated by eGFP fluorescence intensity in reactive astrocytes of ventral horn of lumbar cord in BAC ALDH1L1 eGFP X SOD1 G93A mice. Inset: GFAP immunostaining. Scale bar, 50μm D, Increased APC staining in astrocytes of lumbar cord of end-stage BAC ALDH1L1 eGFP X SOD1 G93A mice. Red arrows: ALDH1+ astrocytes; Yellow arrows: APC+ cells. Scale bar, 20μm. E, Quantitative analysis of the overlap of APC immunoreactivity with eGFP reporter in astrocytes from lumbar spinal cord of end stage BAC ALDH1L1 eGFP X SOD1 G93A mice. Cells were counted from 2-4 fields/slide for 6-7 slides cord for 3 mice.
In addition, an astroglial transgenic reporter mouse that overexpresses a tdTomato reporter driven by a 8.3kb EAAT2 promoter fragment was generated, which selectively labels the majority (70-80%) of astrocytes, suggesting that this 8.3kb EAAT2 promoter is sufficient to drive a strong and selective expression of a gene of interest in astrocytes. On the other hand, the incomplete labeling of a subset of astrocytes in vivo in these transgenic mice also indicates other regulatory mechanisms are operative in controlling GLT1 astroglial expression. Epigenetic regulation of GLT1, through DNA methylation, is an additional mechanism recently shown to regulate the GLT1 promoter activity (Yang et al. 2010). Microscopic visualization of process morphology of astrocytes in this mouse could provide a detailed analysis of astrocyte changes in various physiological and pathological conditions.
Taken together, we provide a comparative analysis of two unique astroglial reporters in vivo, employing BAC ALDH1L1 eGFP and EAAT2-tdTomato transgenic mice. Using these tools we were able to perform a comparative analysis of the molecular properties of astrocytes collected from these astroglial reporter mice. Both of these astroglia reporter mice will be useful in vivo tools to study the molecular and morphological properties of astrocytes in different physiological and pathological conditions.
Supplementary Material
Immunostaining of oligodentracyte and astrocyte markers in BAC ALDH1L1 eGFP mice A, Occasional overlap of eGFP expression and oligodentrocyte marker (olig2 and APC) immunoreactivity in cortex of BAC ALDH1L1 eGFP mice. Examples of overlapped cells were highlighted with yellow arrows in III and VI. White arrows indicate the corresponding eGFP fluorescence and Olig2 or APC immunoreactivity in I, II, IV, and V. Scale bar, 20μm B, High overlap of eGFP expression and astrocyte marker (ALDH1L1 and GFAP) in cortex of BAC ALDH1L1 eGFP mice. Examples of eGFP+/ALDH1L1− or eGFP+/GFAP− cells are highlighted by the white arrows in I and IV. Examples of eGFP−/ALDH1+ or eGFP−/GFAP+ cells are highlighted by yellow arrows in II and V. Scale bar, 20μm C, Expression of eGFP reporter selectively overlaps with GFAP immunoreactivity but not with other major CNS cell type markers in spinal cord of BAC ALDH1L1 eGFP mice. Representative images from P25 lumbar spinal cord sections are shown. Scale bar, 50μm D, Direct comparison of eGFP and tdTomato reporter expression in various astroglial reporter mice. Representative cortex region from each astroglial reporter mouse line is presented. Scale bar, 50μm
ALDH1L1 and GLT1 are co-expressed in the same astroglial cells A, Highly similar expression levels of astrocyte enriched genes in both ALDH1L1+ and GLT1+ astrocytes. Expression signals were normalized and averaged from duplicated arrays. Astrocyte enriched genes were selected from the established CNS cell type specific database (Cahoy et al. 2008). □: ALDH1L1+ cells; ■: GLT1+ cells B, Immunostaining of ALDH1L1 in the cortex of BAC GLT1 eGFP mice. Extensive overlap of eGFP expression and ALDH1L1 immunoreactivity was observed in cortical astrocytes. Scale bar: 50μm
Up-regulation of ALDH1L1 expression in acute injury condition in BAC ALDH1L1 eGFP mice A, Increased ALDH1L1 promoter activity indicated by increased eGFP intensity in reactive astrocytes following acute stab injury (7d post lesion). Stab lesion region was highlighted by the white circle. Reactive astrocytes surrounding the lesion area was shown by the increased GFAP immunoreactivity. Scale bar: 20μm. B, Quantitative analysis of the eGFP fluorescence intensity following stab injury Fluorescence intensity of eGFP in reactive astrocytes surrounding the injury area and uninjured area were measured in Image J (20-30 astrocytes /slide/3 mice, *** P<0.001, mean ± SEM, Student’s test)
Increased overlap between eGFP expression and APC immunoreactivity in spinal cord astrocytes of GFAP eGFP X SOD1 G93A mice A, APC immunostaining in astrocytes of lumbar cord of end-stage GFAP eGFP X SOD1 G93A mice. Red arrows: GFAP+ astrocytes; Yellow arrows: APC+ cells. Scale bar, 20μm. B, Quantitative analysis of the overlap of APC immunoreactivity with eGFP reporter in astrocytes of lumbar cord of end stage GFAP eGFP X SOD1 G93A mice. Cells were counted from 2-4 fields/slide for 6-7 slides cord for 3 mice.
Acknowledgements
This work was supported by NIH NS33958 (JDR), the Muscular Dystrophy Association (YY), and the Robert Packard Center for ALS Research. The work is dedicated to the memory of Johns Hopkins undergraduate Miriam Frankl.
References
- Anthony TE, Heintz N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. J Comp Neurol. 2007;500(2):368–83. doi: 10.1002/cne.21179. [DOI] [PubMed] [Google Scholar]
- Buffo A, Rite I, Tripathi P, Lepier A, Colak D, Horn AP, Mori T, Gotz M. Origin and progeny of reactive gliosis: A source of multipotent cells in the injured brain. Proc Natl Acad Sci U S A. 2008;105(9):3581–6. doi: 10.1073/pnas.0709002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134(10):1887–99. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–62. doi: 10.1016/j.cell.2008.10.029. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Fellin T, Haydon PG. The tripartite synapse: roles for gliotransmission in health and disease. Trends Mol Med. 2007;13(2):54–63. doi: 10.1016/j.molmed.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–48. doi: 10.1016/j.cell.2008.10.028. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2(12):861–70. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Kimelberg HK. Astrocyte Heterogeneity or Homogeneity. In: Parpura VHP, editor. Astrocytes in (Patho)Physiology of the Nervous System. Springer; New York: 2009. pp. 1–27. [Google Scholar]
- Krupenko SA. FDH: an aldehyde dehydrogenase fusion enzyme in folate metabolism. Chem Biol Interact. 2009;178(1-3):84–93. doi: 10.1016/j.cbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovatt D, Sonnewald U, Waagepetersen HS, Schousboe A, He W, Lin JH, Han X, Takano T, Wang S, Sim FJ. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J Neurosci. 2007;27(45):12255–66. doi: 10.1523/JNEUROSCI.3404-07.2007. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CA, Novitch BG, Goldman JE. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J Neurosci. 2005;25(32):7289–98. doi: 10.1523/JNEUROSCI.1924-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neymeyer V, Tephly TR, Miller MW. Folate and 10-formyltetrahydrofolate dehydrogenase (FDH) expression in the central nervous system of the mature rat. Brain Res. 1997;766(1-2):195–204. doi: 10.1016/s0006-8993(97)00528-3. [DOI] [PubMed] [Google Scholar]
- Regan MR, Huang YH, Kim YS, Dykes-Hoberg MI, Jin L, Watkins AM, Bergles DE, Rothstein JD. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci. 2007;27(25):6607–19. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32(12):638–47. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gozen O, Vidensky S, Robinson MB, Rothstein JD. Epigenetic regulation of neuron-dependent induction of astroglial synaptic protein GLT1. Glia. 2010;58(3):277–86. doi: 10.1002/glia.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gozen O, Watkins A, Lorenzini I, Lepore A, Gao Y, Vidensky S, Brennan J, Poulsen D, Won Park J. Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron. 2009;61(6):880–94. doi: 10.1016/j.neuron.2009.02.010. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunostaining of oligodentracyte and astrocyte markers in BAC ALDH1L1 eGFP mice A, Occasional overlap of eGFP expression and oligodentrocyte marker (olig2 and APC) immunoreactivity in cortex of BAC ALDH1L1 eGFP mice. Examples of overlapped cells were highlighted with yellow arrows in III and VI. White arrows indicate the corresponding eGFP fluorescence and Olig2 or APC immunoreactivity in I, II, IV, and V. Scale bar, 20μm B, High overlap of eGFP expression and astrocyte marker (ALDH1L1 and GFAP) in cortex of BAC ALDH1L1 eGFP mice. Examples of eGFP+/ALDH1L1− or eGFP+/GFAP− cells are highlighted by the white arrows in I and IV. Examples of eGFP−/ALDH1+ or eGFP−/GFAP+ cells are highlighted by yellow arrows in II and V. Scale bar, 20μm C, Expression of eGFP reporter selectively overlaps with GFAP immunoreactivity but not with other major CNS cell type markers in spinal cord of BAC ALDH1L1 eGFP mice. Representative images from P25 lumbar spinal cord sections are shown. Scale bar, 50μm D, Direct comparison of eGFP and tdTomato reporter expression in various astroglial reporter mice. Representative cortex region from each astroglial reporter mouse line is presented. Scale bar, 50μm
ALDH1L1 and GLT1 are co-expressed in the same astroglial cells A, Highly similar expression levels of astrocyte enriched genes in both ALDH1L1+ and GLT1+ astrocytes. Expression signals were normalized and averaged from duplicated arrays. Astrocyte enriched genes were selected from the established CNS cell type specific database (Cahoy et al. 2008). □: ALDH1L1+ cells; ■: GLT1+ cells B, Immunostaining of ALDH1L1 in the cortex of BAC GLT1 eGFP mice. Extensive overlap of eGFP expression and ALDH1L1 immunoreactivity was observed in cortical astrocytes. Scale bar: 50μm
Up-regulation of ALDH1L1 expression in acute injury condition in BAC ALDH1L1 eGFP mice A, Increased ALDH1L1 promoter activity indicated by increased eGFP intensity in reactive astrocytes following acute stab injury (7d post lesion). Stab lesion region was highlighted by the white circle. Reactive astrocytes surrounding the lesion area was shown by the increased GFAP immunoreactivity. Scale bar: 20μm. B, Quantitative analysis of the eGFP fluorescence intensity following stab injury Fluorescence intensity of eGFP in reactive astrocytes surrounding the injury area and uninjured area were measured in Image J (20-30 astrocytes /slide/3 mice, *** P<0.001, mean ± SEM, Student’s test)
Increased overlap between eGFP expression and APC immunoreactivity in spinal cord astrocytes of GFAP eGFP X SOD1 G93A mice A, APC immunostaining in astrocytes of lumbar cord of end-stage GFAP eGFP X SOD1 G93A mice. Red arrows: GFAP+ astrocytes; Yellow arrows: APC+ cells. Scale bar, 20μm. B, Quantitative analysis of the overlap of APC immunoreactivity with eGFP reporter in astrocytes of lumbar cord of end stage GFAP eGFP X SOD1 G93A mice. Cells were counted from 2-4 fields/slide for 6-7 slides cord for 3 mice.