Abstract
The apparent diffusion coefficient (ADC) determined from MR diffusion tensor imaging (DTI) has shown promise for distinguishing World Health Organization grade II astrocytoma (AS) from the more prognostically favorable grade II oligodendroglioma (OD). Since mixed oligoastrocytomas (OAs) with codeletions in chromosomes 1p and 19q confer prognoses similar to those of OD, we questioned whether a previously determined ADC-based criterion for distinguishing OD and AS would hold on an independent set of gliomas that included OA with codeleted or intact 1p/19q chromosomes. We also questioned whether the ADC is associated with the tumor microstructure. ADC colormaps generated from presurgical DTI scans were used to guide the collection of biopsies from each tumor. The median normalized ADC distinguished OD from AS with 91% sensitivity and 92% specificity. 1p/19q codeleted OAs were always classified as ODs, while 1p/19q intact OAs were always classified as ASs. There were positive associations between the ADC and both the SMI-31 score of axonal disruption and the fraction of tumor cells in the biopsies. The ADC of OD and 1p/19q codeleted OA was more associated with tumor fraction, while the ADC of AS and 1p/19q intact OA was more associated with SMI-31 score. We conclude that our previously determined threshold median ADC can distinguish grade II OD and AS on a new patient cohort and that the distinctions extend to OA with codeleted and intact 1p/19q chromosomes. Further, the ADC in grade II gliomas is associated with the fraction of tumor cells and degree of axonal disruption in tumor subregions.
Keywords: apparent diffusion coefficient, diffusion-weighted imaging, grade II gliomas, histopathology, image-guided biopsy, 1p/19q codeletion
The 2 major histologic subtypes of World Health Organization (WHO) grade II glioma, oligodendroglioma (OD) and astrocytoma (AS), have distinct biological characteristics and differ in response to therapy and patient outcome. The prognosis for patients with OD is significantly better than that for patients with AS, averaging survival times of 10 years or more versus 4 years, respectively.1,2 This is due partly to their superior response to chemotherapy3–6 compared with AS. Oligoastrocytomas (OAs) are infiltrating gliomas that are variably composed of cell populations with both astrocytic and oligodendroglial histopathologic phenotypes and have a more heterogeneous biologic behavior that comprises the spectrum from AS to OD. The molecular features of OA can aid in the prediction of tumor behavior and patient outcome. Codeletions in the 1p and 19q chromosomal arms occur in 90% of OD and 50% of OA and are associated with improved survival and response to therapy7,8 compared with AS. The biologic factors responsible for the chemosensitivity of tumors with 1p/19q codeletion are unknown; however, studies have suggested that the 1p/19q status may be associated with the differences in the infiltrative growth patterns of gliomas.9,10 Positive staining on immunohistochemical assays of the p53 tumor suppressor protein indicates abnormal p53 function and is most often present in AS, followed by OA, and is least frequent in OD. Immunopositivity of p53 is rarely observed in tumors with a 1p/19q codeletion11,12 and is therefore sometimes considered to be an indicator of poorer prognosis for patients with grade II diffuse-type glioma.
Currently, the standard of practice for distinguishing among the glioma subtypes is histopathological analysis of tissue biopsy, which necessitates an invasive procedure and is prone to tissue sampling error.13,14 Diffuse-type gliomas are spatially heterogeneous tumors, and the accuracy of glioma classification and grading through biopsies is highly dependent on the tumor regions that are sampled. This is particularly true for WHO grade II diffuse-type gliomas, which typically do not show a contrast-enhancing region on MRI that can be used as a target for obtaining a diagnostic biopsy. Defining noninvasive techniques that are capable of differentiating these various histologic types of diffuse-type low-grade gliomas would be an important complement to tissue analysis and a valuable surrogate diagnostic technique. This is especially true for mixed OA tumors that may be more heterogeneous across tumor subregions. A noninvasive method for predicting tumor aggressiveness and/or response to therapy is also critical for managing patients with inoperable tumors, such as brainstem gliomas; patients with postsurgical residual disease; and particularly patients with mixed diffuse-type gliomas, where the residual tumor after surgery may not have the same characteristics as the resected portion of tumor.
Microscopic molecular movement of water in tumors reflects tissue properties that include varying levels of structural alterations, tumor cellularity, and vasogenic edema. MR diffusion tensor imaging (DTI) uses strong magnetic field gradients applied in multiple directions to probe the structure of biologic tissues at a microscopic level by measuring the Brownian motion of water molecules; it has therefore been used for in vivo tissue characterization.15 Two diffusion parameters calculated from DTI are isotropic mean diffusivity, also known as the apparent diffusion coefficient (ADC), and fractional anisotropy (FA), which is a measure of directional diffusivity. Regions rich in cerebrospinal fluid such as the ventricles and cortex appear bright on ADC image maps, reflecting the nonrestricted isotropic water movement in those areas. Tumors allow for more isotropic water movement than the highly structured parenchyma of normal brain and they may also contain regions of edema; thus grade II gliomas often have higher ADC values than normal white matter. In contrast, normal white matter appears brighter than tumors on FA image maps due to the directional movement of water along white matter structures in normal brain that are often disrupted or absent in tumors.
Several groups have reported an inverse correlation between ADC and cell density within glial tumors,16–18, suggesting that cell density is the main determinant of ADC. However, all of the cited studies included a mixture of low- and high-grade gliomas within their study cohorts. In contrast, our group and others have reported that grade II AS has a significantly higher ADC than grade II OD,19,20 even though the cell density of grade II gliomas is uniformly low irrespective of histologic subtype. We also found a trend toward lower FA in grade II AS than in grade II OD, but the results were not as consistent as they were for ADC.19
We hypothesized that other microstructural properties, such as cellular architecture, neuronal integrity, and the degree or pattern of tumor infiltration into normal regions, may influence the diffusion measurements in tumors with relatively low cellularity. A clearer understanding of the microstructural factors that affect the ADC and FA may improve the interpretation of MR diffusion data for grade II gliomas.
The goal of this study was to (1) test a previously determined threshold median ADC value for distinguishing OD and AS on a new test set of grade II diffuse-type gliomas and (2) compare the ADC and FA with microstructural tissue properties at specific biopsy locations within the tumors. Scored, blinded pathological assessment was made of the histologic subtype, fraction of tumor cells to total number of cells, and degree of axonal degradation at one or more biopsy locations within each tumor, and these data were compared with presurgical MR diffusion parameters at that same location.
Materials and Methods
Study Population
A total of 30 patients with newly diagnosed nonenhancing diffuse-type grade II glioma were included in this study. Final tissue diagnosis was based upon histologic examination of clinical and research biopsies using criteria defined by WHO neuropathological criteria.21 Twelve patients had OD (8 female, 4 male) and ranged in age from 29 to 62 years, with a mean of 45 years. Eleven patients had AS (3 female, 8 male) and ranged in age from 29 to 48 years with a mean of 40 years. Seven patients had OA (7 female, 0 male) and ranged in age from 23 to 42 with a mean of 35 years. Imaging studies were performed as standard of care and patients provided informed consent for the biopsy portion as approved by the Committee on Human Research at our institution.
Conventional MRI
MR exams were performed with a 3T GE Signa Echospeed scanner (GE Healthcare Technologies), using an 8-channel head coil. The MRI examination included axial T1-weighted postgadolinium 3D inversion recovery spoiled gradient echo (IRSPGR) images (repetition time [TR] = 9 ms; echo time [TE] = 2 ms; inversion time [TI] = 400 ms; slice thickness = 1.5 mm; matrix = 256 × 256; field of vision [FOV] = 251 × 251 mm2; flip angle = 40°), axial T2-weighted 3D fast spin echo (FSE) (TR = 3667 ms, TE = 100 ms, slice thickness = 1.5 mm, matrix = 256 × 256, FOV = 261 × 261 mm2) and/or axial XETA T2 fluid attenuated inversion recovery (FLAIR) (TR = 10 000 ms, TE = 130 ms, TI = 2200 ms, slice thickness = 1.5 mm, matrix = 256 × 256, FOV = 241 × 241 mm2). After each examination, the images were transferred to a Sun Ultra 10 workstation (Sun Microsystems) for postprocessing.
The FSE or FLAIR was aligned to the postgadolinium SPGR using software developed in our laboratory.22 The normal-appearing white matter (NAWM) mask was segmented using FAST (FMRIB's [Functional Magnetic Resonance Imaging of the Brain] Automated Segmentation Tool) Software on the T2-weighted FSE image.23
Diffusion-weighted Imaging
Patients were scanned with a 6-directional diffusion weighted echo-planar imaging (EPI) sequence (TR = 7000 ms, TE = 63 ms, matrix size = 256 × 256, slice thickness = 3 mm, b = 1000 s/mm2, number of excitations = 4). ADC and FA maps were calculated on a pixel-by-pixel basis using software developed in-house, based on published algorithms.24 Diffusion images were registered to anatomical imaging by rigidly aligning the T2-weighted (b = 0) diffusion image to the T2-weighted FSE and applying the transformation to the ADC and FA maps. Normalized ADC (nADC) and normalized FA (nFA) maps were generated by dividing the diffusion image maps by the median ADC or FA value within the NAWM mask.
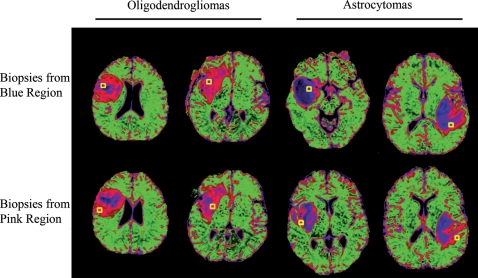
Colormaps were generated from nADC histograms of previously acquired DTI data in a training set of subjects with diffuse-type grade II gliomas25 (Fig. 1). As previously described, the colors are based on the nADC values within the nonenhancing lesion of patients with grade II OD (pink), the NAWM of patients with grade II OD and AS (green), and the nonenhancing lesion of patients with grade II AS (blue). Because of the higher nADC values observed in AS, the blue regions represent higher nADC values than the pink regions.
Fig. 1.
nADC colormaps of grade II oligodendroglioma (OD) and grade II astrocytoma (AS). Two left columns: ODs showing characteristic small central blue regions and extensive pink regions. Two right columns: ASs showing characteristic extensive central blue regions and thin pink rims. A yellow square represents the biopsy location from the blue and pink region.
nADC-guided Biopsy
Surgeons identified candidates for this study from patients with nonenhancing lesions suspected to be low-grade diffuse-type gliomas scheduled to undergo surgical resections. After 10 or 11 fiducials were positioned on the patient's skull, the MRI protocol described in detail above was acquired. After postprocessing of the images, 2–4 biopsy targets were chosen, based on identifying a blue and pink region from the nADC colormaps whenever possible. Biopsy targets were visually overlaid on the axial T2-weighted FSE displayed on the BrainLab VectorVision neuronavigation system but could be overlaid onto any of the anatomical images during surgery. The biopsy target was always a cylinder with a diameter and height of 6 mm to accommodate the 5-mm accuracy of the surgical planning methodology.26
In the operating room, the fiducials were registered to the BrainLab System. Two sets of biopsies were extracted during the course of the surgery: (1) clinical biopsies, extracted from tumor regions chosen by the surgeon that were sent to the clinical pathology service for diagnostic assessment, and (2) nADC-guided biopsies, extracted from regions within or near the target locations shown on the BrainLab system that were used for this research study only. The locations of the biopsy samples obtained using diffusion MR image guidance were recorded during the course of their extraction by using the Screen Save feature of the surgical navigation system. Once extracted, the biopsy specimens were fixed in formalin and sent to the University of California–San Francisco neuropathology service for histopathologic analysis.
Histopathology
Histologic analyses were used to classify the overall tumor histopatholgic subtype, to detect the molecular biomarkers, to score the tumor fraction (relative number of tumor cells per total cells), and to score the axonal integrity at specific tumor subregions. For the overall tumor classification, the clinical diagnosis from the non-nADC-guided biopsies was used to classify the tumors as AS, OD, or OA.
The prognosis for gliomas that harbor codeletions in the 1p and 19q chromosomes is similar to that of OD, while the prognosis for gliomas with intact 1p and 19q chromosomes is similar to that for AS. To determine which of the gliomas had the cytogenetic deletions in the 1p and 19q chromosomes, fluorescence in situ hybridization (FISH) was performed on either a diagnostic or nADC-guided biopsy from each tumor. An adjacent hematoxylin and eosin (H&E) stained section of the case was reviewed to evaluate tumor and normal tissue morphology and to identify the appropriate area of FISH analysis. In cases where FISH could not be performed for technical reasons, an immunohistochemical assay of p53 tumor suppressor protein was performed. Studies have shown that it is rare that both p53 immunoreactivity and 1p/19q codeletion are present in the same WHO grade II glioma.11,12 Therefore, gliomas that were immunopositive for p53 were assumed to lack the classic chemosensitivity and improved outcome attributed to oligodendroglial tumors.
The nADC-guided biopsies were histologically classified according to the clinical diagnosis of the tumor from which they were extracted due to the more comprehensive assessment of the entire tumor that is performed during the clinical diagnostic procedure. For example, tissue stains such as glial fibrillary acidic protein and vimentin are often used in conjunction with H&E during the clinical diagnostic procedure to help the pathologists discriminate astrocytic and oligodendroglial features. The purpose of collecting the nADC-guided biopsies was to more directly evaluate the cellular and microstructural properties of subregions within the tumors for comparison with the MRI diffusion measures in the same regions. Histopathologic H&E stains were examined to determine the contribution of tumor cells to the overall cellularity of the specimen. Biopsies with few or no tumor cells present were classified as “low tumor fraction,” and those in which the bulk of the specimen contained infiltrative tumor were classified as “high tumor fraction.” The immunohistochemical stain SMI-31 was used to assess loss and disruption of neuronal processes (primarily axonal) by scoring on a scale from 0 to 3, where no disruption of normal structures = 0 (normal axonal pattern and high levels of SMI-31 immunoreactivity) and maximal loss and disruption of intrinsic neuronal structures = 3 (inconspicuous normal pattern and only rare SMI-31 immunoreactive structures). All histopathologic assessments of the nADC-guided biopsies were performed by the same experienced neuropathologist (S.R.V.), who was blinded to patient diagnosis and imaging results.
Data Processing and Statistical Analysis
An in-house semi-automated segmentation method was used to define the T2 hyperintense region on the T2-weighted FSE or FLAIR image. The segmented T2 lesion was then overlaid on the nADC maps to calculate the median nADC within each tumor. The tumors were classified as AS-like if the median nADC within the T2 lesion was greater than or equal to 1.8 and as OD-like if the nADC was less than 1.8. The median nADC threshold of 1.8 was found to distinguish the 2 glioma subtypes in a previously published study on a training set of grade II glioma patients.19 Two receiver operating characteristics (ROC) analyses were performed to test the discriminatory capability (area under the curve [AUC]) of the median nADC value within the T2 lesion for distinguishing AS and OD tumors in the new patient set (test set). The first analysis was performed on the subset of AS and OD tumors only, to test the sensitivity and specificity of the previously determined median nADC of 1.8 to distinguish the 2 histologic subtypes. The second ROC analysis was performed on the entire cohort to test the capability of the median nADC for distinguishing tumors with OD-like features from those with AS-like features based on their histologic subtype, 1p/19q status, and p53 immunostaining status.
We then compared the median nADC value, median nFA value, tumor fraction, and SMI score for each nADC-guided biopsy. All reported values are mean ± standard deviation (SD) unless otherwise noted. A random effects model with Patient as a random effect was used to test the association between the SMI and MR diffusion measures at the biopsy locations. This model was chosen to account for the multiple biopsies obtained from each patient. A mixed effects model with Patient as a random effect and Tumor Fraction as the fixed effect was used to compare the MR diffusion values in the low and high tumor fraction biopsies. Fisher's exact tests were used to test the association between the biopsied region on nADC colormaps (pink or blue) and the tissue fraction and SMI staining in the same regions. All statistics were performed using the IBM Predictive Analytics SoftWare statistics package. An alpha level of P< .05 was used for all tests of significance.
Results
ADC-based Tumor Classification
In our previous study, logistic regression analysis on a training set of 39 grade II AS and OD gliomas showed that the 2 types could be classified with 87% accuracy using a threshold median nADC value within the T2 lesion of 1.8. To validate these results, we tested the discriminatory power of the same nADC threshold on an independent test cohort of gliomas. The test cohort consisted of patients with either OD (n = 12), AS, (n = 11), or OA (n = 7). Since gliomas with 1p/19q chromosomal codeletions behave like OD and those with p53 immunoreactivity behave like AS, we also noted either variable for each patient (Table 1).
Table 1.
Glioma histologic subtype, median nADC, 1p/19q chromosomal codeletion, and p53 immunoreactivity information in increasing order of the median nADC values. Assay was not performed
| Patient Number | Subtype | Median nADC | 1p/19q (deletion = 1, no deletion = 0) | p53 (positive = 1, negative = 0) |
|---|---|---|---|---|
| 1 | OD | 1.35 | 1 | 0 |
| 2 | OA | 1.40 | 1 | 0 |
| 3 | OD | 1.49 | 1 | 0 |
| 4 | OD | 1.50 | 1 | – |
| 5 | OD | 1.51 | – | 1 |
| 6 | OD | 1.52 | 1 | 0 |
| 7 | OA | 1.54 | 1 | 0 |
| 8 | OA | 1.55 | 1 | 0 |
| 9 | OD | 1.59 | 1 | 0 |
| 10 | AS | 1.59 | – | 1 |
| 11 | OD | 1.60 | 1 | 0 |
| 12 | OD | 1.61 | 1 | – |
| 13 | OD | 1.61 | – | 0 |
| 14 | OD | 1.63 | 1 | 0 |
| 15 | OD | 1.70 | 1 | 0 |
| 16 | OD | 1.80 | 1 | 0 |
| 17 | AS | 1.85 | 0 | 1 |
| 18 | AS | 1.86 | – | 1 |
| 19 | AS | 1.87 | – | 0 |
| 20 | OA | 1.96 | 0 | 1 |
| 21 | AS | 2.03 | – | 1 |
| 22 | AS | 2.14 | – | 1 |
| 23 | OA | 2.18 | 0 | 1 |
| 24 | OA | 2.18 | 0 | – |
| 25 | AS | 2.21 | – | 1 |
| 26 | AS | 2.29 | – | 0 |
| 27 | OA | 2.33 | 0 | – |
| 28 | AS | 2.35 | – | 1 |
| 29 | AS | 2.40 | – | 1 |
| 30 | AS | 2.42 | – | 0 |
Using a median nADC of less than 1.8 as the criterion for OD, 11 of the 12 ODs were correctly categorized (Table 2). The other OD had a median nADC value equal to 1.8. Assessment of 1p/19q chromosomal status was performed on 10 of the 12 ODs, and all 10 ODs showed a codeletion of 1p/19q, including the one that was misclassified.
Table 2.
Number of each glioma subtype with nADC values above and below the 1.8 threshold
| nADC | OD | AS | OA (1p-/19q-)* |
|---|---|---|---|
| <1.8 | 11 | 1 | 3 (3) |
| ≥1.8 | 1 | 10 | 4 (0) |
*The number of mixed OA tumors with codeletion in chromosomes 1p and 19q (1p-/19q-).
Using a median nADC greater than or equal to 1.8 as the criterion for AS, 10 of the 11 ASs were correctly categorized (Table 2). The other AS had a median nADC value of 1.59. Assessment of p53 was performed on all 11 ASs, and 8 were found to be immunoreactive, including the one that was misclassified.
Using the same criteria as above to classify the OAs, 3 of the 7 OAs were classified as ODs and all had 1p/19q chromosomal codeletions. None of the 4 OAs classified as AS had 1p/19q codeletions.
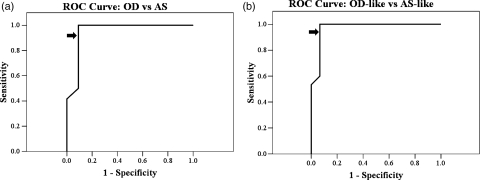
We performed 2 ROC analyses to determine the accuracy of using the median nADC to classify nonenhancing gliomas. Based on the AS and OD gliomas only (n = 23), the AUC of the ROC was 0.95 (90% confidence interval [CI] = 0.85 and 1.00), indicating a high degree of separation between the nADC values from AS and OD (Fig. 2a). Using a threshold median nADC of 1.8, the ODs were distinguished from the ASs with 92% sensitivity and 91% specificity. We then performed an ROC analysis on the entire cohort (n = 30) to determine whether the median nADC value could be used to distinguish the more prognostically favorable OD-like gliomas from the less prognostically favorable AS-like gliomas. The OD-like group comprised tumors with OD histology or OA histology with 1p/19q chromosomal codeletions. The AUC ROC was 0.97 (90% CI = 0.91 and 1.00), again indicating a high degree of separation between the AS-like and OD-like gliomas. Using the same threshold median nADC of 1.8, OD-like gliomas were distinguished from AS-like gliomas with 93% sensitivity and 93% specificity.
Fig. 2.
ROC curves. (a) nADC discrimination of OD from the group of AS and OD tumors (AUC = 0.95). (b) nADC discrimination of OD-like from the group of AS-like and OD-like tumors (AUC = 0.97). Diagonal segments are produced by ties. Arrows indicate the position of the nADC = 1.8 value along the curve.
Analysis of nADC, nFA, and Tissue Microstructure at Biopsy Locations
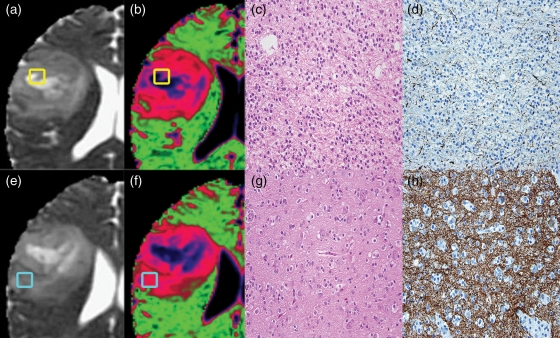
A total of 45 nADC-guided biopsies from 21 of the 30 patients were available for analysis. Either 2 biopsies (n = 18 patients) or 3 biopsies (n = 3 patients) were collected from each tumor. We assessed the median nADC, median nFA, SMI score, and tumor fraction at each biopsy location. An example of the nADC colormaps, H&E, and SMI-31 staining of intact axons in one of the OD cases from Figure 1 is shown in Figure 3.
Fig. 3.
Two biopsies from a patient with a grade II oligodendroglioma overlayed on (a & e) ADC map and (b & f) nADC colormap with (c & g) H&E and (d & h) SMI-31 stains. The yellow biopsy (a–d) is within a blue colormap region showing high nADC, high tumor fraction, and high axonal disruption. The light blue biopsy (e–h) is within a pink colormap region showing lower nADC, low tumor fraction, and minimal axonal disruption.
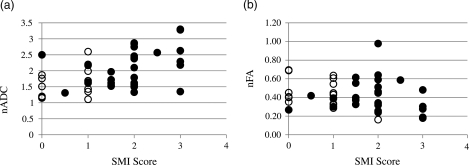
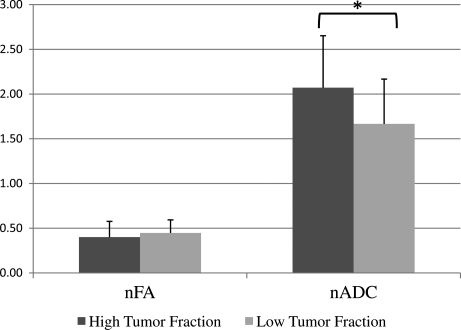
Figure 4 shows plots of the SMI-31 score versus (A) nADC and (B) nFA at the 45 biopsy locations. A random effects model of the data that accounted for the multiple biopsies collected from each patient indicated a significant positive association between nADC and the SMI-31 axonal disruption score (P = 0.008, n = 45) but no association between nFA and the SMI-31 score (P = 0.216, n = 45). We examined the nADC and nFA of the high and low tumor fraction groups. As shown in Figure 5, nADC values were higher (P = 0.004) in the high tumor fraction biopsies (2.07 ± 0.58) relative to the low tumor fraction biopsies (1.67 ± 0.50), but there was no difference in the nFA values (P = 0.166) of the 2 groups (0.40 ± 0.18 versus 0.45 ± 0.15).
Fig. 4.
Plots of SMI-31 score of axonal disruption and (a) nADC (P = .008) and (b) nFA (P = .216) at biopsy locations. Open circles = low tumor fraction, closed circles = high tumor fraction.
Fig. 5.
Mean and standard deviation of nADC and nFA in biopsies with low and high tumor fraction.
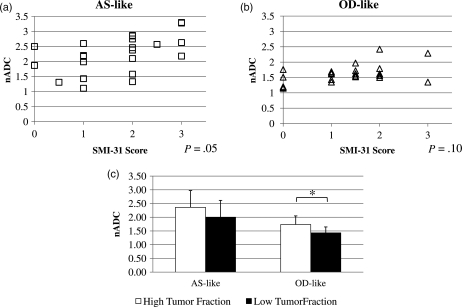
Next, biopsies were grouped by histologic subtype, and a random effects model was used to determine whether the nADC associations with SMI score and tumor fraction existed for each subtype. As in the above analysis, biopsies were deemed OD-like if they came from tumors that had OD histology (n = 15) or OA histology with a 1p/19q co-deletion (n = 8). Biopsies were deemed AS-like if they came from tumors with AS histology (n = 16) or had OA histology with intact 1p/19q chromosomes (n = 6). Figure 6a and b show that the positive association between nADC and SMI-31 score held for the AS-like biopsies (P = 0.053) but not for the OD-like biopsies (P = 0.099). Figure 6c shows that the nADC of the high tumor fraction biopsies from both subtypes were higher than that of the low tumor fraction biopsies; however, only the OD-like biopsies reached statistical significance (AS-like: P = 0.058, OD-like: P = 0.039).
Fig. 6.
Plots of normalized ADC and SMI score of axonal disruption for biopsies from (a) AS-like and (b) OD-like gliomas. (c) Mean and standard deviation of nADC in AS-like and OD-like glioma biopsies with low and high tumor fractions. *P< .05.
We investigated the underlying microstructural properties of the pink (lower nADC values) and blue (higher nADC values) tumor regions on the nADC colormaps. Table 3 shows the distribution of biopsies with high and low tumor fraction within the 2 groups. The blue biopsies came predominantly (79%) from regions with high tumor fraction. However, the pink biopsies were equally as likely (50%) to come from regions with low or high tumor fraction. A Fisher's exact test showed a trend but no statistically significant association (P = 0.065) between the colormap tumor region and tumor fraction. Table 3 also shows the number of biopsies with an SMI-31 score of 2 or greater (high SMI) and the number of biopsies with an SMI-31 score less than 2 (low SMI) within the 2 groups. The blue biopsies came predominantly (68%) from regions with high SMI, and the pink biopsies came predominantly (73%) from regions with low SMI. A Fisher's exact test showed a significant association (P = 0.008) between the colormap tumor region and SMI. The blue and pink regions, respectively, on the nADC colormaps are more predictive of high and low SMI score than high and low tumor fraction.
Table 3.
Tissue properties of pink and blue regions on nADC colormaps. Fisher's exact tests show that the colormap region is predictive of SMI (P = .008) but not tumor fraction (P = .076). Low SMI: SMI-31 score <2. High SMI: SMI-31 score ≥2
| Region on nADC Colormap | Low Tumor Fraction | High Tumor Fraction | Low SMI | High SMI |
|---|---|---|---|---|
| Blue (n = 19) | 4 | 15 | 6 | 13 |
| Pink (n = 26) | 13 | 13 | 19 | 7 |
| P = .065 | P = .008 | |||
To compare the histologic properties of the pink and blue biopsies from the OD-like and AS-like tumors, we categorized the biopsies by histology, tumor fraction, and SMI-31 score (Table 4). Biopsies from OD-like gliomas were primarily from pink colormap regions (20/23) due to the predominance of lower nADC values that characterized OD-like gliomas (Fig. 1). Fifteen of the 20 pink biopsies had low SMI, 10 of which also had low tumor fraction. The remaining 5 pink biopsies had both high SMI and high tumor fraction. Taken together with the above analyses, these results suggest that the lower (pink) nADC values observed in OD-like gliomas arose from a predominance of regions with low SMI in those tumors. The fact that regions with low SMI also tended to have low tumor fractions further contributed to the low nADC values in OD-like gliomas. The 3 biopsies from the small central blue regions of OD-like gliomas all had high tumor fraction, 2 of which also had high SMI. The combined high tumor fraction and high SMI is consistent with the higher ADC values in the blue colormap regions.
Table 4.
Counts and microstructural features of pink and blue biopsies extracted from AS-like and OD-like gliomas. Biopsies from OD-like gliomas were typically from pink colormap regions with low SMI and low tumor fraction, while biopsies from AS-like gliomas were typically from blue colormap regions with high SMI and high tumor fraction
| AS-like |
OD-like |
||||
|---|---|---|---|---|---|
| Pink | Blue | Pink | Blue | ||
| Low SMI | Low Tumor Fraction | 3 | 3 | 10 | 0 |
| High Tumor Fraction | 1 | 2 | 5 | 1 | |
| High SMI | Low Tumor Fraction | 0 | 1 | 0 | 0 |
| High Tumor Fraction | 2 | 10 | 5 | 2 | |
| Total | 6 | 16 | 20 | 3 | |
Contrary to the findings for OD-like gliomas, the biopsies from AS-like gliomas came predominantly (16/22) from blue colormap regions, due to the large area of high ADC values that characterize AS-like gliomas (Fig. 1). Eleven of the 16 blue biopsies had high SMI, 10 of which also had high tumor fraction. The remaining 5 blue biopsies had low SMI, 3 of which had low tumor fraction and 2 with high tumor fraction. These results suggest that the high ADC values in AS-like gliomas arose from the predominance of high SMI values, the majority of which also had high tumor fraction. Four of the 6 biopsies from the thin pink rim of the AS-like gliomas had low SMI, 3 of which also had low tumor fraction. The combined low SMI and low tumor fraction is consistent with the lower ADC values of the pink biopsies.
Discussion
Diffuse-type WHO grade II gliomas are heterogeneous tumors that generally do not enhance on postgadolinium T1-weighted MRIs. The grade II nonenhancing glioma population in this study served as the test data set for the nADC threshold for distinguishing grade II OD from grade II AS that was determined using logistic regression on a training set of patients in a previous study.19 The current study also compared the MRI diffusion parameters and microstructural tissue properties of nADC-guided biopsies collected from pink (low nADC) and blue (high nADC) regions on nADC colormaps.25
ROC analysis indicated a strong distinction between the nADC values of OD and AS in this new patient cohort, which is consistent with the high classification accuracy that was found using logistic regression on the training set of patients in our previous study. Using the previously determined threshold of 1.8 median nADC within the T2 lesion, we were able to distinguish AS from OD in this new patient cohort with high sensitivity and specificity. These results further validate the use of the median nADC within the T2 lesion to classify diffuse-type gliomas that are suspected of having grade II histology. Nonenhancing gliomas may also be of mixed histology (eg, OA) and have a biologic behavior that comprises the spectrum from AS to OD. We therefore performed a second ROC analysis of the entire patient cohort to determine whether the OA gliomas with molecular characteristics that were OD-like would be classified with the OD gliomas, based on their median nADC values. Again, there was a strong distinction between OD-like gliomas, which were defined as having an oligodendroglial component in conjunction with 1p/19q chromosomal codeletions, and AS-like gliomas, which did not have these histologic and chromosomal features. This finding was a key outcome of the study because of the more favorable prognosis for patients harboring gliomas with OD-like features. It suggests that the median nADC may be a useful prognostic marker for unresectable nonenhancing tumor.
All of the mixed OAs that had nADC values similar to OD (<1.8) had deletions in chromosomes 1p and19q, while gliomas with nADC values similar to AS (≥1.8) showed no 1p/19q codeletions. OA with intact 1p/19q also appeared similar to AS on nADC colormaps, having a large central blue region and a sharp, smooth transition to a thin pink rim (data not shown). This is similar to observations by other groups: a sharp, smooth border along with homogeneous signal intensity on T1- and T2-weighted MRI was associated with intact (nondeleted) 1p/19q chromosomes.27,28 In contrast, the typical pattern on nADC colormaps of OD and 1p/19q codeleted OA gliomas was a small central blue region surrounded by the more extensive and homogeneous pink region.
Normalized ADC values were higher in the high tumor fraction biopsies relative to the low tumor fraction biopsies. This appears to be contrary to previous papers suggesting lower nADC values in more highly cellular tumor regions.16 However, when one considers that the low tumor fraction biopsies comprised primarily normal brain, while the high tumor fraction biopsies comprised primarily tumor, the results are consistent with the higher ADC values that are typically reported for tumor versus normal brain. Indeed, 5 of the 17 low tumor fraction biopsies contained no discernible tumor cells at all (data not shown). This not only explains the lower nADC values in the low tumor fraction biopsies but also underscores the need for additional imaging methods, such as MR spectroscopy,29 to help distinguish tumors from nontumors within the hyperintense lesion on T2-weighted images. In addition to the positive association between nADC values and tumor burden, we also found a positive association between nADC values and SMI-31 scores of axonal disruption. Taken together, both an increase in the number of tumor cells and an increase in the degree of axonal disruption can result in an increase in the nADC value within a region of tumor.
There was no difference in the nFA value of low and high tumor fraction biopsies and no association between nFA and SMI-31 score. The lack of a clear relationship between nFA and the underlying tumor microstructure may be due partly to the inability to fully characterize the anisotropic water movement with the 6-direction diffusion tensor sequence that we used for this study. Of the 2 tissue properties that we measured, however, the SMI-31 score of axonal disruption appeared to have the strongest influence on the nFA.
Separating the biopsies by subtype, the association between nADC and SMI-31 score was observed in only the AS-like biopsies. This was due in part to the larger range and higher values of SMI-31 scores observed in AS-like biopsies compared with OD-like biopsies. Although both subtypes exhibited the positive association between nADC and tumor fraction, statistical significance was reached only in the OD-like biopsies. This was due in part to the more equal distribution of biopsies with high and low tumor fraction among the OD-like biopsies compared with the AS-like biopsies. These findings reflected the different patterns of infiltration of AS and OD that were observed in this study. In AS, engulfed normal structures were often disrupted, particularly in regions where there was a predominance of tumor cells. However, in OD the underlying normal structures were often maintained irrespective of the number of tumor cells in a given region.
Examination of the biopsies from the pink (high nADC) and blue (low nADC) colormap regions showed that there was a strong association between the colormap region and SMI-31 score and a weak (not statistically significant) association between the colormap region and tumor fraction. The biopsies from OD-like gliomas were overwhelmingly pink, consistent with their characteristic appearance on colormaps, which was primarily pink with a small blue region in the center of the tumor. The pink OD-like biopsies had predominantly a combination of low SMI-31 scores and low tissue fractions, while the blue OD-like biopsies all had high tissue fraction. This parallels the 2 results showing that (1) the SMI-31 score was strongly associated with colormap region, and (2) the tissue fraction was associated with nADC in OD-like gliomas. In comparison, the biopsies from the AS-like gliomas were primarily blue, consistent with their characteristic appearance on colormaps, which had a large central blue region transitioning rapidly to a thin pink rim. The blue AS-like biopsies had predominantly a combination of high SMI-31 scores and high tissue fractions, while the pink AS-like biopsies primarily had low SMI-31. Again, these results parallel the 2 findings that (1) the SMI-31 score was strongly associated with colormap region, and (2) the SMI-31 score was associated with nADC in AS-like tumors.
In conclusion, the median nADC value within the T2 lesion could be used to classify the histology of the OD and AS in a new cohort of diffuse-type grade II gliomas, validating the findings from our previous study on a training cohort. Mixed OAs harboring the favorable prognostic marker—1p/19q chromosomal codeletion—could also be classified by their median nADC values. The ADC values within tumor subregions were associated primarily with the degree of disruption of neuronal processes and less so with the fraction of tumor cells, particularly in AS and mixed OA with intact 1p/19q chromosomes. In OD and mixed OA with 1p/19q chromosomal codeletions, the ADC is less influenced by the degree of neuronal disruption and is more associated with the fraction of tumor cells in a given region. These results suggest that ADC variations in grade II gliomas are prognostically significant and are influenced by the arrangement and density of neuronal processes and tumor cells rather than by overall cell density. More studies are needed to determine whether the observed associations hold among nonenhancing gliomas of higher histologic grade and how the inclusion of additional imaging parameters can improve the noninvasive diagnosis of nonenhancing gliomsa of all histologic grades and subtypes.
Conflict of interest statement. None declared.
Funding
National Institutes of Health (R01 CA11604 to T.R.M., K.J.S., and C.P.C.); National Institutes of Health (P50 CA097257 to I.S.K., S.R.V., S.J.N., S.M.C., and S.C.)
Acknowledgments
The authors would like to thank Niles Bruce and Bert Jimenez of the Department of Radiology at UCSF for their assistance with data acquisition, and Dr. John Kornak of the Biostatistics Core of the UCSF Clinical and Translational Science Institute for his assistance with the data analysis.
References
- 1.Olson JD, Riedel E, DeAngelis LM. Long-term outcome of low-grade oligodendroglioma and mixed glioma. Neurology. 2000;54(7):1442–1448. doi: 10.1212/wnl.54.7.1442. [DOI] [PubMed] [Google Scholar]
- 2.Shaw EG, Scheithauer BW, O'Fallon JR, Tazelaar HD, Davis DH. Oligodendrogliomas: the Mayo Clinic experience. J Neurosurg. 1992;76(3):428–434. doi: 10.3171/jns.1992.76.3.0428. doi:10.3171/jns.1992.76.3.0428. [DOI] [PubMed] [Google Scholar]
- 3.Fortin D, Macdonald DR, Stitt L, Cairncross JG. PCV for oligodendroglial tumors: in search of prognostic factors for response and survival. Can J Neurol Sc. 2001;28(3):215–223. doi: 10.1017/s0317167100001359. [DOI] [PubMed] [Google Scholar]
- 4.Glass J, Hochberg FH, Gruber ML, Louis DN, Smith D, Rattner B. The treatment of oligodendrogliomas and mixed oligodendroglioma-astrocytomas with PCV chemotherapy. Journal of Neurosurgery. 1992;76(5):741–745. doi: 10.3171/jns.1992.76.5.0741. doi:10.3171/jns.1992.76.5.0741. [DOI] [PubMed] [Google Scholar]
- 5.Kitange GJ, Smith JS, Jenkins RB. Genetic alterations and chemotherapeutic response in human diffuse gliomas. Expert Rev Anticancer Ther. 2001;1(4):595–605. doi: 10.1586/14737140.1.4.595. doi:10.1586/14737140.1.4.595. [DOI] [PubMed] [Google Scholar]
- 6.Mason WP, Krol GS, DeAngelis LM. Low-grade oligodendroglioma responds to chemotherapy. Neurology. 1996;46(1):203–207. doi: 10.1212/wnl.46.1.203. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann C, von Deimling A. Molecular pathology of oligodendroglial tumors. Recent Results Cancer Res. 2009;171:25–49. doi: 10.1007/978-3-540-31206-2_2. doi:10.1007/978-3-540-31206-2_2. [DOI] [PubMed] [Google Scholar]
- 8.Jaeckle KA, Ballman KV, Rao RD, Jenkins RB, Buckner JC. Current strategies in treatment of oligodendroglioma: evolution of molecular signatures of response. J Clin Oncol. 2006;24(8):1246–1252. doi: 10.1200/JCO.2005.04.9874. doi:10.1200/JCO.2005.04.9874. [DOI] [PubMed] [Google Scholar]
- 9.Mukasa A, Ueki K, Ge X, et al. Selective expression of a subset of neuronal genes in oligodendroglioma with chromosome 1p loss. Brain Pathol. 2004;14(1):34–42. doi: 10.1111/j.1750-3639.2004.tb00495.x. doi:10.1111/j.1750-3639.2004.tb00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palfi S, Swanson KR, De Bouard S, et al. Correlation of in vitro infiltration with glioma histological type in organotypic brain slices. Br J Cancer. 2004;91(4):745–752. doi: 10.1038/sj.bjc.6602048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maintz D, Fiedler K, Koopmann J, et al. Molecular genetic evidence for subtypes of oligoastrocytomas. J Neuropathol Exp Neurol. 1997;56(10):1098–1104. doi: 10.1097/00005072-199710000-00003. doi:10.1097/00005072-199710000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol (Berl) 2004;108(1):49–56. doi: 10.1007/s00401-004-0861-z. doi:10.1007/s00401-004-0861-z. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki H, Zlatescu MC, Betensky RA, et al. Histopathological-molecular genetic correlations in referral pathologist-diagnosed low-grade “oligodendroglioma.”. J Neuropathol Exp Neurol. 2002;61(1):58–63. doi: 10.1093/jnen/61.1.58. [DOI] [PubMed] [Google Scholar]
- 14.van den Bent MJ. New perspectives for the diagnosis and treatment of oligodendroglioma. Expert Rev Anticancer Ther. 2001;1(3):348–356. doi: 10.1586/14737140.1.3.348. doi:10.1586/14737140.1.3.348. [DOI] [PubMed] [Google Scholar]
- 15.Le Bihan DJ. Differentiation of benign versus pathologic compression fractures with diffusion-weighted MR imaging: a closer step toward the “holy grail” of tissue characterization? Radiology. 1998;207(2):305–307. doi: 10.1148/radiology.207.2.9577472. [DOI] [PubMed] [Google Scholar]
- 16.Gupta RK, Cloughesy TF, Sinha U, et al. Relationships between choline magnetic resonance spectroscopy, apparent diffusion coefficient and quantitative histopathology in human glioma. J Neurooncol. 2000;50(3):215–226. doi: 10.1023/a:1006431120031. doi:10.1023/A:1006431120031. [DOI] [PubMed] [Google Scholar]
- 17.Kono K, Inoue Y, Nakayama K, et al. The role of diffusion-weighted imaging in patients with brain tumors. AJNR Am J Neuroradiol. 2001;22(6):1081–1088. [PMC free article] [PubMed] [Google Scholar]
- 18.Sugahara T, Korogi Y, Kochi M, et al. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging. 1999;9(1):53–60. doi: 10.1002/(sici)1522-2586(199901)9:1<53::aid-jmri7>3.0.co;2-2. doi:10.1002/(SICI)1522-2586(199901)9:1<53::AID-JMRI7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Khayal IS, McKnight TR, McGue C, et al. Apparent diffusion coefficient and fractional anisotropy of newly diagnosed grade II gliomas. NMR Biomed. 2009;22(4):449–455. doi: 10.1002/nbm.1357. doi:10.1002/nbm.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tozer DJ, Jager HR, Danchaivijitr N, et al. Apparent diffusion coefficient histograms may predict low-grade glioma subtype. NMR Biomed. 2007;20(1):49–57. doi: 10.1002/nbm.1091. doi:10.1002/nbm.1091. [DOI] [PubMed] [Google Scholar]
- 21.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. doi:10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson SJ, Nalbandian AB, Proctor E, Vigneron DB. Registration of images from sequential MR studies of the brain. Journal of Magnetic Resonance Imaging. 1994;4(6):877–883. doi: 10.1002/jmri.1880040621. doi:10.1002/jmri.1880040621. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. doi:10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 24.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B. 1996;111(3):209–219. doi: 10.1006/jmrb.1996.0086. doi:10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 25.Khayal IS, Nelson SJ. Characterization of low-grade gliomas using RGB color maps derived from ADC histograms. J Magn Reson Imaging. 2009;30(1):209–213. doi: 10.1002/jmri.21810. doi:10.1002/jmri.21810. [DOI] [PubMed] [Google Scholar]
- 26.Keles GE, Lamborn KR, Berger MS. Coregistration accuracy and detection of brain shift using intraoperative sononavigation during resection of hemispheric tumors. Neurosurgery. 2003;53(3):556–562. doi: 10.1227/01.neu.0000080949.44837.4c. discussion 562–554 doi:10.1227/01.NEU.0000080949.44837.4C. [DOI] [PubMed] [Google Scholar]
- 27.Jenkinson MD, du Plessis DG, Smith TS, Joyce KA, Warnke PC, Walker C. Histological growth patterns and genotype in oligodendroglial tumours: correlation with MRI features. Brain. 2006;129(Pt 7):1884–1891. doi: 10.1093/brain/awl108. doi:10.1093/brain/awl108. [DOI] [PubMed] [Google Scholar]
- 28.Megyesi JF, Kachur E, Lee DH, et al. Imaging correlates of molecular signatures in oligodendrogliomas. Clin Cancer Res. 2004;10(13):4303–4306. doi: 10.1158/1078-0432.CCR-04-0209. doi:10.1158/1078-0432.CCR-04-0209. [DOI] [PubMed] [Google Scholar]
- 29.McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. Journal of Magnetic Resonance Imaging. 2001;13(2):167–177. doi: 10.1002/1522-2586(200102)13:2<167::aid-jmri1026>3.0.co;2-k. doi:10.1002/1522-2586(200102)13:2<167::AID-JMRI1026>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]