Abstract
Putative cancer stem cells have been identified in glioblastomas and are associated with radio- and chemo-resistance. Further knowledge about these cells is thus highly warranted for the development of better glioblastoma therapies.
Gene expression analyses of 11 high-grade glioma cultures identified 2 subsets, designated type A and type B cultures. The type A cultures displayed high expression of CXCR4, SOX2, EAAT1, and GFAP and low expression of CNP, PDGFRB, CXCL12, and extracellular matrix proteins. Clinical significance of the 2 types was indicated by the expression of type A– and type B–defining genes in different clinical glioblastoma samples. Classification of glioblastomas with type A– and type B–defining genes generated 2 groups of tumors composed predominantly of the classical, neural, and/or proneural subsets and the mesenchymal subset, respectively. Furthermore, tumors with EGFR mutations were enriched in the group of type A samples. Type A cultures possessed a higher capacity to form xenograft tumors and neurospheres and displayed low or no sensitivity to monotreatment with PDGF- and IGF-1–receptor inhibitors but were efficiently growth inhibited by combination treatment with low doses of these 2 inhibitors. Furthermore, siRNA-induced downregulation of SOX2 reduced sphere formation of type A cultures, decreased expression of type A–defining genes, and conferred sensitivity to monotreatment with PDGF- and IGF-1–receptor inhibitors.
The present study thus describes a tumor- and neurosphere-forming SOX2-dependent subset of glioblastoma cultures characterized by a gene expression signature similar to that of the recently described classical, proneural, and/or neural subsets of glioblastoma. The findings that resistance to PDGF- and IGF-1–receptor inhibitors is related to SOX2 expression and can be overcome by combination treatment should be considered in ongoing efforts to develop novel stem cell–targeting therapies.
Keywords: cancer stem cell, glioma, SOX2, subset
According to the cancer stem cell hypothesis, the bulk of tumor cells are derived from a smaller subset of cells that have a series of stem-like phenotypic traits, including self-renewal, skewed cell division, and potential for multilineage differentiation.1,2 During past years, tentative cancer stem cells have been isolated from many types of solid tumors.3–8 However, the molecular characteristics of cancer stem cells are still poorly defined, and it also remains unclear from which cells they originate.
In the case of brain tumors, multiple studies have reported the isolation of brain cancer stem cells from glioblastomas and medulloblastomas.3,4 The tentative glioma stem cells display high capacity for self-renewal, limited differentiation, and a disturbed karyotype and were mainly isolated because of their expression of CD133. It was reported that CD133+ cells implanted into mouse brain can recapitulate the histology of the original tumor, implying that the original tumors were indeed derived from these cells.3,4,9,10 Investigation of CD133 expression in paired, newly diagnosed, and recurrent gliomas also revealed an increase in the CD133+ population in recurrent tumors, suggesting a role for these cells in tumor progression after treatment. However, both CD133+ and CD133- glioma stem cells were later described, and CD133- brain tumor cells also formed tumors in orthotopic transplantation experiments.11–13 Tumorigenic clones of CD133+ and CD133- cancer cells coexist in the same tumor, and capacities for self-renewal and tumor initiation need not be restricted to a uniform, infrequent subpopulation of cells.11,14 A particular dependence of brain tumor stem cells on host factors provided by the special microenvironment of the perivascular space has also been suggested.15
With use of lineage-specific conditional knockout of tumor-suppressor genes in mouse brain, it was recently shown that highly malignant astrocytic brain tumors can originate from the neural stem cells residing in the subventricular zone of the lateral ventricle wall and in the subgranular zone of the dentate gyrus.16–18 Of importance, tumor suppressor gene knockout (p53/Pten/Nf1) was associated with an increased capacity of cells to grow as neurospheres, supporting the significance of this in vitro phenotype as a feature of cancer stem cells.
Brain tumor stem cells have been compared with regard to growth factor dependency and sensitivity to irradiation.19 Other data have suggested unique responses of cancer stem cells to manipulations of certain signaling pathways, including the bone morphogenetic protein (BMP), BMI1 polycomb ring finger oncogene, and hedgehog-GLI pathways; L1 cell adhesion molecule; cMYC; peroxisome proliferator-activated receptor gamma; and Notch.20–27 Autocrine transforming growth factor beta signaling was reported to maintain stemness of glioma-initiating cells via sex determining region Y-box 4 (SOX4) and sex determining region Y-box 2 (SOX2).28
In previous studies, we characterized a panel of glioblastoma cultures with regard to their sensitivity to imatinib and NVP-AEW541. These low-molecular-weight tyrosine kinase inhibitors were selected on the basis of their ability to target platelet-derived growth factor receptors (PDGFRs) and insulin-like growth factor–1 receptor (IGF-1R) signaling, respectively. The target profile and specificities of these compounds have been described elsewhere in detail.29,30 PDGFR signaling has been implied in glioblastoma biology through studies based on analyses of human tumor tissue, of cultured glioblastoma cells, and of mouse glioblastoma models.31–35 Furthermore, imatinib and hydroxyurea have demonstrated promising activity in phase II studies on recurrent glioblastoma.36,37 More recently, a negative phase III study with the same combination was reported in an unselected group of patients with recurrent glioblastoma,38 suggesting the need for combination therapies and improved patient selection. IGF1-R signaling has been described in glioblastoma, and findings from various preclinical studies suggest favorable combination effects when IGF1-R inhibitors have been combined with other receptor tyrosine kinase (RTK)-targeting agents.39–43
In the present study, we further characterized a set of 11 high-grade glioma cultures. This effort has led to the identification of a SOX2-dependent subset of neurosphere- and tumor-forming glioma cells with distinct sensitivity to a combination of tyrosine kinase inhibitors targeting PDGFRs and IGF-1R.
Results
Identification of 2 Gene Expression–Based Subsets of High Grade Glioma Cultures
Previously published gene expression data from a panel of glioblastoma cultures were combined with published44,45 and novel data on the sensitivity of these cells to imatinib and the IGF-1R inhibitor NVP-AEW541 (Supplemental Fig. 1). As shown in Fig. 1A, these analyses revealed an association between one of the gene expression–based clusters and low sensitivity to monotreatment with either imatinib or NVP-AEW541.
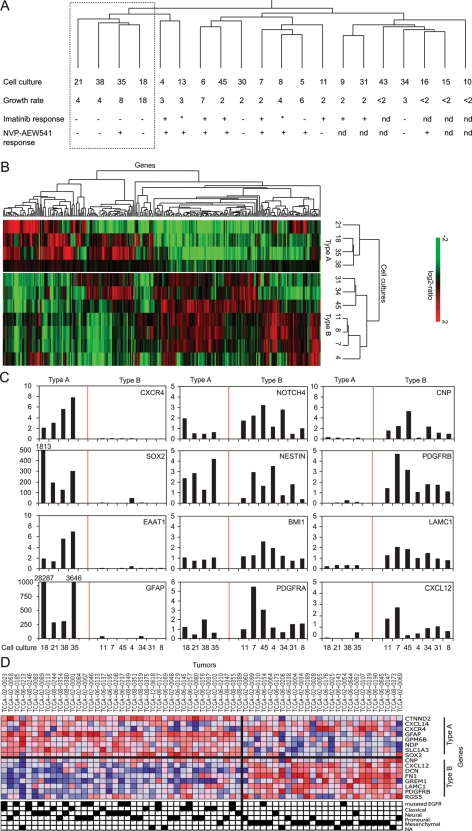
Fig. 1.
Identification of 2 subsets of high-grade glioma cultures, type A and type B, based on gene expression. (A) Unsupervised hierarchical clustering of 20 high-grade glioma cultures, their growth rate as doublings per 4 days, and imatinib and NVP-AEW541 sensitivity. Response of >40% growth inhibition to drug treatment is indicated with +, <20% is indicated as –, and 20%–40% is indicated with *. 1 μM imatinib or 3 μM NVP-AEW541 was used in these experiments. (B) Hierarchical clustering of a subset of 11 cell cultures divides the cultures into 2 distinct groups defined as type A cultures (21, 18, 35, and 38) and type B cultures (31, 34, 45, 11, 8, 7, and 4). (C) Expression levels, as determined by qPCR, of 8 stem/progenitor cell and differentiation markers that display differential expression between type A and type B cultures are shown in the left and right graphs. The middle graph shows the relative expression of 4 genes that did not differ clearly between type A and type B cultures. Shortened bars have their relative expression level stated above them. (D) Supervised analysis of expression data from TCGA glioma samples by type A– and type B–defining genes distinctly divides the samples to match previous data in which the type A group consists of classical, proneural, and/or neural tumors and is enriched with EGFR-mutated samples, and type B consists of gliomas with mesenchymal features (Fisher's exact test, EGFR-mutated, P = .02; mesenchymal subset, P<.001). Color code for expression is red for high expression and blue for low expression.
This group of low-sensitivity cultures (cultures 21, 18, 35, and 38) also formed a cluster in a second hierarchical clustering analysis, based on the expression of 285 differentially expressed genes, in an analysis performed on the 11 cultures that were still available for experimental studies (Fig. 1B). Cluster robustness analysis revealed high robustness with R- and D-index values of 1 and 0, respectively.46 The same 2 groups were formed when clustering was performed on the basis of the expression of 553 or 1339 features filtered by less stringent criteria (data not shown).
For continued analyses, the low-sensitivity group of cells was tentatively designated type A cultures, whereas the other group of cells was referred to as type B cultures (Fig. 1B).
Type A Cultures are GFAP Positive, Whereas Type B Cultures Display Mesenchymal Features
A significance analysis of microarrays (SAM) analysis was performed to investigate differences in gene expression between the type A and type B cultures and identified 34 genes with >3-fold difference in expression between the 2 groups (Supplemental Table 1A). The dominating categories were genes encoding secreted soluble proteins, extracellular matrix proteins, and transcription factors.
The most striking observation was that all 6 differentially expressed genes that code for extracellular matrix proteins showed higher expression in type B cultures (see Supplemental Table 1A for details). Among the genes encoding secreted soluble proteins, 4 were highly expressed in type A cultures (Norris disease pseudoglioma [Norrin], chemokine C-X-C motif ligand 14 [CXCL14], CD70, tumor necrosis factor ligand super family member 7 [TNFSF7], and pleiotrophin [PTN]), whereas 5 gene products were more abundant in type B cultures (gremlin 1 [CKTSF1B1], insulin-like growth factor binding protein 3 [IGFBP3], insulin-like growth factor binding protein 7 [IGFBP7], stroma-derived factor 1 [SDF1 or CXCL12], and wingless-type MMTV integration site family member 5A [WNT5A]). Among genes encoding transcriptional regulators only, SRY-box 9 (SOX9) was more highly expressed in type A, whereas 5 transcriptional regulators (Kruppel-like factor 4 [KLF4], necdin homolog [NDN], forkhead box F2 [FOXF2], pleiomorphic adenoma gene-like 1 [PLAGL1], nuclear receptor subfamily 2 group F member 1 [NR2F1]) were highly expressed in type B cultures. Another striking observation was that expression of glial fibrillary acidic protein (GFAP) was a unique feature of type A cultures.
In summary, type B cultures showed high expression of genes encoding extracellular matrix proteins, suggesting mesenchymal features of this subset. This description was also supported by their high expression of regulator of G protein signaling 5 (RGS5), which has been described as a marker for brain pericytes and perivascular smooth muscle cells.47 Type A cultures, in contrast, were characterized by high expression of GFAP. Of interest, GFAP expression is also a feature of normal subventricular zone neuroepithelial stem cells in mice48 and of early progenitors.49
Gene Set Enrichment Analysis Identifies Candidate Mechanisms Regulating Type A and Type B Expression Characteristics
Gene set enrichment analysis (GSEA) was used to analyze differences in transcription factor profiles between type A and type B cultures. The top 5 transcription factor sets were Friend leukemia virus integration 1 (FLI1), SMAD family member 1 (SMAD1), jun B proto-oncogene (JUNB), nuclear factor I/C (NFIC), and Spi-C transcription factor (SPIC), and their target genes were highly expressed in type B cells. OCT4 (POU5F1) was the seventh significant transcription factor gene set, and these target genes were higher expressed in type A cells (data not shown). A microRNA gene set analysis was also performed. Genes in the mir-486, mir-203, and mir-703 regulated gene sets were highly expressed in type B cells, mir-467 and mir-519 regulated gene sets showed higher expression in type A cells.
In summary, GSEA suggests that genes characterizing type A and type B cells are regulated by different mechanisms, potentially involving transcription factors and microRNAs.
Type A and Type B Cultures Vary with Regard to Expression of Lineage Markers
The differential expression of developmental regulatory genes, such as CKTSF1B1 (BMP-inhibitor gremlin 1), the frizzled ligand NDP (Norrin), WNT5A, SOX9, and KLF4 and of GFAP indicated that the 2 types of glioma cultures possibly represent derivatives of different developmental lineages. Because only a few markers of adult neuroepithelial stem cells and their differentiated progeny were available in the microarray data set, we performed quantitative polymerase chain reaction (qPCR) analyses to test a set of 17 genes, including genes previously implicated as markers for neuroepithelial stem cells and different derivatives of these cells (Supplemental Table 2). Hierarchical clustering based on qPCR data of these genes yielded a subdivision composed of 2 sets of cultures identical with the 2 sets produced by the array data (Supplemental Fig. 2).
Eight of the 17 genes showed an expression pattern that clearly differed between the type A and type B cultures (Fig. 1C, Supplemental Table 3). Chemokine C-X-C receptor 4 (CXCR4), SOX2, excitatory amino acid transporter 1 (EAAT1), and GFAP were more highly expressed in type A cultures, whereas 2′,3′-cyclic nucleotide 3′ phosphodiesterase (CNP), PDGFRB, laminin gamma 1 (LAMC1), and CXCL12 were more highly expressed in type B cultures. Two suggested neuroepithelial stem cell markers, NESTIN and BMI1, did not show any clear difference between type A and type B cultures (Fig. 1C, Supplemental Table 3) and neither did CD133 (data not shown). In contrast, SOX2 was highly expressed in type A cultures. Finally, type B cultures showed higher expression of genes associated with microglial and perivascular cells (LAMC1 and PDGFRB) and a gene associated with the oligodendrocyte lineage (CNP).
Type A and Type B Profiles Identify Molecular Subsets of Glioblastoma
To investigate the clinical significance of the 2 gene expression–defined subsets of cultures, the expression profiles of the type-defining genes were analyzed in human glioblastoma samples. For this purpose, 8 differentially expressed lineage-related genes identified by qPCR (Fig. 1C) and the top 4 upregulated or downregulated genes identified in the SAM analysis (Supplemental Table 1A) were used to analyze 3 publicly available gene expression data sets.50–52 Two of the data sets were analyzed by hierarchical clustering to visualize the expression pattern of the type-defining genes in glioma tissue. Data from the Cancer Genome Atlas (TCGA) were analyzed in a supervised fashion to identify novel correlations to genetic events and gene expression–based subsets of human glioblastomas.52
Hierarchical clustering analysis of a set of 14 high-grade gliomas demonstrated coexpression of type A genes and type B genes.51 In this data set, expression data could only be retrieved for 13 of the 16 type-defining genes. Type A and type B genes were completely segregated between 2 cluster arms, with the exception of CXCR4 and RGS5 (Supplemental Fig. 3A). The same analysis of a larger set of human glioblastoma-derived gene expression data that consisted of 49 glioma samples generated 3 cluster groups of coexpressed genes (Supplemental Fig. 3B). The first branch was composed of only type B genes, and the other 2 contained only type A genes. The tumor samples appeared to be nonrandomly distributed in the 3 different cluster branches with respect to the described proliferative, mesenchymal, and proneural tumor subsets. Branch 1 contained 10 samples of the proliferative and 14 samples of the mesenchymal subset; branch 2 contained 6 proliferative, 5 proneural, and 3 mesenchymal samples; and branch 3 contained 8 samples of the proliferative and 3 samples of the mesenchymal subset. Statistical analyses confirmed significant associations between the cluster branches and the molecular glioblastoma subsets.50
Because the type-defining genes cocluster in tissue samples, they were used for a supervised analysis of expression data from the TCGA data set of glioma samples.52 In this analysis of 138 samples, 39 were determined to be of type A character, 26 of type B, and 73 of neither. These A and B sets of tumor samples were then compared with regard to mutated or amplified genes and distribution of the 4 gene expression–based glioblastoma subsets: classical, neural, proneural, and mesenchymal.53–55 The type A group was found to be enriched with samples carrying mutated and/or amplified epidermal growth factor receptor (EGFR) (Fig. 1D and not shown) and the samples were a mixture of the classical, neural, and/or proneural subsets, whereas the type B group was strongly enriched with samples of the mesenchymal subset (Fig. 1D). Neither the type A nor the type B subset showed any significant enrichment of tumors with isocitrate dehydrogenase 1 (IDH1) mutations or the methylator phenotype; however, 2 of 3 samples with SOX2 gene amplification were in the type A group, and the third was among the nonseparated tumor samples.
Together, these results demonstrate that the selected type A– and type B–defining genes are expressed in different sets of human tumors rather than coexpressed in the same tumors. The analyses also demonstrate that the type A– and type B–defining genes, when used in unsupervised clustering analyses or as classifier genes, divide tumor sets into different clusters that display a nonrandom content of previously described molecular glioblastoma subsets.50,53
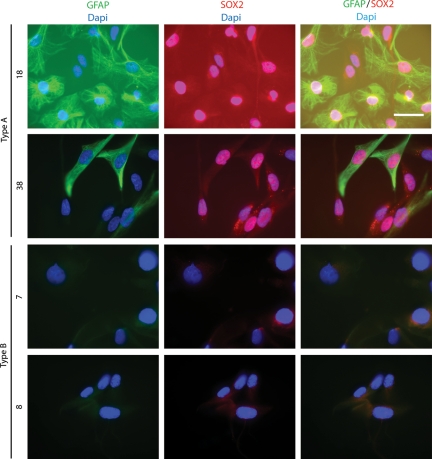
Immunofluorescence Staining Demonstrates Expression of GFAP and SOX2 in Type A Cells
The qPCR analyses suggested that high GFAP and SOX2 expression were features of type A cultures (Fig. 1C). To further explore this finding, immunofluorescence staining of GFAP and SOX2 was performed on 2 type A and 2 type B cultures.
GFAP+ cells were found only in type A cultures. It was also noted that GFAP was restricted to 10%–80% of cells in early passages and to 1%–5% of cells in later passages (data not shown). By immunofluorescence staining, it was observed that nuclear SOX2 protein was present in both GFAP+ and GFAP- cells of type A cultures (Fig. 2). SOX2 was not detected in type B cultures (Fig. 2).
Fig. 2.
Immunofluorescence staining demonstrates that the presence of GFAP+ and SOX2+ cells is restricted to type A cultures. Immunofluorescence staining of GFAP and SOX2 was performed on type A cultures 18 and 38 and type B cultures 7 and 8. Presence of GFAP and SOX2 was restricted to type A cultures 18 and 38. Double immunofluorescence analysis of SOX2 and GFAP in cells of cultures 18 and 38 demonstrated that SOX2 is present in a majority of the cells, whereas GFAP expression varies. GFAP staining is colored green, SOX2 red, and nuclear DAPI blue. Scale bar is 50 μm.
This experiment thus confirmed that type A and type B cultures differ with regard to GFAP and SOX2 expression. The restricted expression of GFAP to a subset of cells is compatible with these cells dividing asymmetrically to generate progeny that retain or lose GFAP expression. However, the presence of SOX2 in most cells of the type A cultures indicates a tissue stem and/or progenitor cell phenotype of a majority of these cells.
Type A Cultures Display Increased Ability to Form Tumors in vivo and Neurospheres in vitro
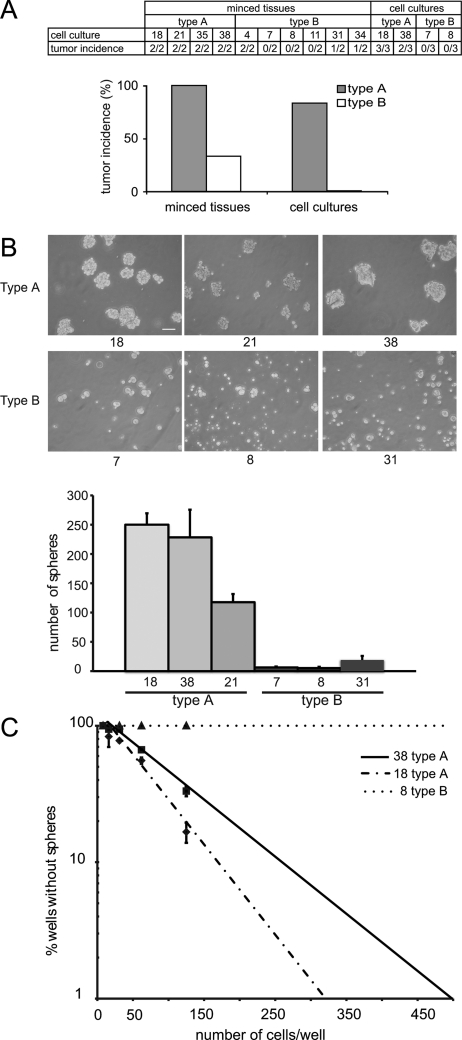
Pieces of the original tumor tissue were used for in vivo subcutaneous tumor growth in severe combined immune deficiency (SCID) mice. All 8 tissue preparations of tumors from which type A cultures were derived formed tumors in vivo (Fig. 3A). In contrast, tumor formation was only observed with 4 of the 12 tissue preparations of tumors representing the type B cultures (Fig. 3A).
Fig. 3.
Type A cultures show higher tumorigenic potential and higher ability to form spheres and to self-renew. (A) Tumor formation in SCID mice after subcutaneous injection of original minced tumor tissues and of glioma cultures, representing type A and type B subsets. Results are given as number of mice with visible tumor per number of injected mice. (B) The ability to form tumor spheres after 8 days of growth in neurosphere medium was tested for type A cultures 18, 21, and 38 and type B cultures 7, 8, and 31. Type A cultures, but not type B cultures, formed a high number of tumor spheres. Scale bar is 100 μm. (C) Limiting dilution assays after plating of cells at limiting dilution in 200 μL media showed a measurable fraction of sphere-forming cells in type A cultures 18 and 38, but not in the type B culture 8.
Furthermore, cultures 18 and 38 (type A) and cultures 7 and 8 (type B) were used for subcutaneous xenograft assays in SCID mice. Both type A cultures resulted in tumors, but not the type B cultures (Fig. 3A).
As an additional method to examine cancer stem cell properties of the cultures, neurosphere assays were performed. Type A and type B cultures displayed different abilities to form spheres (Fig. 3B and C). Type A cultures generally formed a higher number of spheres, and these were also larger than the spheres formed by type B cultures (Fig. 3B). Similar results were observed when analyzing the ability to form serial spheres (data not shown).
To further address the self-renewal capacity of the glioma cultures, limiting dilution assays were applied to 2 cultures of type A and 1 culture of type B. The fraction of sphere-forming cells was calculated to be 1.2% and 0.8% in cultures 18 and 38, respectively (Fig. 3C). In this assay format, the fraction of sphere-forming cells in the type B culture 8 was below the detection level (Fig. 3C). Thus, the type A and type B cultures differed with regard to tumor- and sphere-forming capacities, which are more pronounced in the type A cultures.
The Sphere-forming Phenotype of Type A Cultures is SOX2 Dependent
One of the molecular features that most clearly discriminated between type A and type B cultures was the expression of SOX2, which is known as a progenitor cell transcription factor capable of contributing stem cell characteristics to somatic cells.56,57 We therefore tested whether the cancer stem cell features of type A cultures were dependent on SOX2.
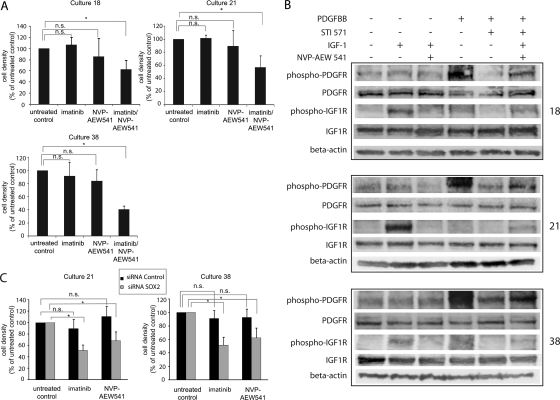
The differential expression of SOX2 between type A and type B cultures was confirmed by immunoblotting (Fig. 4A and data not shown). siRNA-mediated SOX2 downregulation in cultures 18 and 21, using a pool of 4 siRNAs, resulted in a decreased growth rate of the cultures, which was most evident for culture 18 (Fig. 4A and B). Moreover, the expression of type A–defining genes (CXCR4, EAAT1, and GFAP) was decreased in both cultures (Fig. 4C). Of importance, SOX2 siRNA–treated cells also displayed significantly reduced neurosphere formation capacity, with fewer and smaller spheres, compared with control siRNA-treated cells (Fig. 4D). Similar results were obtained with an independent SOX2 siRNA from another provider (Supplemental Fig. 5).
Fig. 4.
siRNA-mediated downregulation of SOX2 in type A cultures. (A) Immunoblotting analyses of SOX2 in cells transfected with control or SOX2 siRNA. (B) Type A cultures 18 and 21 have lower growth rate after siRNA-mediated downregulation of SOX2. Cell density was determined after 72 h. Results are derived from 2 experiments with 10 replicates per condition. Error bars indicate standard deviation. (C) siRNA-mediated SOX2 downregulation in type A cultures 18 and 21 leads to lower expression levels of type A genes measured by qPCR. (D) SOX2 siRNA treatment reduces tumor sphere formation. The effect is illustrated by phase contrast microscopy (10 x magnification) and was quantified by counting the number of spheres ≥50 μm/well. All neurosphere formation assays were repeated 4 times. The mean number of spheres in SOX2 siRNA–treated culture 18 was 102, compared with 243 in control siRNA-treated cultures (P≤.05). Corresponding numbers for culture 21 were 89 and 150 (P≤.05). Scale bar is 200 μm.
From these data, we conclude that the neurosphere-forming ability and the defining gene expression signature of type A cultures are dependent on SOX2.
Type A Cultures are Highly Susceptible to Combination Treatment with Imatinib and NVP-AEW541
One possible explanation for the reduced sensitivity of type A cultures to monotreatment with the 2 tyrosine-kinase inhibitors (Fig. 1A) could be a higher extent of redundancy in growth factor signaling in type A cultures. It was therefore analyzed whether the growth-inhibitory response could be enhanced by combination treatment with the 2 drugs. For these analyses, 3 of the type A cultures—18, 21, and 38—were selected.
In agreement with previous studies, all 3 cultures showed no or low response to treatment with 0.25 μM of imatinib or 0.1 μM of NVP-AEW541 (Fig. 5A). Of interest, all 3 cultures displayed a significant growth inhibition when treated with a combination of 0.25 μM of imatinib and 0.1 μM of NVP-AEW541 (Fig. 5A). Ability of the drugs to block PDGF and IGF-1receptors at these concentrations was confirmed in an independent experiment monitoring receptor phosphorylation after combination treatment with 0.25 μM of imatinib and 0.1 μM of NVP-AEW541 (Fig. 5B). No evidence for synergistic effects of the 2 drugs was detected when the combination treatment was applied to 2 different type B cultures (data not shown).
Fig. 5.
Type A cultures are growth inhibited by combination treatment with imatinib and NVP-AEW541 and display sensitivity to monotreatment after SOX2 downregulation. (A) Type A cultures 18, 21, and 38 were untreated (control) or treated with 0.25 μM imatinib, 0.1 μM NVP-AEW541, or the combination of 2 drugs at the same concentrations. Drug sensitivity is expressed as cell density (percentage of untreated control). All 3 cultures demonstrated strong growth-inhibitory response after combination treatment. Asterisks (*) represent P< .001, and “n.s.” represents nonsignificant P values. Effects on cell growth were determined after 4 days of drug exposure. Media and drug was changed after 2 days of culture. The data shown are derived from 3 independent experiments: 1 performed with duplicate and 2 with quadruplicate measurements. (B) Immunoblotting analyses of PDGFR and IGF-1R tyrosine phosphorylation after monotreatment with 1 μM imatinib or NVP-AEW541 or combination treatment with 0.25 μM imatinib and 0.1 μM NVP-AEW541. A total of 100 ng/mL PDGF-BB or 50 ng/mL IGF-1 was used to stimulate the cells for 10 min at 37°C. (C) Cell cultures 21 and 38 were transfected with either SOX2 siRNA or control siRNA. Drug treatment with either 1 μM imatinib or 1 μM NVP-AEW541 was started 24 h after transfection and was maintained for 72 h. Cultures with SOX2 downregulation demonstrated significant growth reduction after treatment with either drug. Results are derived from 2 experiments. Error bars indicate standard deviation. Asterisks (*) represent P≤.01, and “n.s.” represents nonsignificant.
Together, these results demonstrated that type A cultures display a strong sensitivity to combination treatment with low doses of imatinib and NVP-AEW541.
SOX2 Downregulation Increases the Sensitivity of Type A Cultures to Imatinib and NVP-AEW541
To investigate whether SOX2 had any influence on sensitivity to monotreatment with either of the 2 drugs in the low-sensitivity type A cultures, SOX2 down-regulation experiments were performed. For these experiments, type A cell culture 18 was excluded because of the significant growth inhibition observed after SOX2 downregulation (Fig. 4B). SOX2 downregulation in cultures 21 and 38 was confirmed by SOX2 immunoblotting (Supplemental Fig. 4).
In agreement with previous analyses, the control cells displayed lack of significant growth reduction after treatment with 1 μM of imatinib or NVP-AEW541 (Fig. 5C). Of interest, the SOX2 downregulation conferred a significant imatinib and NVP-AEW541 sensitivity to both cultures (Fig. 5C).
Together, these findings identify a previously unrecognized impact of SOX2 on sensitivity to PDGFR and IGF-1R targeting. The mechanism behind this is not known. On the basis of preliminary findings, we cannot exclude an effect on tyrosine kinase receptor levels and/or activities. qPCR analyses of PDGFRs and IGF-1R after SOX2 siRNA treatment showed that IGF-1R mRNA levels were significantly downregulated in cultures 18 and 21 but only marginally affected in culture 38 (data not shown). Effects on PDGFR mRNA levels were less distinct.
Discussion
The present study defines 2 novel phenotypically distinct subsets of high-grade glioma cell cultures. Both types of cultures have certain characteristics of neural stem cells, because they both express the established stem cell–related proteins NESTIN and BMI1. The characteristics describing the 2 subsets are summarized in Table 1. The features defining type A cultures include higher expression of GFAP, SOX2, SOX9, EAAT1, and CXCR4; tumorigenic potential in vivo; a prominent ability for neurosphere formation in vitro; and reduced sensitivity to tyrosine kinase inhibitors targeting PDGF- or IGF-1–receptors. Moreover, siRNA treatment showed that the phenotype of type A cultures is dependent on SOX2. Type B cultures are characterized by high expression of genes encoding extracellular matrix proteins and the mesenchymal marker RGS5 and genes associated with microglia (PDGFRB and LAMC1) and oligodendrocytes (CNP). Clinical significance of these 2 types of cultures is indicated by the findings that type A genes and type B genes display coordinated expression and are present in different tumor samples when analyzed in 2 independent sets of human glioblastoma tumors; furthermore, when used as classifier genes, they divide a set of tumors into groups enriched in samples of different molecular subtypes.
Table 1.
Characteristics defining type A and type B high grade glioma cultures
| Variable | Type A cultures | Type B cultures |
|---|---|---|
| expression of neural stem cell markers, NESTIN and BMI1 | high | high |
| expression of GFAP, SOX2 and SOX9 | high | low |
| expression of mesenchymal markers | low | high |
| tumorigenic potential | high | low |
| neurosphere growth and self-renewal capacity | high | low |
| SOX2-dependent stem cell phenotype | yes | n.a. |
| sensitivity to mono-treatment with imatinib gene, or NVP-AEW541 | low | high |
| sensitivity to combination-treatment with imatinib and NVP-AEW541 | high | n.a. |
| SOX2-dependent resistance to mono-treatment with imatinib or NVP-AEW541 | yes | n.a. |
| TCGA related tumors with EGFR gene amplification and/or mutation | yes | no |
| TCGA related tumors with classical/pro-neural/neural gene expression signature | yes | no |
| TCGA related tumors with mesenchymal gene expression signature | no | yes |
n. a., not applicable.
The overall profiles of type A and type B cultures, with high expression of genes coding for mesenchymal proteins as one characteristic of type B cultures, is indicative of relationships to recently described subsets, including the classical, neural, proneural, and mesenchymal glioblastoma subsets identified on the basis of gene expression analyses of clinical samples in the TCGA project.50,53,58 A supervised analysis of the TCGA data set, based on the type-defining genes, separated the tumor samples into a mixed group of the classical, neural, and/or proneural subsets and another group of the mesenchymal subset. The type A tumor group was also enriched in EGFR gene–mutated and/or–amplified samples. High SOX2 expression is known to characterize the proneural subset, and in the supervised analysis of TCGA samples, we found 2 of 3 SOX2-amplified tumor samples among the type A tumors and the third among the unseparated samples. In an initial unsupervised hierarchical clustering analysis using the TCGA data set, the type A genes were split into 2 cluster branches, whereas the type B genes were kept together in 1 branch. The type A cluster contained all 8 IDH1 gene-mutated and the methylator samples, known to also be included in the proneural subset (not shown). There was, however, a selection against these IDH1-mutant and/or methylator samples in the supervised analysis because neither the A nor the B type group contained any of these samples, although they contained other samples of the proneural subset (Fig. 1D). Thus, large fractions of the classical, proneural, and/or neural glioblastoma subsets, including samples with SOX2 or EGFR gene amplifications, but not the IDH1 gene-mutated and/or methylator samples, were identified by this classifier.
Several of the suggested microRNAs and transcription factor gene sets distinguishing type A and type B cultures (Supplemental Table 1B) have been implicated in glioma biology. Mir-425, mir-451, and mir-486 have been reported to be highly expressed in glioma cells. Introduction of these into glioma cells inhibited neurosphere formation. Of interest, mir-451 was shown to have a cooperative effect with imatinib.59 OCT4 (POU5F1) is here shown to be associated with type A cultures, similar to some of its target genes, and is known to affect colony formation capacity and proliferation of glioma cells.60 In addition, OCT4 has the potential to reprogram fibroblasts into pluripotent stem cells in combination with SOX2, Kruppel-like factor 4 (KLF4), and c-MYC.56 These data suggest highly relevant mechanisms that also could be involved in generating the differences in phenotypes observed between type A and type B cultures.
Among the 11 cultures of the present study, 4 type A cultures and 6 type B cultures have previously been characterized with regard to phosphatase and tensin homolog (PTEN) protein expression, phosphoinositide-3-kinase (PI3K) mutations, and serum sensitivity of serine/threonine protein kinase Akt (Akt) phosphorylation.44 Aberrations of this pathway were found in both type A and type B cultures, suggesting heterogeneity with regard to mutational activation of growth factor pathways in the 2 subsets.
The contrasting characteristics between type A and type B cultures and/or tumors are relevant in the context of other recent studies on brain cancer stem cells. The GFAP expression and SOX2 dependency of type A cultures suggest a relationship to the cancer-forming cells identified in studies in which brain tumors were induced in mice by inactivation of tumor suppressors in neural stem cells.16–18 On the other hand, the expression of PDGFRB and RGS5 in type B cultures suggests similarities to the CD133+/NESTIN+ glioblastoma cells that were identified in perivascular niches.15 Furthermore, EGFR expression defined a set of glioblastoma tumor–initiating cells.61 Because several types of tentative glioblastoma stem cells have been described in the literature, a potential topic for future studies will be to explore the possibility that type A and type B cells coexist in tumors and constitute cell types with complementary functions. This type of tumor-promoting intratumoral heterogeneity of malignant cells is attracting increasing interest, and experimental support for this notion was recently provided in a study on glioma62 and in a genetic model of small cell lung cancer.63 However, the fact that the type A– and type B–defining gene sets also define distinct subsets of tumors in vivo favors the idea that the 2 stem-like cancer cell populations described here are present in and determine the phenotype of different sets of tumors rather than occur mixed in the same tumor. Furthermore, in a recent analysis of human glioma samples, we found that SOX2, together with pluripotency factors OCT4 and Nanog, was quite homogenously expressed in glioblastoma tumors.64
Studies of cancer stem cells are spurred by expectations to improve therapy. In this context, it is noteworthy that the type A cultures identified in this study displayed a consistent high sensitivity to combination treatment with inhibitors of PDGF and IGF-1 receptors, whereas monotreatment was not efficient. Coactivation of multiple tyrosine kinases is a well-known phenomenon in glioma.65,66 Clinical trials with imatinib for high-grade gliomas have, in general, not yielded major positive results.36,67,68 The present work should prompt studies on whether the small fraction of responsive tumors have characteristics of type A or type B cultures. The finding that resistance among type A cultures to PDGF- and IGF-I–receptor inhibitors can be overcome by combination treatment suggests that this combination is highly interesting for ongoing efforts to develop novel stem cell–targeting therapies. Of note, type A cultures displayed high expression of the chemokine receptor CXCR4. Inhibitors of CXCR4 have already demonstrated growth inhibitory effects in various glioma model systems69 and have effects on proliferation of glioblastoma progenitor cells.70 The finding that the type A phenotype depends on high levels of SOX2 suggests that SOX2 is a potential target for treatment. Such initiatives are also spurred by the very recent identification of SOX2 as an important oncogene in lung and esophageal squamous cell carcinoma.71 SOX2 is universally expressed in neuroglial tumors72 and together with other pluripotency genes in glioblastoma.64 It was indicated that silencing of SOX2 in glioblastoma may affect proliferation and tumorigenicity.73
Material and Methods
Establishment of Cell Cultures and Analysis of Sensitivity to Receptor Tyrosine Kinase Inhibitors
The establishment of the 20 high-grade glioma cultures used here and analyses of their sensitivity to imatinib and NVP-AEW541 has been described elsewhere.44,45 Immunoblotting analyses to monitor effects of drugs on receptor tyrosine phosphorylation were done as described elsewhere.44,45
Generation of Gene Expression Data
Experiments followed standard and published procedures (see Supplemental Material). qPCR analyses were performed on cDNA isolated from the glioblastoma cultures and analyzed with an ABI Prism 7500 (Applied Biosystems). Expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. Microarray analyses relied on previously published gene expression data,45 which has been deposited at EMBL-EBI Array Express with accession number E-MEXP-1063. The passage number of the glioma cell cultures used for the gene expression analysis ranged from 5 to 10. Cluster analysis of glioblastoma tumor samples used publicly available datasets50,51 (data is accessible at http://www-genome.wi.mit.edu/cancer/pub/glioma and through GEO at accession number GDS1816 and at the TCGA data portal).
Immunofluorescence Staining
Primary antibodies used were mouse monoclonal anti-GFAP (Sigma) and rabbit anti-SOX2 polyclonal antibody (Chemicon ab5603). Bound antibodies were visualized by fluorophore-conjugated secondary antibodies (Dako, GE Healthcare, Abcam). Cells were analyzed using a Zeiss Axioplan 2 microscope, and images were collected with AxioCam HRm Camera and Axiovision software, version 4.2 (further details in Supplemental Material).
Glioma Cell Neurosphere Assay
Glioma cells were plated in nonadherent 6-well plates (NUNC) at ∼3000 cells/cm2 in neurosphere medium (#05751 Neurocult NA-S proliferation human kit; StemCell Technologies), supplemented with 20 ng/mL EGF (Invitrogen), 20 ng/mL bFGF (Invitrogen), and 2 μg/mL heparin (Sigma). Growth factors were resupplemented every second day. Images were taken after 8 days, and spheres ≥50 μm in diameter were counted. For analyses of serial sphere formation, spheres were split with TrypLE Express (Invitrogen) when they reached >100 μm in size, after which cells were resuspended and reseeded.
Limiting Dilution Assay
Limiting dilution assay was performed as described elsewhere.3,74 Cells were split and seeded in 96-well plates with dilutions ranging from 1000 cells/well to 1 cell/well in 200 μL NeuroCult media (StemCell Technologies) supplemented as described above. Supplements were added to cultures every second day. After 7 days, the wells without spheres were counted and plotted against the number of cells added per well. The number of cells needed to form one sphere, which represented the proportion of stem cells in the entire culture population, was determined from the point at which the line crossed at the 0.37 level.74
Test of in vivo Tumorigenic Potential
In vivo tumorigenic potential of the primary tumor material was analyzed as described in Supplemental Material. Time to the development of visible tumor was registered, as was time when tumor size reached 100 mm3, 500 mm3, and 1 cm3.
For xenograft assays of selected type A and type B cultures, 500 000 cells were injected subcutaneously into the low flank of SCID mice. Three mice were used for each cell culture. Tumor size was measured and monitored daily until reaching a volume of ∼500 mm3. Then, animals were killed, and tumors were collected (further details in Supplemental Material).
Detection of SOX2 by Immunoblotting
SOX2 protein levels were determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblotting of cell lysates. Immunoblotting signals were quantified using the AIDA software, version 3.10.039 (Raytest). See Supplemental Material for further details.
Analyses of Neurosphere Formation, Growth Rate, and Gene Expression After siRNA-Mediated Downregulation of SOX2
siRNA-mediated downregulation of SOX2 was done in accordance with standard procedures (see Supplemental Material).
Drug Treatment with Imatinib and NVP-AEW541 of Cells Pretreated or not with SOX2 siRNA
Procedures for preparation of drug stock solutions, siRNA-mediated downregulation, and immunoblotting of SOX2 are described in detail in Supplemental Material.
For analyses of effects on drug sensitivity, cells were seeded at a density of 5000 cells per well in 96-well plates, and were transfected with either SOX2 siRNA or control siRNA on the next day. After 24 h, the cells were retransfected and treated with 1 μM of imatinib or 1 μM NVP-AEW541 for 72 h.
Supplementary Material
Conflict of interest statement. Monica Nistér and Arne Östman have received research grants from Novartis. Arne Östman and Daniel Hägerstrand have received honoraria for consulting services for Novartis. The other authors have no conflicts of interest to report.
Funding
This work was supported by the Swedish Cancer Society (Cancerfonden), the Swedish Childhood Cancer Foundation (Barncancerfonden), the Swedish Research Council (Vetenskapsrådet), the Stockholm Cancer Society (Cancerföreningen i Stockholm), Karolinska Institutet, and Novartis (to M.N. and A.Ö.).
Supplementary Material
Acknowledgments
M.N. and A.Ö. laboratories members are acknowledged for critical discussions throughout the project. We thank Dr. Anna Eriksson-Hedrén for valuable discussions and technical support and Mrs Marianne Kastemar for initial help with cell cultures. Francesco Hofman, Carlos Garcia-Echeverria, and Elisabeth Buchdunger are acknowledged for initial discussions and for providing drugs. We would also like to express our gratitude to the TCGA, Heidi Phillips, Ken Aldape, Catherine Nutt, and Todd Golub for making gene expression data publicly available. Ethical approvals had been obtained for animal experiments (C 207/1) and for use of human samples (UpS98415).
References
- 1.Polyak K, Hahn WC. Roots and stems: stem cells in cancer. Nat Med. 2006;12:296–300. doi: 10.1038/nm1379. doi:10.1038/nm1379. [DOI] [PubMed] [Google Scholar]
- 2.Tan BT, Park CY, Ailles LE, Weissman IL. The cancer stem cell hypothesis: a work in progress. Lab Invest. 2006;86:1203–1207. doi: 10.1038/labinvest.3700488. doi:10.1038/labinvest.3700488. [DOI] [PubMed] [Google Scholar]
- 3.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 4.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. doi:10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 5.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. doi:10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. doi:10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. doi:10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. doi:10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 9.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. doi:10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 10.Tunici P, Bissola L, Lualdi E, et al. Genetic alterations and in vivo tumorigenicity of neurospheres derived from an adult glioblastoma. Mol Cancer. 2004;3:25. doi: 10.1186/1476-4598-3-25. doi:10.1186/1476-4598-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. doi:10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 12.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. doi:10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 13.Sakariassen PO, Prestegarden L, Wang J, et al. Angiogenesis-independent tumor growth mediated by stem-like cancer cells. Proc Natl Acad Sci USA. 2006;103:16466–16471. doi: 10.1073/pnas.0607668103. doi:10.1073/pnas.0607668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X, Shen G, Yang X, Liu W. Most C6 cells are cancer stem cells: evidence from clonal and population analyses. Cancer Res. 2007;67:3691–3697. doi: 10.1158/0008-5472.CAN-06-3912. doi:10.1158/0008-5472.CAN-06-3912. [DOI] [PubMed] [Google Scholar]
- 15.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. doi:10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Alcantara Llaguno S, Chen J, Kwon CH, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. doi:10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng H, Ying H, Yan H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. doi:10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Yang J, Zheng H, et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. doi:10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. doi:10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 20.Seidel S, Garvalov BK, Wirta V, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133:983–995. doi: 10.1093/brain/awq042. doi:10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 21.Piccirillo SG, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. doi:10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 22.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. doi:10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan X, Matsui W, Khaki L, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66:7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. doi:10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 24.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. doi:10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 25.Bao S, Wu Q, Li Z, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. doi:10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Wang H, Li Z, et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS ONE. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. doi:10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chearwae W, Bright JJ. PPARgamma agonists inhibit growth and expansion of CD133+ brain tumour stem cells. Br J Cancer. 2008;99:2044–2053. doi: 10.1038/sj.bjc.6604786. doi:10.1038/sj.bjc.6604786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. doi:10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Buchdunger E, Zimmermann J, Mett H, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–104. [PubMed] [Google Scholar]
- 30.Garcia-Echeverria C, Pearson MA, Marti A, et al. In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell. 2004;5:231–239. doi: 10.1016/s1535-6108(04)00051-0. doi:10.1016/S1535-6108(04)00051-0. [DOI] [PubMed] [Google Scholar]
- 31.Hermanson M, Funa K, Hartman M, et al. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52:3213–3219. [PubMed] [Google Scholar]
- 32.Shawver LK, Schwartz DP, Mann E, et al. Inhibition of platelet-derived growth factor-mediated signal transduction and tumor growth by N-[4-(trifluoromethyl)-phenyl]5-methylisoxazole-4-carboxamide. Clin Cancer Res. 1997;3:1167–1177. [PubMed] [Google Scholar]
- 33.Kilic T, Alberta JA, Zdunek PR, et al. Intracranial inhibition of platelet-derived growth factor-mediated glioblastoma cell growth by an orally active kinase inhibitor of the 2-phenylaminopyrimidine class. Cancer Res. 2000;60:5143–5150. [PubMed] [Google Scholar]
- 34.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. doi:10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhrbom L, Hesselager G, Nister M, Westermark B. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res. 1998;58:5275–5279. [PubMed] [Google Scholar]
- 36.Reardon DA, Egorin MJ, Quinn JA, et al. Phase II study of imatinib mesylate plus hydroxyurea in adults with recurrent glioblastoma multiforme. J Clin Oncol. 2005;23:9359–9368. doi: 10.1200/JCO.2005.03.2185. doi:10.1200/JCO.2005.03.2185. [DOI] [PubMed] [Google Scholar]
- 37.Reardon DA, Dresemann G, Taillibert S, et al. Multicentre phase II studies evaluating imatinib plus hydroxyurea in patients with progressive glioblastoma. Br J Cancer. 2009;101:1995–2004. doi: 10.1038/sj.bjc.6605411. doi:10.1038/sj.bjc.6605411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dresemann G, Weller M, Rosenthal MA, et al. Imatinib in combination with hydroxyurea versus hydroxyurea alone as oral therapy in patients with progressive pretreated glioblastoma resistant to standard dose temozolomide. J Neurooncol. 2010;96:393–402. doi: 10.1007/s11060-009-9976-3. doi:10.1007/s11060-009-9976-3. [DOI] [PubMed] [Google Scholar]
- 39.Esparis-Ogando A, Ocana A, Rodriguez-Barrueco R, Ferreira L, Borges J, Pandiella A. Synergic antitumoral effect of an IGF-IR inhibitor and trastuzumab on HER2-overexpressing breast cancer cells. Ann Oncol. 2008;19:1860–1869. doi: 10.1093/annonc/mdn406. doi:10.1093/annonc/mdn406. [DOI] [PubMed] [Google Scholar]
- 40.Cunningham MP, Thomas H, Marks C, Green M, Fan Z, Modjtahedi H. Co-targeting the EGFR and IGF-IR with anti-EGFR monoclonal antibody ICR62 and the IGF-IR tyrosine kinase inhibitor NVP-AEW541 in colorectal cancer cells. Int J Oncol. 2008;33:1107–1113. [PubMed] [Google Scholar]
- 41.Desbois-Mouthon C, Baron A, Blivet-Van Eggelpoel MJ, et al. Insulin-like growth factor-1 receptor inhibition induces a resistance mechanism via the epidermal growth factor receptor/HER3/AKT signaling pathway: rational basis for cotargeting insulin-like growth factor-1 receptor and epidermal growth factor receptor in hepatocellular carcinoma. Clin Cancer Res. 2009;15:5445–5456. doi: 10.1158/1078-0432.CCR-08-2980. doi:10.1158/1078-0432.CCR-08-2980. [DOI] [PubMed] [Google Scholar]
- 42.Chakraborty AK, Liang K, DiGiovanna MP. Co-targeting insulin-like growth factor I receptor and HER2: dramatic effects of HER2 inhibitors on nonoverexpressing breast cancer. Cancer Res. 2008;68:1538–1545. doi: 10.1158/0008-5472.CAN-07-5935. doi:10.1158/0008-5472.CAN-07-5935. [DOI] [PubMed] [Google Scholar]
- 43.Warshamana-Greene GS, Litz J, Buchdunger E, Hofmann F, Garcia-Echeverria C, Krystal GW. The insulin-like growth factor-I (IGF-I) receptor kinase inhibitor NVP-ADW742, in combination with STI571, delineates a spectrum of dependence of small cell lung cancer on IGF-I and stem cell factor signaling. Mol Cancer Ther. 2004;3:527–535. [PubMed] [Google Scholar]
- 44.Hagerstrand D, Lindh MB, Pena C, et al. PI3K/PTEN/Akt pathway status affects the sensitivity of high-grade glioma cell cultures to the insulin-like growth factor-1 receptor inhibitor NVP-AEW541. Neuro Oncol. 2010;12:967–975. doi: 10.1093/neuonc/noq029. doi:10.1093/neuonc/noq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagerstrand D, Hesselager G, Achterberg S, et al. Characterization of an imatinib-sensitive subset of high-grade human glioma cultures. Oncogene. 2006;25:4913–4922. doi: 10.1038/sj.onc.1209497. doi:10.1038/sj.onc.1209497. [DOI] [PubMed] [Google Scholar]
- 46.McShane LM, Radmacher MD, Freidlin B, Yu R, Li MC, Simon R. Methods for assessing reproducibility of clustering patterns observed in analyses of microarray data. Bioinformatics. 2002;18:1462–1469. doi: 10.1093/bioinformatics/18.11.1462. doi:10.1093/bioinformatics/18.11.1462. [DOI] [PubMed] [Google Scholar]
- 47.Cho H, Kozasa T, Bondjers C, Betsholtz C, Kehrl JH. Pericyte-specific expression of Rgs5: implications for PDGF and EDG receptor signaling during vascular maturation. Faseb J. 2003;17:440–442. doi: 10.1096/fj.02-0340fje. [DOI] [PubMed] [Google Scholar]
- 48.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. doi:10.1016/S0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 49.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 50.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. doi:10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 51.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 52.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. doi:10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. doi:10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. doi:10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. doi:10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 57.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. doi:10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gunther HS, Schmidt NO, Phillips HS, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–2909. doi: 10.1038/sj.onc.1210949. doi:10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 59.Gal H, Pandi G, Kanner AA, et al. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem Biophys Res Commun. 2008;376:86–90. doi: 10.1016/j.bbrc.2008.08.107. doi:10.1016/j.bbrc.2008.08.107. [DOI] [PubMed] [Google Scholar]
- 60.Du Z, Jia D, Liu S, et al. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009;57:724–733. doi: 10.1002/glia.20800. doi:10.1002/glia.20800. [DOI] [PubMed] [Google Scholar]
- 61.Mazzoleni S, Politi LS, Pala M, et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. doi:10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- 62.Inda MM, Bonavia R, Mukasa A, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–1745. doi: 10.1101/gad.1890510. doi:10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calbo J, van Montfort E, Proost N, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell. 2011;19:244–256. doi: 10.1016/j.ccr.2010.12.021. doi:10.1016/j.ccr.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 64.Holmberg J, He X, Peredo I, et al. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS ONE. 2011;6:e18454. doi: 10.1371/journal.pone.0018454. doi:10.1371/journal.pone.0018454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. doi:10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 66.Huang PH, Mukasa A, Bonavia R, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. doi:10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wen PY, Yung WK, Lamborn KR, et al. Phase I/II study of imatinib mesylate for recurrent malignant gliomas: North American Brain Tumor Consortium Study 99–08. Clin Cancer Res. 2006;12:4899–4907. doi: 10.1158/1078-0432.CCR-06-0773. doi:10.1158/1078-0432.CCR-06-0773. [DOI] [PubMed] [Google Scholar]
- 68.Dresemann G. Imatinib and hydroxyurea in pretreated progressive glioblastoma multiforme: a patient series. Ann Oncol. 2005;16:1702–1708. doi: 10.1093/annonc/mdi317. doi:10.1093/annonc/mdi317. [DOI] [PubMed] [Google Scholar]
- 69.Rubin JB, Kung AL, Klein RS, et al. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci USA. 2003;100:13513–13518. doi: 10.1073/pnas.2235846100. doi:10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ehtesham M, Mapara KY, Stevenson CB, Thompson RC. CXCR4 mediates the proliferation of glioblastoma progenitor cells. Cancer Lett. 2009;274:305–312. doi: 10.1016/j.canlet.2008.09.034. doi:10.1016/j.canlet.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. doi:10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Phi JH, Park SH, Kim SK, et al. Sox2 expression in brain tumors: a reflection of the neuroglial differentiation pathway. Am J Surg Pathol. 2008;32:103–112. doi: 10.1097/PAS.0b013e31812f6ba6. doi:10.1097/PAS.0b013e31812f6ba6. [DOI] [PubMed] [Google Scholar]
- 73.Gangemi RM, Griffero F, Marubbi D, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. doi:10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 74.Tropepe V, Sibilia M, Ciruna BG, Rossant J, Wagner EF, van der Kooy D. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev Biol. 1999;208:166–188. doi: 10.1006/dbio.1998.9192. doi:10.1006/dbio.1998.9192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.